Ulosarcina terrestrica gen. nov., sp. nov., a New Ulvophycean Sarcinoid Alga from the Russian Far East
Abstract
1. Introduction
2. Results
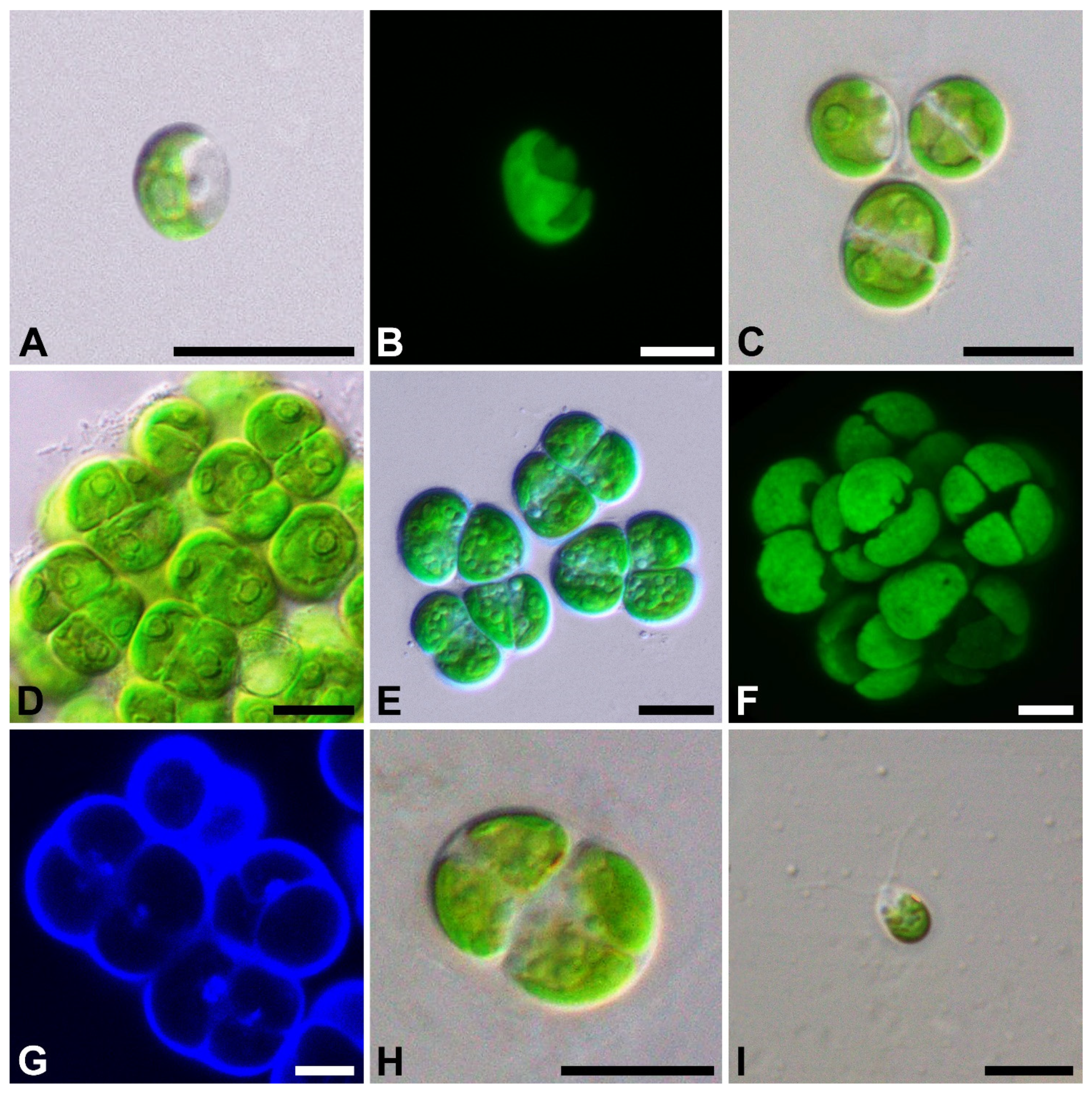
2.1. Taxonomic Treatment
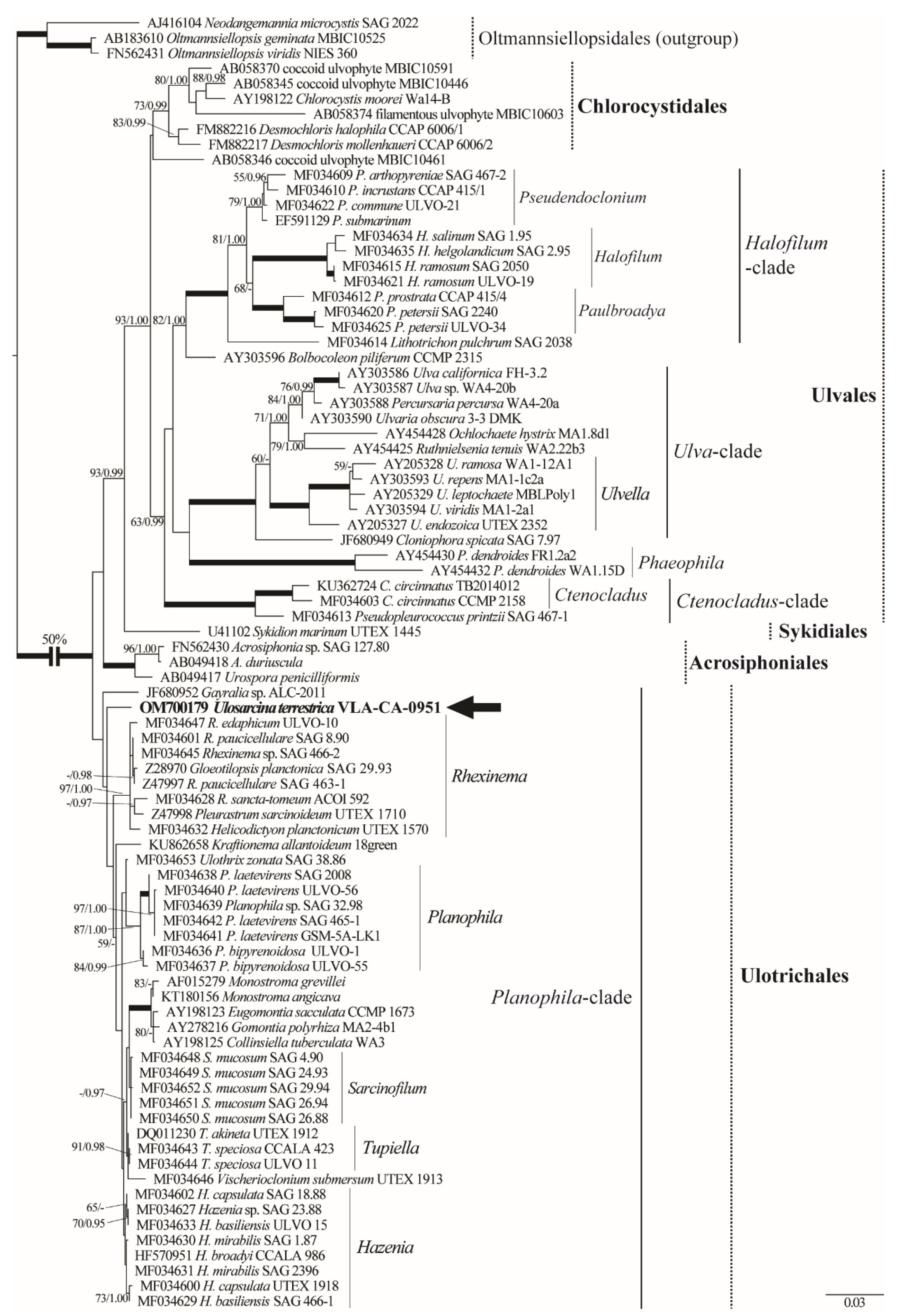
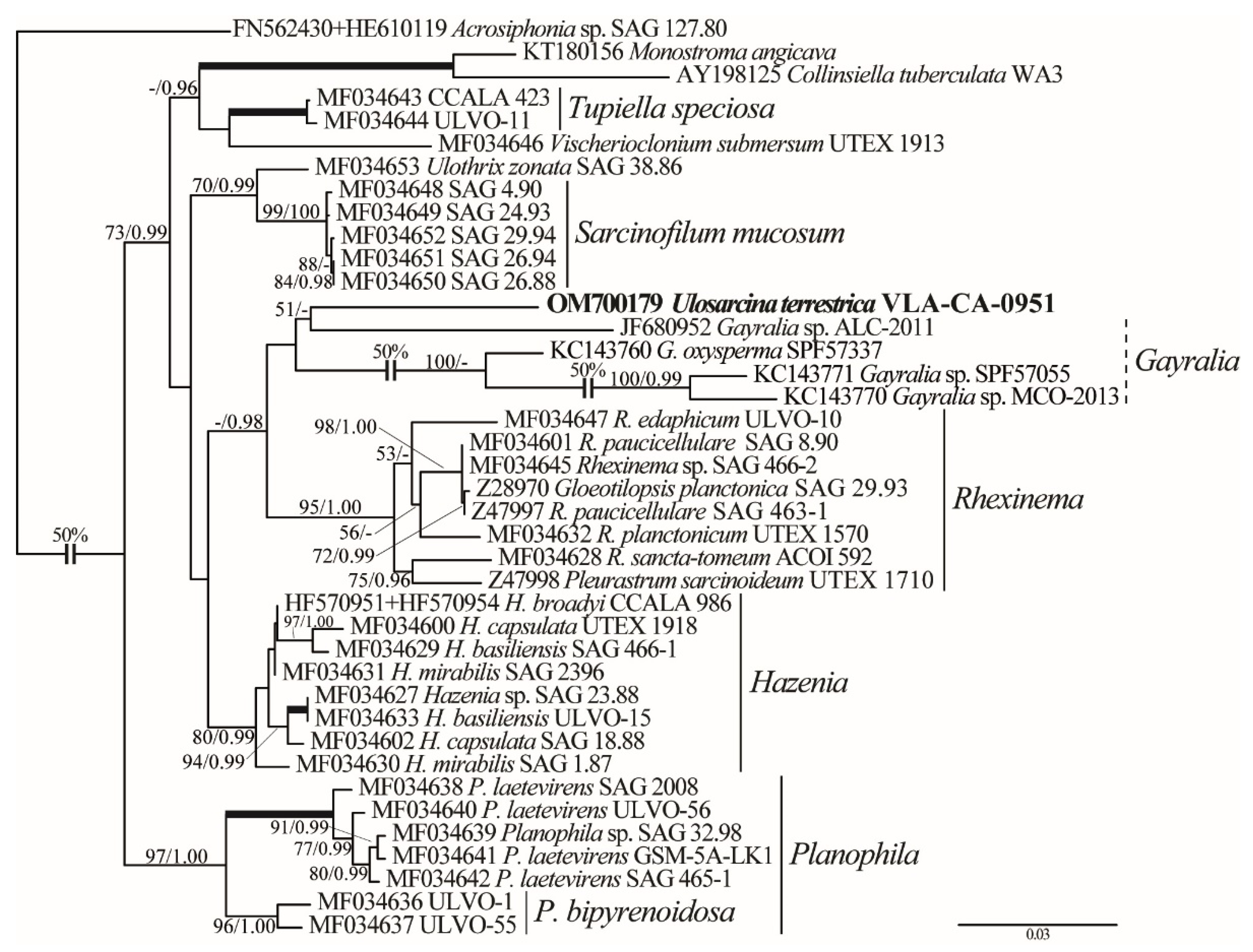
2.2. Phylogenetic Analyses
3. Discussion
4. Materials and Methods
4.1. Strain Origin, Culture Conditions, and Light Microscopy
4.2. DNA Extraction, Amplification, and Sequencing
4.3. Alignment, Secondary Structure Modeling, and Datasets
4.4. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friedl, T.; Rybalka, N. Systematics of the Green Algae: A Brief Introduction to the Current Status. In Progress in Botany; Lüttge, U., Beyschlag, W., Francis, D., Cushman, J., Eds.; Springer: Heidelberg, Germany, 2012; Volume 73, pp. 252–280. [Google Scholar] [CrossRef]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and Molecular Evolution of the Green Algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-Wide Electronic Publication, National University of Ireland, Galway. 2022. Available online: https://www.algaebase.org (accessed on 10 October 2022).
- Darienko, T.; Pröschold, T. Toward a Monograph of Non-Marine Ulvophyceae Using an Integrative Approach (Molecular Phylogeny and Systematics of Terrestrial Ulvophyceae II.). Phytotaxa 2017, 324, 1–41. [Google Scholar] [CrossRef]
- Darienko, T.; Rad-Menéndez, C.; Campbell, C.N.; Pröschold, T. Molecular Phylogeny of Unicellular Marine Coccoid Green Algae Revealed New Insights into the Systematics of the Ulvophyceae (Chlorophyta). Microorganisms 2021, 9, 1586. [Google Scholar] [CrossRef]
- Wetherbee, R.; Verbruggen, H. Kraftionema allantoideum, a New Genus and Family of Ulotrichales (Chlorophyta) Adapted for Survival in High Intertidal Pools. J. Phycol. 2016, 52, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Škaloud, P.; Rindi, F.; Boedeker, C.; Leliaert, F. Süßwasserflora von Mitteleuropa. Freshwater Flora of Central Europe. Bd 13. Chlorophyta: Ulvophyceae; Krienitz, L., Ed.; Springer Spektrum: Berlin, Germany, 2018; pp. 1–288. ISBN 978-3-662-55495-1. [Google Scholar]
- Kuzyakhmetov, G.G.; Dubovik, I.E. Metody Izucheniya Pochvennyh Vodorosley [Methods for Studying Soil Algae]; Izdatelstvo RIO BashGU: Ufa, Russia, 2001. (In Russian) [Google Scholar]
- Andersen, R.A. Algal Culturing Techniques; Elsevier Academic Press: Burlington, MA, USA, 2005; ISBN 0-12-088426-7. [Google Scholar]
- McFadden, G.I.; Melkonian, M. Use of Hepes buffer for microalgal culture media and fixation for electron microscopy. Phycologia 1986, 25, 551–557. [Google Scholar] [CrossRef]
- Abdullin, S.R.; Nikulin, A.Y.; Bagmet, V.B.; Nikulin, V.Y.; Gontcharov, A.A. New cyanobacterium Aliterella vladivostokensis sp. nov. (Aliterellaceae, Chroococcidiopsidales), isolated from temperate monsoon climate zone (Vladivostok, Russia). Phytotaxa 2021, 527, 221–233. [Google Scholar] [CrossRef]
- López-García, P.; Philippe, H.; Gail, F.; Moreira, D. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl. Acad. Sci. USA 2003, 100, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Marin, B.; Palm, A.; Klingberg, M.; Melkonian, M. Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 2003, 154, 99–145. [Google Scholar] [CrossRef] [PubMed]
- Mikhailyuk, T.; Lukešová, A.; Glaser, K.; Holzinger, A.; Obwegeser, S.; Nyporko, S.; Friedl, T.; Karsten, U. New Taxa of Streptophyte Algae (Streptophyta) from Terrestrial Habitats Revealed Using an Integrative Approach. Protist 2018, 169, 406–431. [Google Scholar] [CrossRef] [PubMed]
- Edgcomb, V.P.; Kysela, D.T.; Teske, A.; de Vera Gomez, A.; Sogin, M.L. Benthic Eukaryotic Diversity in the Guaymas Basin Hydrothermal Vent Environment. Proc. Natl. Acad. Sci. USA 2002, 99, 7658–7662. [Google Scholar] [CrossRef] [PubMed]
- Hoef-Emden, K.; Melkonian, M. Revision of the Genus Cryptomonas (Cryptophyceae): A Combination of Molecular Phylogeny and Morphology Provides Insights into a Long-Hidden Dimorphism. Protist 2003, 154, 371–409. [Google Scholar] [CrossRef] [PubMed]
- Bonfield, J.K.; Smith, K.F.; Staden, R. A new DNA sequence assembly program. Nucleic Acids Res. 1995, 23, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Gouy, M.; Gautier, C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 1996, 12, 543–548. [Google Scholar] [CrossRef]
- Coleman, A.W.; Mai, J.C. Ribosomal DNA and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J. Mol. Evol. 1997, 45, 168–177. [Google Scholar] [CrossRef]
- Bast, F. Monostroma: The Jeweled Seaweed for Future; Lap Lambert Academic Publishing: Chisinau, Moldova, 2011; pp. 1–192. ISBN 978-3-8473-2710-3. [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. A New look at the statistical model identification. IEEE Trans. Autom. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gontcharov, A.A.; Nikulin, A.Y.; Nikulin, V.Y.; Allaguvatova, R.Z.; Bagmet, V.B.; Abdullin, S.R. Ulosarcina terrestrica gen. nov., sp. nov., a New Ulvophycean Sarcinoid Alga from the Russian Far East. Plants 2022, 11, 3228. https://doi.org/10.3390/plants11233228
Gontcharov AA, Nikulin AY, Nikulin VY, Allaguvatova RZ, Bagmet VB, Abdullin SR. Ulosarcina terrestrica gen. nov., sp. nov., a New Ulvophycean Sarcinoid Alga from the Russian Far East. Plants. 2022; 11(23):3228. https://doi.org/10.3390/plants11233228
Chicago/Turabian StyleGontcharov, Andrey A., Arthur Yu. Nikulin, Vyacheslav Yu. Nikulin, Rezeda Z. Allaguvatova, Veronika B. Bagmet, and Shamil R. Abdullin. 2022. "Ulosarcina terrestrica gen. nov., sp. nov., a New Ulvophycean Sarcinoid Alga from the Russian Far East" Plants 11, no. 23: 3228. https://doi.org/10.3390/plants11233228
APA StyleGontcharov, A. A., Nikulin, A. Y., Nikulin, V. Y., Allaguvatova, R. Z., Bagmet, V. B., & Abdullin, S. R. (2022). Ulosarcina terrestrica gen. nov., sp. nov., a New Ulvophycean Sarcinoid Alga from the Russian Far East. Plants, 11(23), 3228. https://doi.org/10.3390/plants11233228









