Stability of Transgene Inheritance in Progeny of Field-Grown Pear Trees over a 7-Year Period
Abstract
:1. Introduction
2. Results
2.1. Inheritance of Transgenes in Progeny
2.2. Evaluation of Seed Characteristics
2.3. Stability of Transgene Expression in Progeny
3. Discussion
4. Materials and Methods
4.1. Plant and Seed Material
4.2. Transgene Segregation in Progeny
4.3. Expression of uidA Gene in Progeny
4.4. Statistical Analysis
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pons, E.; Peris, J.E.; Peňa, L. Field performance of transgenic citrus trees: Assessment of the long-term expression of uidA and nptII transgenes and the impact on relevant agronomic and phenotypic characteristics. BMC Biotechnol. 2012, 12, 41. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.L.; Cohen, D.; van den Brink, R.; Morris, B. Assessment of expression and inheritance patterns of three transgenes with the aid of techniques for promoting rapid flowering of transgenic apple trees. Plant Cell Rep. 1999, 18, 727–732. [Google Scholar] [CrossRef]
- Flachowsky, H.; Le Roux, P.M.; Peil, A.; Patocchi, A.; Richter, K.; Hanke, M.V. Application of a high-speed breeding technology to apple (Malus × domestica) based on transgenic early flowering plants and marker-assisted selection. New Phytol. 2011, 192, 364–377. [Google Scholar] [CrossRef]
- Peña, L.; Martin-Trillo, M.; Juarez, J.; Pina, J.A.; Navarro, L.; Martinez-Zapater, J.M. Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechol. 2001, 19, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Flachowsky, H.; Peil, A.; Sopanen, T.; Elo, A.; Hanke, V. Overexpression of BpMADS4 from silver birch (Betula pendula Roth.) induces early flowering in apple (Malus × domestica Borkh.). Plant Breed. 2007, 126, 137–145. [Google Scholar] [CrossRef]
- Matsuda, N.; Ikeda, K.; Kurosaka, M.; Takashina, T.; Isuzugawa, K.; Endo, N.; Omura, M. Early flowering phenotype in transgenic pears (Pyrus communis L.) expressing the CiFT gene. J. Jpn. Soc. Hort. Sci. 2009, 78, 410–416. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, C.; Dardick, C.; Callahan, A.; Scorza, R. Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PLoS ONE 2012, 7, e40715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.Y.; Liu, Y.; Luthe, D.S.; Yuceer, C. Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 2006, 18, 1846–1861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elo, A.; Lemmetyinen, J.; Novak, A.; Keinonen, K.; Porali, I.; Hassinen, M.; Sopanen, T. BpMADS4 has a central role in inflorescence initiation in silver birch (Betula pendula). Physiol. Plant. 2007, 131, 149–158. [Google Scholar] [CrossRef]
- Klocko, A.L.; Ma, C.; Robertson, S.; Esfandiari, E.; Nilsson, O.; Strauss, S.H. FT overexpression induces precocious flowering and normal reproductive development in Eucalyptus. Plant Biotechnol. J. 2016, 14, 808–819. [Google Scholar] [CrossRef]
- Wenzel, S.; Flachowsky, H.; Hanke, M.V. The fast-track breeding approach can be improved by heat-induced expression of the FLOWERING LOCUS T genes from poplar (Populus trichocarpa) in apple (Malus × domestica Borkh.). Plant Cell Tissue Organ Cult. 2013, 115, 127–137. [Google Scholar] [CrossRef]
- Petri, C.; Alburquerque, N.; Faize, M.; Scorza, R.; Dardick, C. Current achievements and future directions in genetic engineering of European plum (Prunus domestica L.). Transgenic Res. 2018, 27, 225–240. [Google Scholar] [CrossRef] [Green Version]
- Endo, T.; Fujii, H.; Omura, M.; Shimada, T. Fast-track breeding system to introduce CTV resistance of trifoliate orange into citrus germplasm, by integrating early flowering transgenic plants with marker-assisted selection. BMC Plant Biol. 2020, 20, 224. [Google Scholar] [CrossRef]
- Lebedev, V.G.; Popova, A.A.; Shestibratov, K.A. Genetic engineering and genome editing for improving nitrogen use efficiency in plants. Cells 2021, 10, 3303. [Google Scholar] [CrossRef]
- Rajeevkumar, S.; Anunanthini, P.; Sathishkumar, R. Epigenetic silencing in transgenic plants. Front. Plant Sci. 2015, 6, 693. [Google Scholar] [CrossRef] [Green Version]
- Ren, Y.C.; Zhang, J.; Liang, H.Y.; Wang, J.M.; Yang, M.S. Inheritance and expression stability of exogenous genes in insect-resistant transgenic poplar. Plant Cell Tissue Organ Cult. 2017, 130, 567–576. [Google Scholar] [CrossRef]
- Ahuja, M.R.; Fladung, M. Integration and inheritance of transgenes in crop plants and trees. Tree Genet. Genomes 2014, 10, 779–790. [Google Scholar] [CrossRef]
- Vain, P.; Afolabi, A.S.; Worland, B.; Snape, J.W. Transgenic behavior in populations of rice plants transformed using a new dual binary vector system: pGreen/pSoup. Theor. Appl. Genet. 2003, 107, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.X.; Fang, Z.Y.; Yang, L.M. Inheritance and expression of Bt cry1Ba3 gene in progeny from transformed cabbage plants. Mol. Biol. Rep. 2020, 47, 2583–2589. [Google Scholar] [CrossRef]
- Yin, Z.; Plader, W.; Malepszy, S. Transgene inheritance in plants. J. Appl. Genet. 2004, 45, 127–144. [Google Scholar] [PubMed]
- Nandakumar, N.; Tyagi, N.K.; Chikkappa, K.; Aprajita, G.; Mohapatra, T.; Prabhu, K.V.; Singh, A.K. Molecular analysis of transgene (psy) inheritance in a golden rice line developed in the genetic background of IR64. J. Plant Biochem. Biotechnol. 2008, 17, 127–132. [Google Scholar] [CrossRef]
- Hensel, G.; Oleszczuk, S.; Daghma, D.E.S.; Zimny, J.; Melzer, M.; Kumlehn, J. Analysis of T-DNA integration and generative segregation in transgenic winter triticale (× Triticosecale Wittmack). BMC Plant Biol. 2012, 12, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, D.J.; Passey, A.J.; Baker, S.A.; Wilson, F.M. Transgenes display stable patterns of expression in apple fruit and Mendelian segregation in the progeny. Nat. Biotechol. 1996, 14, 56–60. [Google Scholar] [CrossRef]
- Eissa, H.F.; Hassanien, S.E.; Ramadan, A.M.; El-Shamy, M.M.; Saleh, O.M.; Shokry, A.M.; Abdelsattar, M.; Morsy, Y.B.; El-Maghraby, M.; Alameldin, H.F.; et al. Developing transgenic wheat to encounter rusts and powdery mildew by overexpressing barley chi26 gene for fungal resistance. Plant Methods 2017, 13, 41. [Google Scholar] [CrossRef]
- Schlathölter, I.; Jänsch, M.; Flachowsky, H.; Broggini, G.A.L.; Hanke, M.-V.; Patocchi, A. Generation of advanced fire blight resistant apple (Malus domestica) selections of the 5th generation within seven years applying the early flowering approach. Planta 2018, 247, 1475–1488. [Google Scholar] [CrossRef] [Green Version]
- Borejsza-Wysocha, E.; Norelli, J.L.; Aldwinckle, H.S.; Malnoy, M. Stable expression and phenotypic impact of attacinE transgene in orchard grown apple trees over a 12 years period. BMC Biotechnol. 2010, 10, 41. [Google Scholar] [CrossRef] [Green Version]
- Newhouse, A.E.; Polin-McGuigan, L.D.; Baier, K.A.; Valletta, K.E.R.; Rottmann, W.H.; Tschaplinski, T.J.; Maynard, C.A.; Powell, W.A. Transgenic American chestnuts show enhanced blight resistance and transmit the trait to T1 progeny. Plant Sci. 2014, 228, 88–97. [Google Scholar] [CrossRef]
- Ladics, G.S.; Bartholomaeus, A.; Bregitzer, P.; Doerrer, N.G.; Gray, A.; Holzhauser, T.; Jordan, M.; Keese, P.; Kok, E.; Macdonald, P.; et al. Genetic basis and detection of unintended effects in genetically modified crop plants. Transgenic Res. 2015, 24, 587–603. [Google Scholar] [CrossRef] [Green Version]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [Green Version]
- Bauer-Panskus, A.; Miyazaki, J.; Kawall, K.; Then, C. Risk assessment of genetically engineered plants that can persist and propagate in the environment. Environ. Sci. Eur. 2020, 32, 32. [Google Scholar] [CrossRef] [Green Version]
- Luo, F.; Norelli, J.L.; Howard, N.P.; Wisniewski, M.; Flachowsky, H.; Hanke, M.-V.; Peace, C. Introgressing blue mold resistance into elite apple germplasm by rapid cycle breeding and foreground and background DNA-informed selection. Tree Genet. Genomes 2020, 16, 28. [Google Scholar] [CrossRef]
- Ravelonandro, M.; Scorza, R.; Briard, P.; Lafargue, B.; Renaud, R. Inheritance of silencing in transgenic plums. Acta Hortic. 2011, 899, 139–144. [Google Scholar] [CrossRef]
- Ravelonandro, M.; Briard, P.; Scorza, R.; Callahan, A.; Zagrai, I.; Kundu, J.K.; Dardick, C. Robust response to Plum pox virus infection via plant biotechnology. Genes 2021, 12, 816. [Google Scholar] [CrossRef]
- Van Nocker, S.; Gardiner, S. Breeding better cultivars, faster: Applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic. Res. 2014, 1, 14022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freiman, A.; Shlizerman, L.; Golobovitch, S.; Yablovitz, Z.; Korchinsky, R.; Cohen, Y.; Samach, A.; Chevreau, E.; Le Roux, P.-M.; Patocchi, A.; et al. Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2. Planta 2012, 235, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M. Pear breeding. In Breeding Plantation Tree Crops: Temperate Species; Mohan Jain, S., Priyadarshan, P.M., Eds.; Springer: New York, NY, USA, 2009; pp. 1–26. [Google Scholar]
- Lebedev, V.G.; Skryabin, K.G.; Dolgov, S.V. Transgenic pear clonal rootstocks resistant to herbicide “Basta”. Acta Hortic. 2002, 596, 193–197. [Google Scholar] [CrossRef]
- Zhu, L.-H.; Li, X.-Y.; Ahlman, A.; Welander, M. The rooting ability of the dwarfing pear rootstock BP10030 (Pyrus communis) was significantly increased by introduction of the rolB gene. Plant Sci. 2003, 165, 829–835. [Google Scholar] [CrossRef]
- Malnoy, M.; Venisse, J.S.; Chevreau, E. Expression of a bacterial effector, harpin N, causes increased resistance to fire blight in Pyrus communis. Tree Genet. Genomes 2005, 1, 41. [Google Scholar] [CrossRef]
- Dolgov, S.V.; Lebedev, V.G.; Firsov, A.P. Pear fruit taste modification by thaumatin II gene expression. Acta Hortic. 2011, 909, 67–73. [Google Scholar] [CrossRef]
- Dong, H.; Wang, C.; Xing, C.; Yang, T.; Yan, J.; Gao, J.; Li, D.; Wang, R.; Blumwald, E.; Zhang, S.; et al. Overexpression of PbrNHX2 gene, a Na+/H+ antiporter gene isolated from Pyrus betulaefolia, confers enhanced tolerance to salt stress via modulating ROS levels. Plant Sci. 2019, 285, 14–25. [Google Scholar] [CrossRef]
- Lebedev, V.G.; Dolgov, S.V. Stability of gus and bar gene expression in transgenic pear clonal rootstock plants during several years. Acta Hortic. 2008, 800, 373–382. [Google Scholar] [CrossRef]
- Ma, M.; Chen, X.; Yin, Y.; Fan, R.; Li, B.; Zhan, Y.; Zeng, F. DNA methylation silences exogenous gene expression in transgenic birch progeny. Front. Plant Sci. 2020, 11, 523748. [Google Scholar] [CrossRef] [PubMed]
- Bregitzer, P.; Tonks, D. Inheritance and expression of transgenes in barley. Crop Sci. 2003, 43, 4–12. [Google Scholar] [CrossRef]
- Tizaoui, K.; Kchouk, M.E. Genetic approaches for studying transgene inheritance and genetic recombination in three successive generations of transformed tobacco. Genet. Mol. Biol. 2012, 35, 640–649. [Google Scholar] [CrossRef] [Green Version]
- Dole, R.; Hoerling, M.; Perlwitz, J.; Eischeid, J.; Pegion, P.; Zhang, T.; Quan, X.-W.; Xu, T.; Murray, D. Was there a basis for anticipating the 2010 Russian heat wave? Geophys. Res. Lett. 2011, 38, L06702. [Google Scholar] [CrossRef] [Green Version]
- Lebedev, V.G. Evaluation of fruit quality and environmental biosafety of transgenic pear trees. In Proceedings of the ESNA XLIII Annual Meeting, Bolzano, Italy, 3–6 September 2014; p. 48. [Google Scholar]
- Kohli, A.; Twyman, R.M.; Abranches, R.; Wegel, E.; Stoger, E.; Christou, P. Transgene integration, organization and interaction in plants. Plant Mol. Biol. 2003, 52, 247–258. [Google Scholar] [CrossRef]
- Cao, Q.-J.; Xia, H.; Yang, X.; Lu, B.-R. Performance of hybrids between weedy rice and insect-resistant transgenic rice under field experiments: Implication for environmental biosafety assessment. J. Integr. Plant Biol. 2009, 51, 1138–1148. [Google Scholar] [CrossRef]
- Wang, W.; Xia, H.; Yang, X.; Xu, T.; Si, H.J.; Cai, X.X.; Wang, F.; Su, J.; Snow, A.A.; Lu, B.-R. A novel 5-enolpyruvoylshikimate-3-phosphate (EPSP) synthase transgene for glyphosate resistance stimulates growth and fecundity in weedy rice (Oryza sativa) without herbicide. New Phytol. 2014, 202, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef]
- Upadhaya, K.; Pandey, H.N.; Law, P.S. The effect of seed mass on germination, seedling survival and growth in Prunus jenkinsii Hook.f. & Thoms. Turk. J. Bot. 2007, 31, 31–36. [Google Scholar]
- Mao, P.; Guo, L.; Gao, Y.; Qi, L.; Cao, B. Effects of seed size and sand burial on germination and early growth of seedlings for coastal Pinus thunbergii Parl. in the Northern Shandong Peninsula, China. Forests 2019, 10, 281. [Google Scholar] [CrossRef] [Green Version]
- Tumpa, K.; Vidakovic, A.; Drvodelic, D.; Šango, M.; Idžojtic, M.; Perkovic, I.; Poljak, I. The effect of seed size on germination and seedling growth in sweet chestnut (Castanea sativa Mill.). Forests 2021, 12, 858. [Google Scholar] [CrossRef]
- Jeon, D.W.; Park, J.R.; Jang, Y.H.; Kim, E.-G.; Ryu, T.; Kim, K.-M. Safety verification of genetically modified rice morphology, hereditary nature, and quality. Environ. Sci. Eur. 2021, 33, 73. [Google Scholar] [CrossRef]
- Lamichaney, A.; Parihar, A.K.; Hazra, K.K.; Dixit, G.P.; Katiyar, P.K.; Singh, D.; Singh, A.K.; Kumar, N.; Singh, N.P. Untangling the influence of heat stress on crop phenology, seed set, seed weight, and germination in field pea (Pisum sativum L.). Front. Plant Sci. 2021, 12, 635868. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.F.; Sun, H.Z.; Wen, H.L.; Du, Y.X.; Zhang, J.; Li, J.Z.; Peng, T.; Zhao, Q.Z. Germination vigour difference of superior and inferior rice grains revealed by physiological and gene expression studies. J. Agric. Sci. 2018, 156, 350–357. [Google Scholar] [CrossRef]
- Fung, R.W.M.; Janssen, B.-J.; Morris, B.A.; Gardner, R.C. Inheritance and expression of transgenes in kiwifruit. N. Z. J. Crop Hort. Sci. 1998, 26, 169–179. [Google Scholar] [CrossRef]
- James, D.J.; Passey, A.J.; Barbara, D.J. Agrobacterium-mediated transformation of the cultivated strawberry (Fragaria × ananassa Duch.) using disarmed binary vectors. Plant Sci. 1990, 69, 79–94. [Google Scholar] [CrossRef]
- Lebedev, V. The rooting of stem cuttings and the stability of uidA gene expression in generative and vegetative progeny of transgenic pear rootstock in the field. Plants 2019, 8, 291. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Cellini, F.; Chesson, A.; Colquhoun, I.; Constable, A.; Davies, H.V.; Engel, K.H.; Gatehouse, A.M.; Karenlampi, S.; Kok, E.J.; Leguay, J.J.; et al. Unintended effects and their detection in genetically modified crops. Food Chem. Toxicol. 2004, 42, 1089–1125. [Google Scholar] [CrossRef]
- Lebedev, V.G.; Dolgov, S.V. The effect of selective agents and a plant intron on transformation efficiency and expression of heterologous genes in pear Pyrus communis L. Rus. J. Genet. 2000, 36, 650–655. [Google Scholar]
- Scott, R.; Draper, J.; Jefferson, R.; Dury, G.; Jacob, L. Analysis of gene organization and expression in plants. In Plant Genetic Transformation and Gene Expression: A Laboratory Manual; Draper, J., Scott, R., Armitage, P., Walden, R., Eds.; Blackwell Scientific Pubs.: Oxford, UK, 1988; pp. 263–339. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]


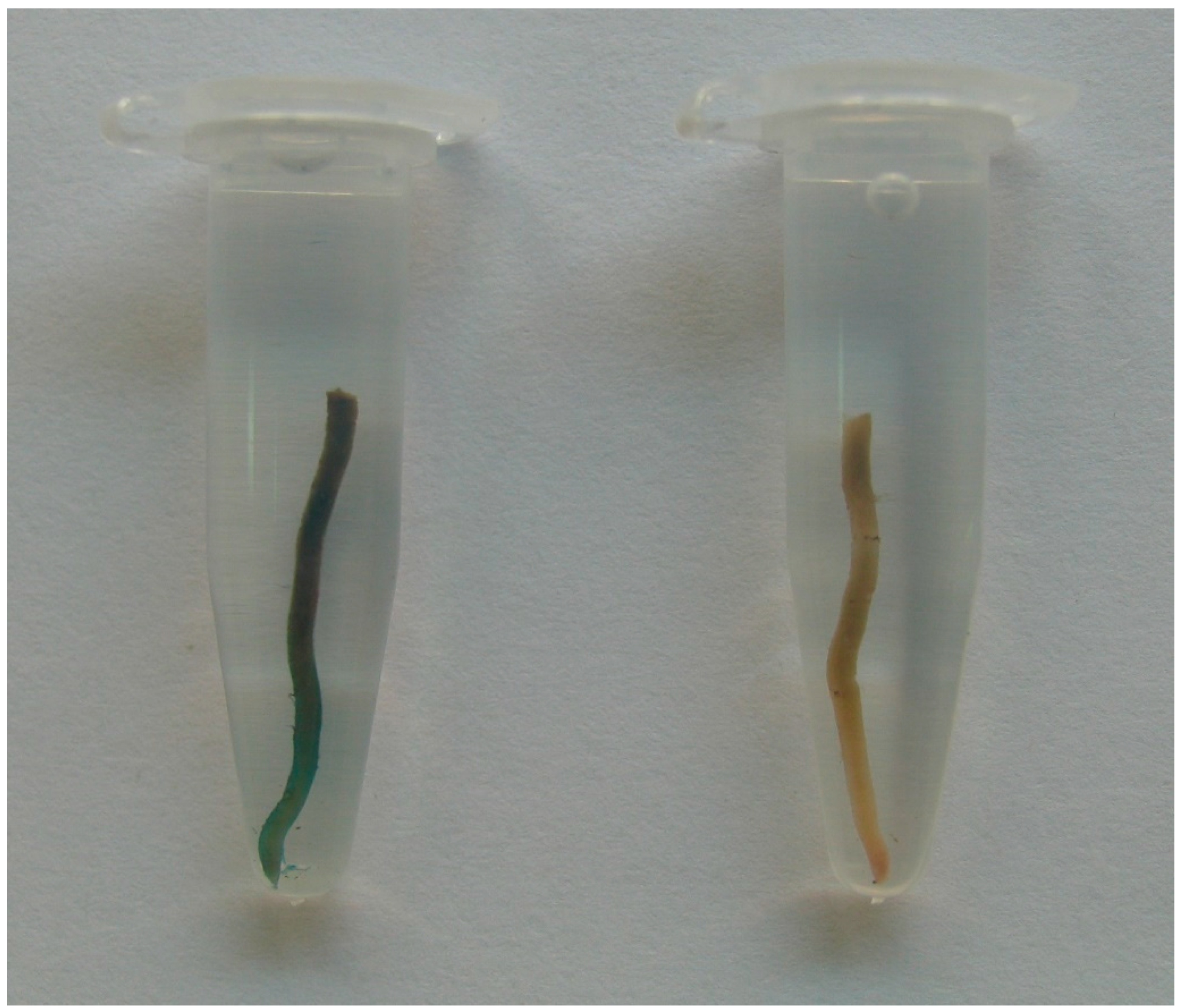

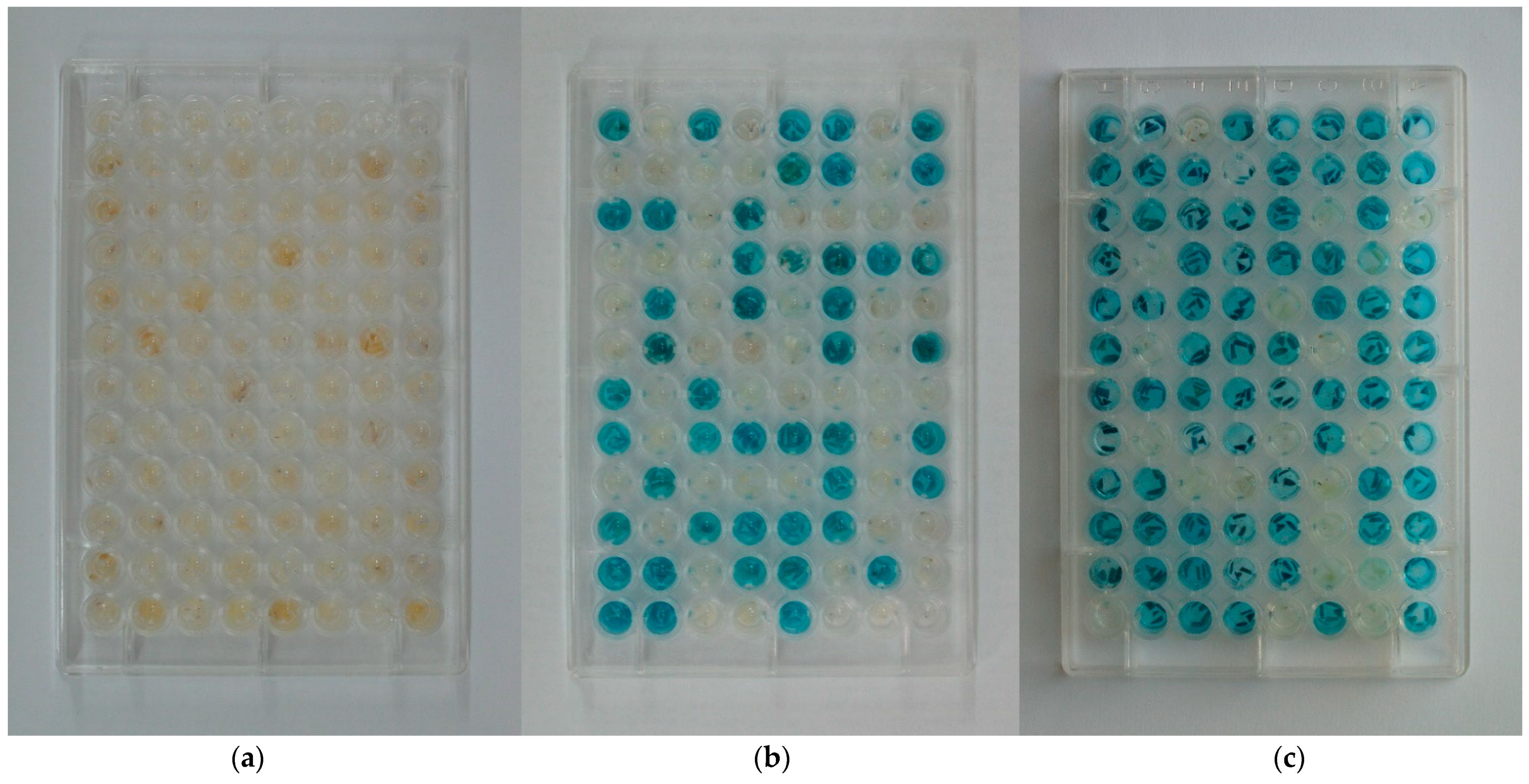
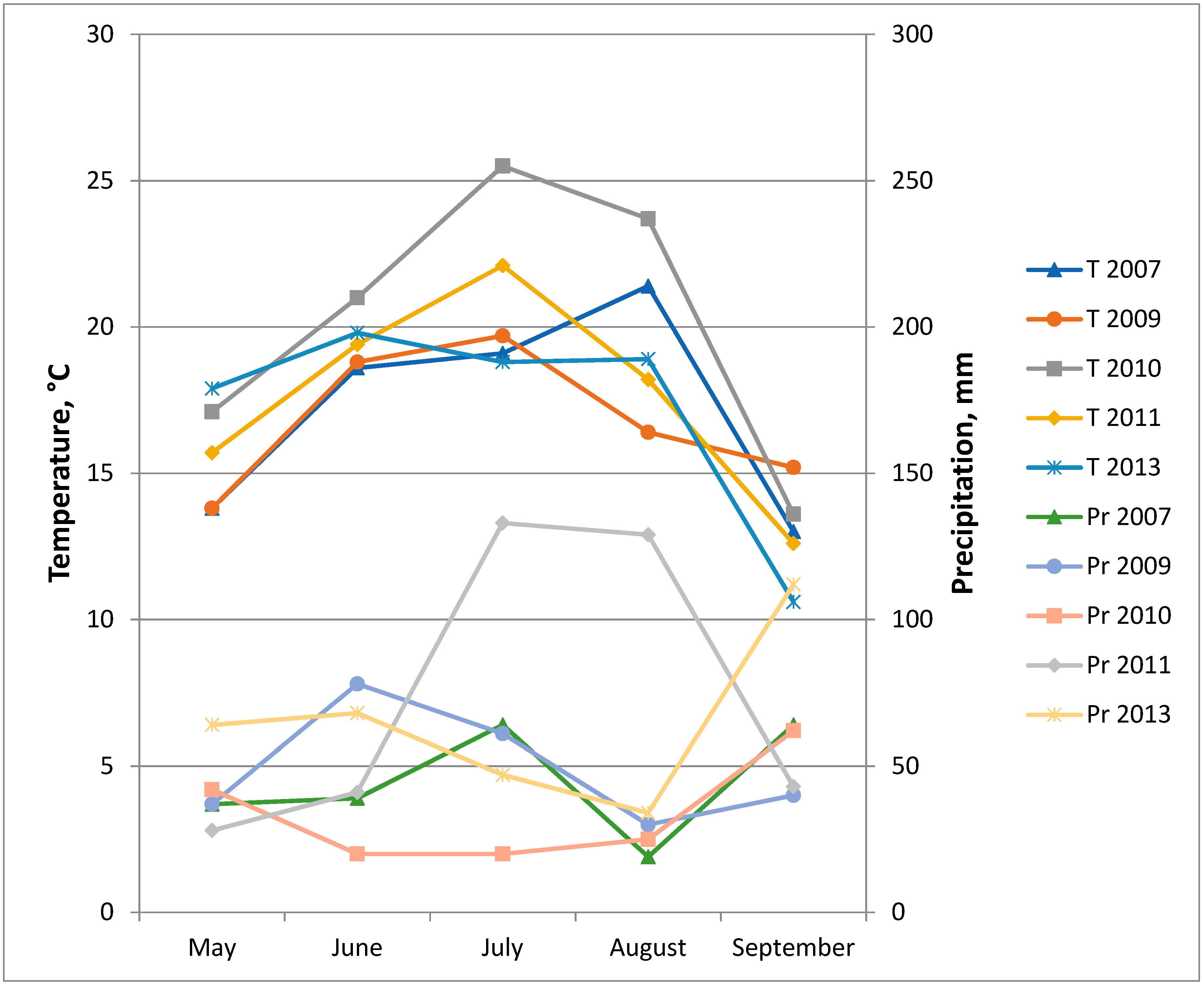
| Transgene | Lines | Ratio | 2007 | 2009 | 2010 | 2011 | 2013 | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Analyzed | GUS+/− | χ2 | GUS+/− | χ2 | GUS+/− | χ2 | GUS+/− | χ2 | GUS+/− | χ2 | GUS+/− | χ2 | ||
| control | GP217 | 0/123 | 0/109 | 0/127 | 0/106 | 0/92 | 0/557 | |||||||
| uidA-int | HB-1 | 3:1 | nd | 78/25 | 0.029 | nd | 101/39 | 0.610 | 38/13 | 0.007 | 217/77 | 0.222 | ||
| HA-2 | 1:1 | nd | nd | nd | nd | 22/33 | 2.200 | 22/33 | 2.200 | |||||
| HA-3 | 1:1 | nd | 69/72 | 0.064 | 39/23 | 4.129 * | 84/66 | 2.160 | 70/63 | 0.368 | 262/224 | 2.971 | ||
| HA-4 | 1:1 | 75/53 | 3.781 | nd | nd | nd | 44/48 | 0.174 | 119/101 | 1.473 | ||||
| HA-5 | 3:1 | 83/26 | 0.076 | nd | nd | nd | nd | 83/26 | 0.076 | |||||
| HA-6 | 1:1 | 114/91 | 2.580 | 58/44 | 1.922 | 104/89 | 1.166 | nd | nd | 276/224 | 5.408 * | |||
| uidA | NII-1 | 3:1 | 148/51 | 0.042 | 88/27 | 0.142 | 147/45 | 0.250 | nd | nd | 383/123 | 0.129 | ||
| NII-2 | 1:1 | nd | 46/51 | 0.258 | 88/104 | 1.333 | 44/76 | 8.533 ** | 26/29 | 0.164 | 204/260 | 6.759 * | ||
| NII-3 | 3:1 | nd | 73/28 | 0.399 | 100/25 | 1.667 | 111/38 | 0.020 | nd | 284/91 | 0.108 | |||
| NIII-2 | 1:1 | 24/25 | 0.020 | 39/28 | 1.806 | 42/54 | 1.500 | 29/48 | 4.688 * | 38/50 | 1.636 | 172/205 | 2.889 | |
| NIII-4 | 1:1 | 25/27 | 0.077 | 83/71 | 0.935 | 73/85 | 0.911 | nd | nd | 181/183 | 0.011 | |||
| NIII-5 | 1:1 | 59/92 | 7.212 ** | 19/28 | 1.723 | 24/23 | 0.021 | nd | nd | 102/143 | 8.126 ** | |||
| NIV-2 | 1:1 | 76/95 | 2.111 | 69/93 | 3.556 | 70/81 | 0.801 | nd | nd | 215/269 | 6.025 * | |||
| Line | 2007 | 2009 | 2010 | 2011 | 2013 |
|---|---|---|---|---|---|
| GP217 | 30.9 | 29.3 | 26.8 | 30.5 | 27.5 |
| HB-1 | nd | 27.6 | nd | 31.2 | 22.4 *** |
| HA-2 | nd | nd | nd | nd | 27.2 |
| HA-3 | nd | 22.7 *** | 24.1 | 26.5 ** | 19.6 *** |
| HA-4 | 30.3 | nd | nd | nd | 28.2 |
| HA-5 | 29.9 | nd | nd | nd | nd |
| HA-6 | 31.2 | 29.4 | 28.1 | 31.3 | nd |
| NII-1 | 30.9 | 28.4 | 27.9 | 30.0 | nd |
| NII-2 | nd | 30.1 | 28.8 | 32.8 | 30.0 * |
| NII-3 | nd | 30.3 | 25.1 | 32.5 | nd |
| NIII-2 | 29.3 | 18.1 *** | 22.4 ** | 30.2 | 23.5 *** |
| NIII-4 | 29.1 | 22.1 *** | 27.0 | 29.6 | nd |
| NIII-5 | 27.1 *** | 22.4 *** | 26.8 | nd | nd |
| NIV-2 | 30.8 | 19.6 *** | 19.6 *** | 31.1 | nd |
| Line | 2007 | 2009 | 2010 | 2011 | 2013 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length | Width | Shape | Length | Width | Shape | Length | Width | Shape | Length | Width | Shape | Length | Width | Shape | |
| GP217 | 8.3 | 4.9 | 1.68 | 8.5 | 5.0 | 1.73 | 8.3 | 4.9 | 1.72 | 8.4 | 4.9 | 1.72 | 8.3 | 4.8 | 1.72 |
| H-1 | nd | nd | nd | 8.4 | 5.0 | 1.69 | nd | nd | nd | 8.3 | 4.9 | 1.72 | 8.0 | 4.8 | 1.69 |
| H-2 | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | 8.3 | 4.9 | 1.69 |
| H-3 | nd | nd | nd | 8.4 | 4.8 | 1.75 | 8.2 | 4.7 | 1.74 | 8.1 * | 4.6 *** | 1.76 | 7.9 | 4.5 * | 1.76 |
| H-4 | 8.1 | 4.7 * | 1.72 | nd | nd | nd | nd | nd | nd | nd | nd | nd | 8.3 | 4.8 | 1.74 |
| H-5 | 8.2 | 4.7 | 1.72 | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| H-6 | 8.4 | 5.0 | 1.69 | 8.4 | 4.8 | 1.76 | 8.3 | 4.9 | 1.72 | 8.3 | 5.0 | 1.67 | nd | nd | nd |
| NII-1 | 8.2 | 5.0 | 1.66 | 8.4 | 4.9 | 1.72 | 8.6 | 5.1 | 1.69 | 8.3 | 5.0 | 1.68 | nd | nd | nd |
| NII-2 | nd | nd | nd | 8.3 * | 4.5 *** | 1.82 * | 8.7 | 4.9 | 1.75 | 8.5 | 5.0 | 1.70 | 8.2 | 4.9 | 1.69 |
| NII-3 | nd | nd | nd | 8.5 | 4.9 | 1.74 | 8.2 | 4.8 | 1.71 | 8.3 | 4.9 | 1.71 | nd | nd | nd |
| NIII-2 | 8.2 | 4.8 | 1.72 | 8.3 * | 4.8 | 1.74 | 8.0 | 4.6 * | 1.73 | 8.4 | 4.9 | 1.73 | 8.0 | 4.7 | 1.71 |
| NIII-4 | 8.4 | 4.9 | 1.71 | 8.3 | 4.8 | 1.74 | 8.4 | 4.9 | 1.72 | 8.5 | 4.8 | 1.77 | nd | nd | nd |
| NIII-5 | 7.7 *** | 4.7 * | 1.66 | 8.3 | 4.7 * | 1.75 | 8.3 | 4.9 | 1.72 | nd | nd | nd | nd | nd | nd |
| NIV-2 | 8.1 | 4.9 | 1.67 | 8.2 * | 4.6 *** | 1.79 | 7.8 ** | 4.5 ** | 1.74 | 8.3 | 4.9 | 1.71 | nd | nd | nd |
| Line | Germination Rate, % | |
|---|---|---|
| 2007 | 2009 | |
| GP217 | 88.6 ± 3.0 | 76.6 ± 7.0 |
| HB-1 | - | 75.2 ± 4.2 |
| HA-2 | - | 84.9 ± 3.7 |
| HA-3 | 89.1 ± 3.6 | - |
| HA-4 | 82.9 ± 1.5 | - |
| HA-6 | 89.7 ± 5.3 | 72.9 ± 5.4 |
| NII-1 | 93.6 ± 3.2 | 74.2 ± 4.6 |
| NII-2 | - | 76.4 ± 5.3 |
| NII-3 | - | 81.5 ± 4.3 |
| NIII-2 | 86.0 ± 3.1 | 77.9 ± 6.9 |
| NIII-4 | 85.3 ± 2.8 | 73.0 ± 4.0 |
| NIII-5 | 88.0 ± 4.6 | 78.3 ± 7.3 |
| NIV-2 | 96.3 ± 1.1 | 73.6 ± 4.0 |
| Gene | Line | Locus | GUS Activity—Average (min–max), pmol 4-MU/min/µg Protein | ||
|---|---|---|---|---|---|
| Number | Crossing 2007 (Field 2010) | Crossing 2008 (Field 2011) | Crossing 2009 (Greenhouse 2010) | ||
| uidA-int | HB-1 | 2 | nd | 72.8 (50.1–100.4) | 11.6 (10.5–12.6) |
| HA-2 | 1 | nd | nd | 2.5 (0.6–5.1) | |
| HA-3 | 1 | 10.4 (1.0–16.8) | nd | nd | |
| HA-4 | 2 | 8.4 (6.9–10.7) | 38.9 (27.6–47.4) | nd | |
| HA-5 | 1 | nd | 18.0 (11.9–29.6) | nd | |
| HA-6 | 1 | 10.5 (1.7–36.0) | nd | 18.3 (7.6–35.4) | |
| uidA | NII-1 | 2 | 5.2 (1.3–14.9) | 35.1 (30.5–41.0) | 14.5 (7.3–25.7) |
| NII-2 | 1 | nd | nd | 5.0 (3.8–7.2) | |
| NII-3 | 2 | nd | 23.0 (13.5–30.6) | 3.7 (2.4–5.3) | |
| NIII-2 | 1 | 4.9 (1.4–6.7) | nd | 2.5 (1.3–4.7) | |
| NIII-4 | 1 | 9.5 (1.4–21.2) | nd | 5.0 (1.7–11.2) | |
| NIII-5 | 1 | 3.0 (0.7–7.9) | 7.3 (1.1–19.4) | nd | |
| NIV-2 | 1 | 2.2 (1.3–3.2) | nd | 8.2 (1.1–19.2) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebedev, V. Stability of Transgene Inheritance in Progeny of Field-Grown Pear Trees over a 7-Year Period. Plants 2022, 11, 151. https://doi.org/10.3390/plants11020151
Lebedev V. Stability of Transgene Inheritance in Progeny of Field-Grown Pear Trees over a 7-Year Period. Plants. 2022; 11(2):151. https://doi.org/10.3390/plants11020151
Chicago/Turabian StyleLebedev, Vadim. 2022. "Stability of Transgene Inheritance in Progeny of Field-Grown Pear Trees over a 7-Year Period" Plants 11, no. 2: 151. https://doi.org/10.3390/plants11020151
APA StyleLebedev, V. (2022). Stability of Transgene Inheritance in Progeny of Field-Grown Pear Trees over a 7-Year Period. Plants, 11(2), 151. https://doi.org/10.3390/plants11020151





