Three-Fluorophore FRET Enables the Analysis of Ternary Protein Association in Living Plant Cells
Abstract
1. Introduction
2. Results
2.1. Physicochemical Properties of the Used Fluorophores
2.2. Calculated Properties of the mTRQ2/mVEN/mRFP Three-Fluorophore FRET System
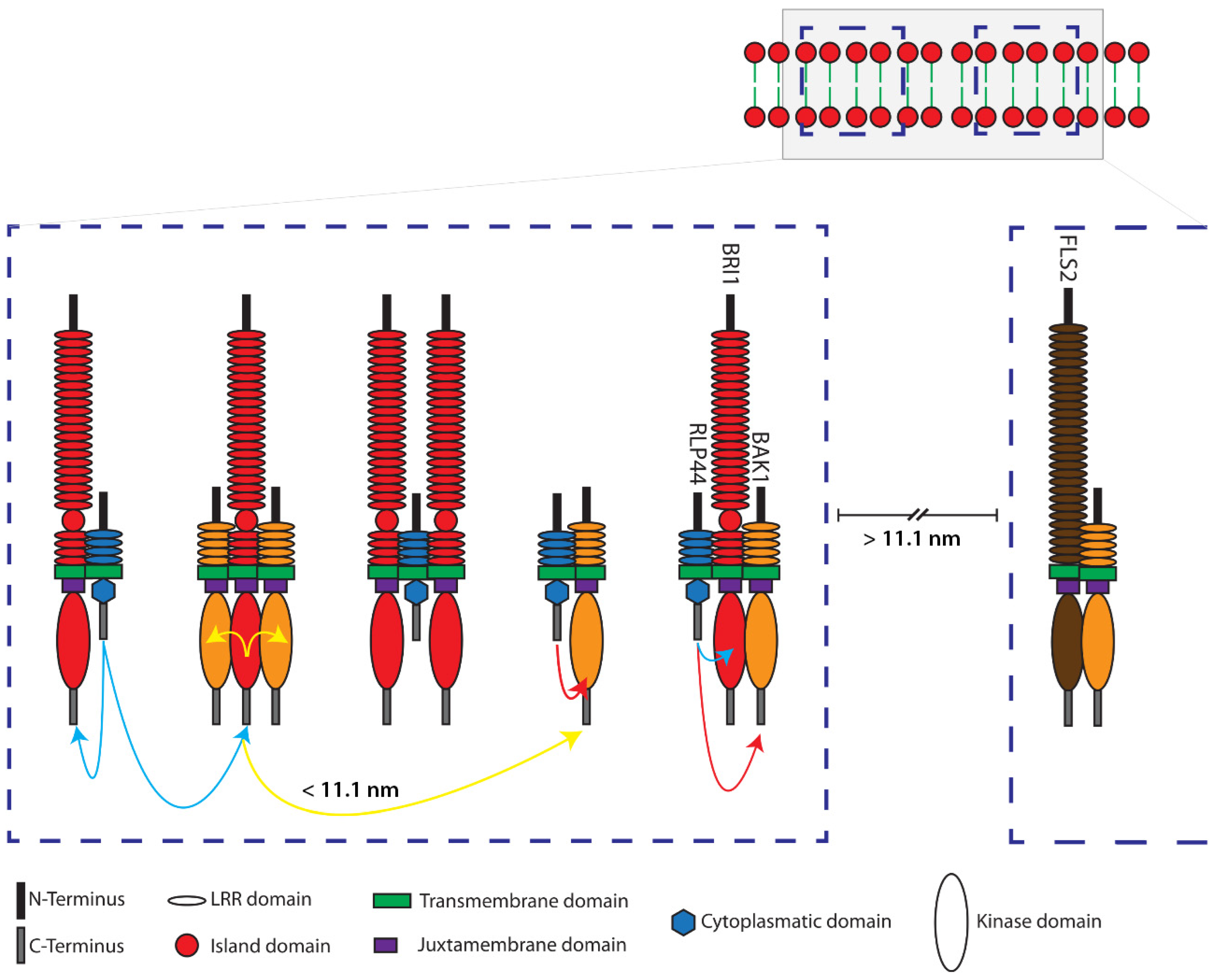
2.3. Structural Simulation of the Arrangement of the Fluorophore-Tagged Proteins for the Estimation of the FRET Range
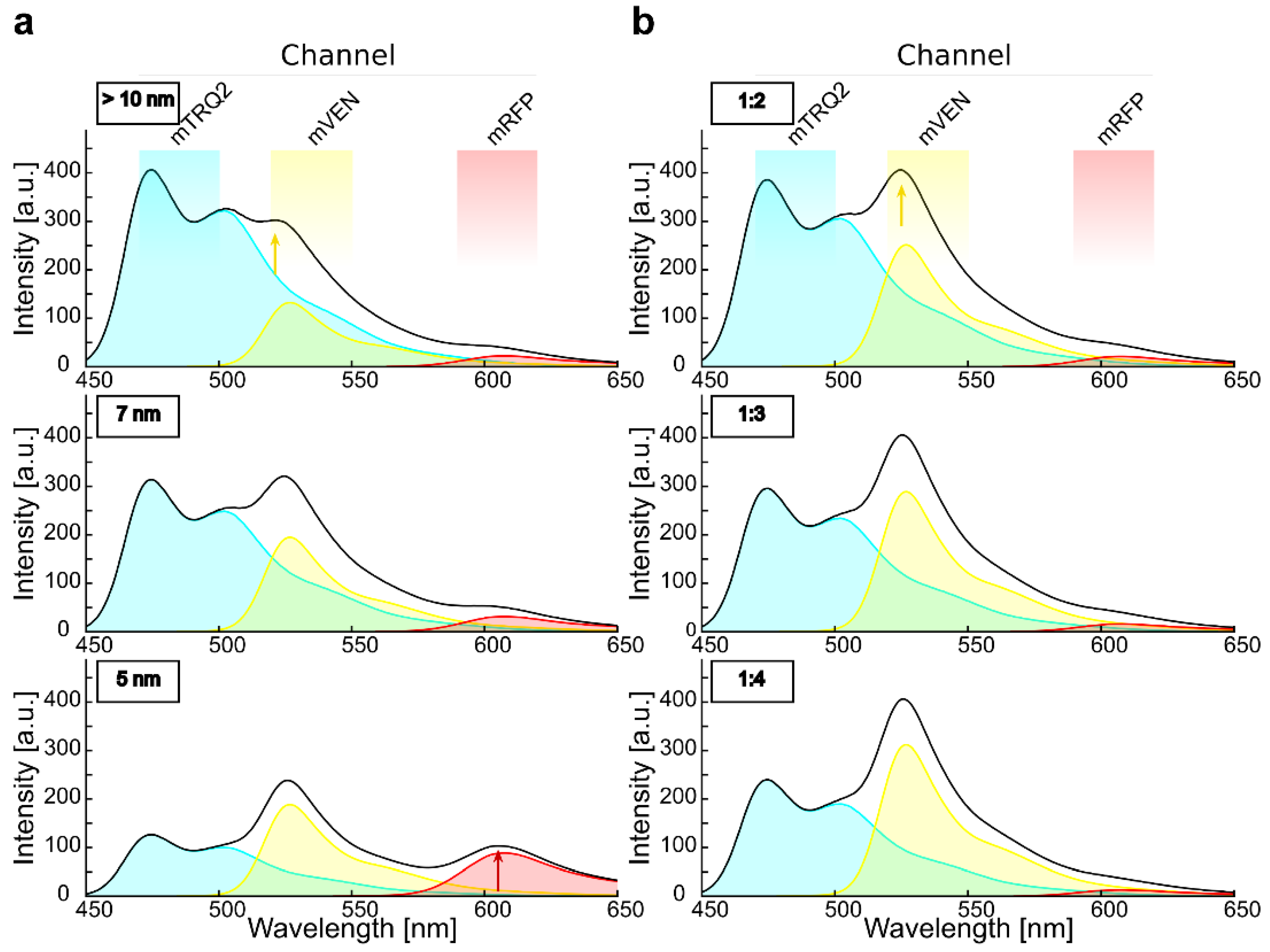
2.4. Calculation of Cross-Excitation and Bleed-Through in Intensity-Based Spectral FRET Measurements
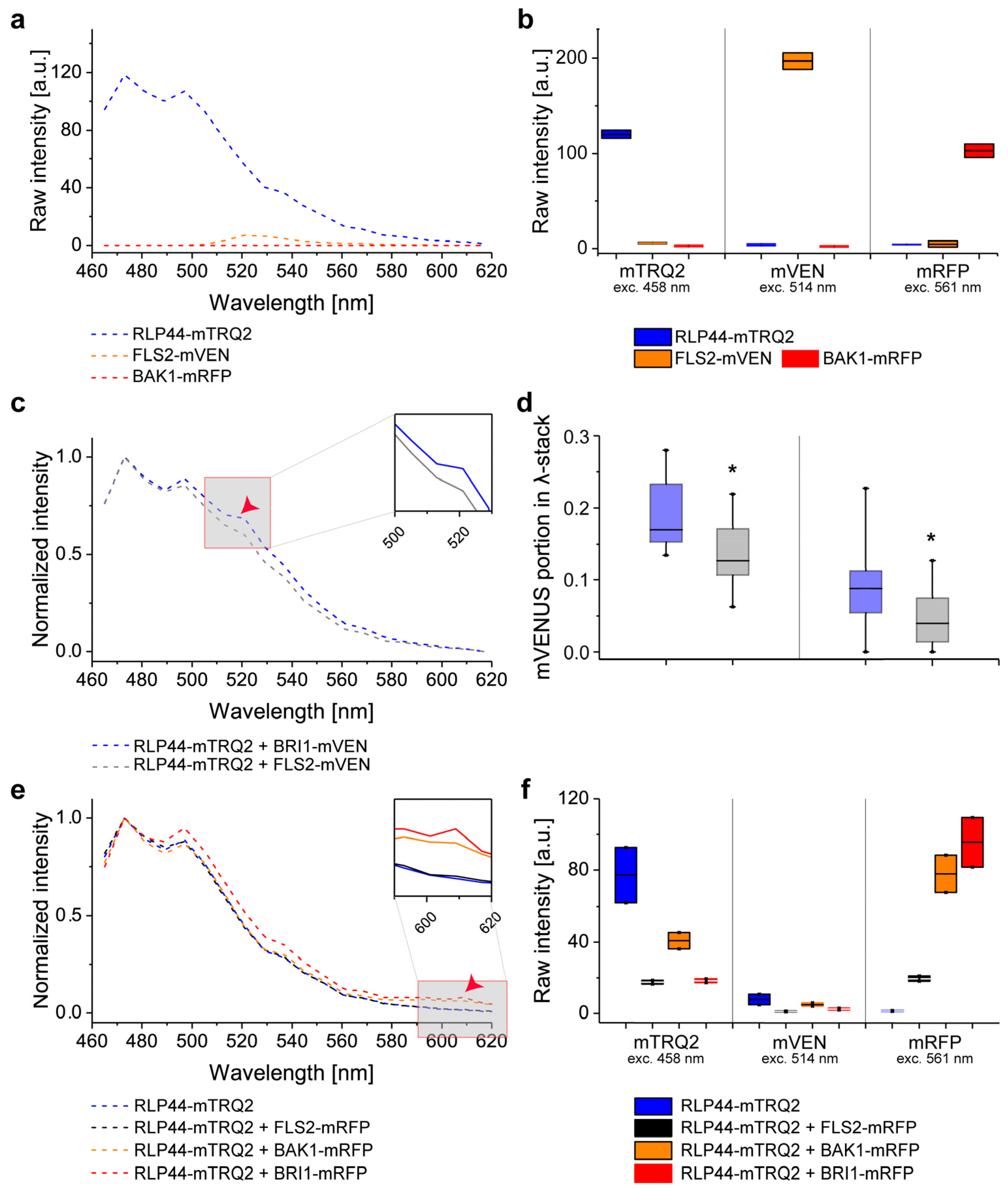
2.5. Experimental Determination of Cross-Excitation and Bleed-Through in Plant Cells
2.6. Intensity-Based FRET Analysis of Dual Protein-Protein Interactions
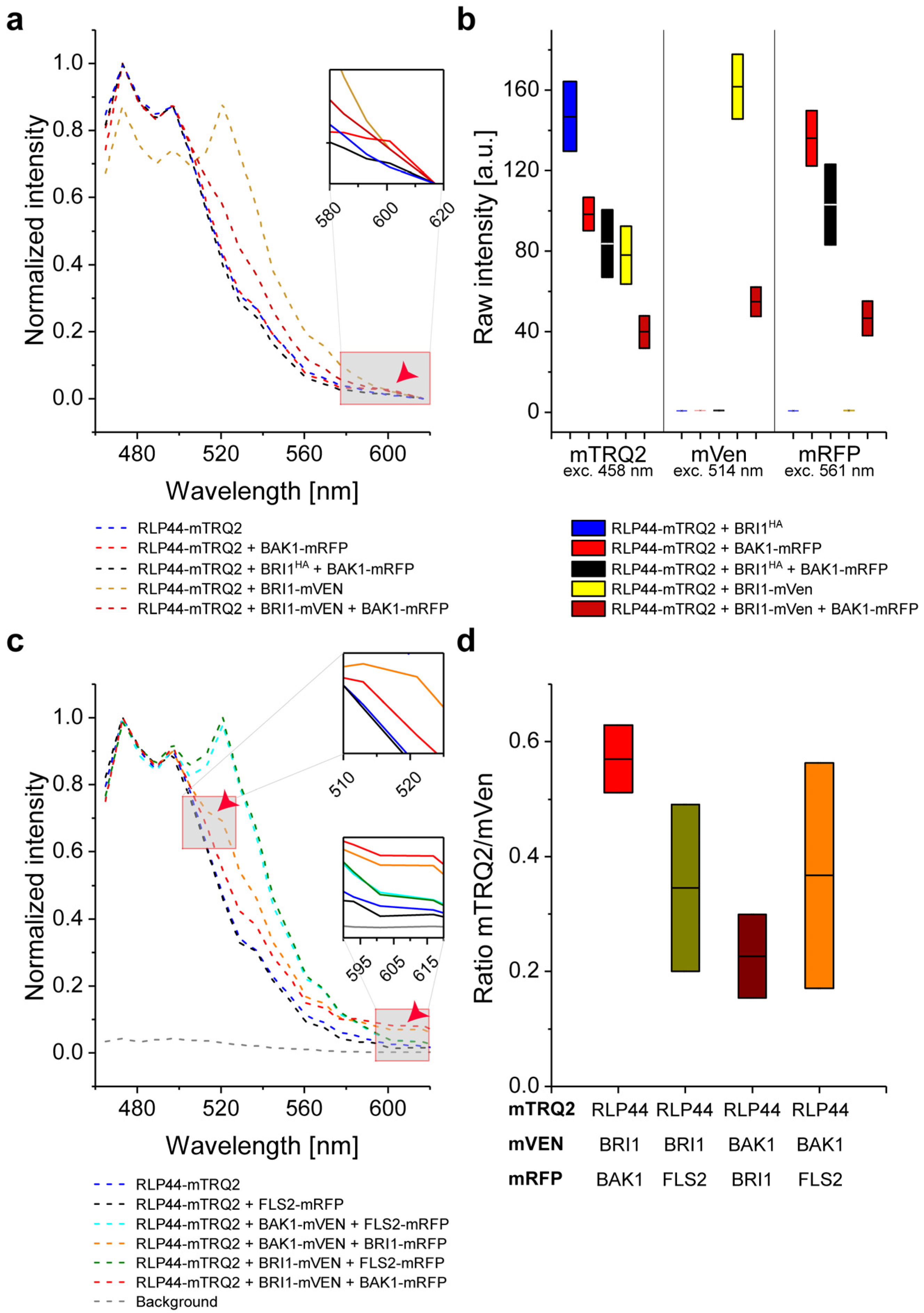
2.7. Intensity-Based FRET Analysis of Ternary Protein Complex Formation
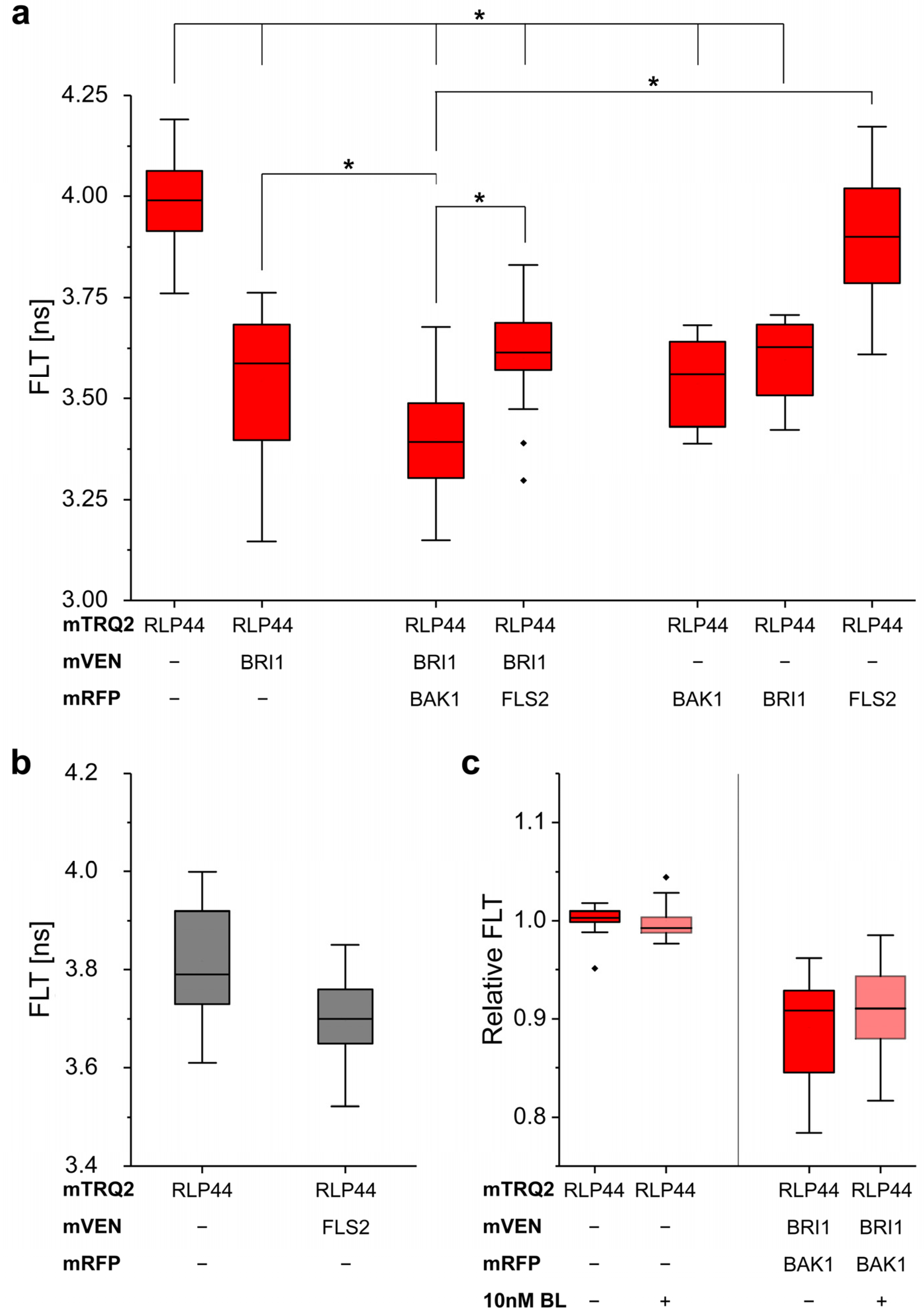
2.8. Measurement of In Vivo RLP44/BRI1/BAK1 Ternary Complex Formation by FRET-FLIM
3. Discussion
4. Material and Methods
4.1. Plasmid Construction
4.2. Localization and FRET-FLIM Studies
4.3. Acquisition of λ-Stacks (Spectra)
4.4. Protein Structures and Sizes
4.5. Statistics
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gou, X.; Li, J. Paired Receptor and Coreceptor Kinases Perceive Extracellular Signals to Control Plant Development. Plant Physiol. 2020, 182, 1667–1681. [Google Scholar] [CrossRef]
- Wolf, S. Deviating from the Beaten Track: New Twists in Brassinosteroid Receptor Function. Int. J. Mol. Sci. 2020, 21, 1561. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- Vert, G.; Chory, J. Downstream nuclear events in brassinosteroid signalling. Nature 2006, 441, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Mora-García, S.; Vert, G.; Yin, Y.; Caño-Delgado, A.; Cheong, H.; Chory, J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004, 18, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-Y.; Li, Y.; Cao, D.-M.; Yang, H.; Oh, E.; Bi, Y.; Zhu, S.; Wang, Z.-Y. The F-box Protein KIB1 Mediates Brassinosteroid-Induced Inactivation and Degradation of GSK3-like Kinases in Arabidopsis. Mol. Cell 2017, 66, 648–657.e4. [Google Scholar] [CrossRef] [PubMed]
- Caesar, K.; Elgass, K.; Chen, Z.; Huppenberger, P.; Witthöft, J.; Schleifenbaum, F.; Blatt, M.R.; Oecking, C.; Harter, K. A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J. 2011, 66, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Witthöft, J.; Caesar, K.; Elgass, K.; Huppenberger, P.; Kilian, J.; Schleifenbaum, F.; Oecking, C.; Harter, K. The activation of the Arabidopsis P-ATPase 1 by the brassinosteroid receptor BRI1 is independent of threonine 948 phosphorylation. Plant Signal. Behav. 2011, 6, 1063–1066. [Google Scholar] [CrossRef][Green Version]
- Wolf, S.; van der Does, D.; Ladwig, F.; Sticht, C.; Kolbeck, A.; Schürholz, A.-K.; Augustin, S.; Keinath, N.; Rausch, T.; Greiner, S.; et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 15261–15266. [Google Scholar] [CrossRef]
- Holzwart, E.; Huerta, A.I.; Glöckner, N.; Gómez, B.G.; Wanke, F.; Augustin, S.; Askani, J.C.; Schürholz, A.-K.; Harter, K.; Wolf, S. BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proc. Natl. Acad. Sci. USA 2018, 115, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Förster, T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys. 1948, 437, 55–75. [Google Scholar] [CrossRef]
- Noomnarm, U.; Clegg, R.M. Fluorescence lifetimes: Fundamentals and interpretations. Photosynth. Res. 2009, 101, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Goedhart, J.; von Stetten, D.; Noirclerc-Savoye, M.; Lelimousin, M.; Joosen, L.; Hink, M.A.; Van Weeren, L.; Gadella, T.W.J., Jr.; Royant, A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 2012, 3, 751. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.M.; Galliardt, H.; Schneider, J.; Barisas, B.G.; Seidel, T. Quantification of Förster resonance energy transfer by monitoring sensitized emission in living plant cells. Front. Plant Sci. 2013, 4, 413. [Google Scholar] [CrossRef]
- Hecker, A.; Wallmeroth, N.; Peter, S.; Blatt, M.R.; Harter, K.; Grefen, C. Binary 2in1 Vectors Improve in Planta (Co)localization and Dynamic Protein Interaction Studies. Plant Physiol. 2015, 168, 776–787. [Google Scholar] [CrossRef]
- Martin, K.J.; McGhee, E.J.; Schwarz, J.P.; Drysdale, M.; Brachmann, S.M.; Stucke, V.; Sansom, O.J.; Anderson, K.I. Accepting from the best donor; analysis of long-lifetime donor fluorescent protein pairings to optimise dynamic FLIM-based FRET experiments. PLoS ONE 2018, 13, e0183585. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, A.; Tsien, R.Y. Cell Biology and Physiology; Thorner, J., Ed.; Academic Press: San Diego, MA, USA, 2000; pp. 472–500. [Google Scholar]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Wouters, F.S. Förster Resonance Energy Transfer and Fluorescence Lifetime Imaging; Wiley-VCH: Weinheim, Germany, 2017. [Google Scholar]
- Großeholz, R.; Feldman-Salit, A.; Wanke, F.; Schulze, S.; Glöckner, N.; Kemmerling, B.; Harter, K.; Kummer, U. Specifying the role of BAK1-interacting receptor-like kinase 3 in brassinosteroid signaling. J. Integr. Plant Biol. 2019, 62, 456–469. [Google Scholar] [CrossRef]
- Sun, Y.; Wallrabe, H.; Booker, C.F.; Day, R.N.; Periasamy, A. Three-color spectral FRET microscopy localizes three interacting proteins in living cells. Biophys. J. 2010, 99, 1274–1283. [Google Scholar] [CrossRef] [PubMed]
- Haustein, E.; Jahnz, M.; Schwille, P. Triple FRET: A tool for studying long-range molecular interactions. ChemPhysChem 2003, 4, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Rekas, A.; Alattia, J.-R.; Nagai, T.; Miyawaki, A.; Ikura, M. Crystal structure of venus, a yellow fluorescent protein with improved maturation and reduced environmental sensitivity. J. Biol. Chem. 2002, 277, 50573–50578. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Ma, Y.; Liu, D.; Wei, X.; Sun, Y.; Chen, X.; Zhao, H.; Zhou, J.; Wang, Z.-Y.; Shui, W.; et al. Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1. Cell Res. 2012, 22, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Bojar, D.; Martinez, J.; Santiago, J.; Rybin, V.; Bayliss, R.; Hothorn, M. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014, 78, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Fasterde novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a Receptor Kinase Pair Mediating Brassinosteroid Signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef]
- Gómez, B.G.; Holzwart, E.; Shi, C.; Lozano-Durán, R.; Wolf, S. Phosphorylation-dependent routing of RLP44 towards brassinosteroid or phytosulfokine signalling. J. Cell Sci. 2021, 134, jcs259134. [Google Scholar] [CrossRef]
- Bücherl, C.A.; Jarsch, I.K.; Schudoma, C.; Segonzac, C.; Mbengue, M.; Robatzek, S.; MacLean, D.; Ott, T.; Zipfel, C. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 2017, 6, e25114. [Google Scholar] [CrossRef]
- Galperin, E.; Verkhusha, V.; Sorkin, A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat. Chem. Biol. 2004, 1, 209–217. [Google Scholar] [CrossRef]
- Hochreiter, B.; Pardo-Garcia, A.P.; Schmid, J.A. Fluorescent proteins as genetically encoded FRET biosensors in life sciences. Sensors 2015, 15, 26281–26314. [Google Scholar] [CrossRef]
- Becker, W. Fluorescence lifetime imaging-techniques and applications. J. Microsc. 2012, 247, 119–136. [Google Scholar] [CrossRef]
- Bunt, G.; Wouters, F.S. FRET from single to multiplexed signaling events. Biophys. Rev. 2017, 9, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Scott, B.L.; Hoppe, A.D. Optimizing fluorescent protein trios for 3-Way FRET imaging of protein interactions in living cells. Sci. Rep. 2015, 5, 10270. [Google Scholar] [CrossRef] [PubMed]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, A. Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Annu. Rev. Biochem. 2011, 80, 357–373. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, A.D.; Scott, B.L.; Welliver, T.P.; Straight, S.W.; Swanson, J.A. N-way FRET microscopy of multiple protein-protein interactions in live cells. PLoS ONE 2013, 8, e64760. [Google Scholar] [CrossRef]
- He, L.; Wu, X.; Simone, J.; Hewgill, D.; Lipsky, P.E. Determination of tumor necrosis factor receptor-associated factor trimerization in living cells by CFP-YFP-mRFP FRET detected by flow cytometry. Nucleic Acids Res. 2005, 33, e61. [Google Scholar] [CrossRef][Green Version]
- Kuo, H.-L.; Ho, P.-C.; Huang, S.-S.; Chang, N.-S. Chasing the signaling run by tri-molecular time-lapse FRET microscopy. Cell Death Discov. 2018, 4, 1–9. [Google Scholar] [CrossRef]
- Pauker, M.H.; Hassan, N.; Noy, E.; Reicher, B.; Barda-Saad, M. Studying the dynamics of SLP-76, Nck, and Vav1 multimolecular complex formation in live human cells with triple-color FRET. Sci. Signal. 2012, 5, rs3. [Google Scholar] [CrossRef]
- Wallrabe, H.; Cai, Y.; Sun, Y.; Periasamy, A.; Luzes, R.; Fang, X.; Kan, H.-M.; Cameron, L.-C.; Schafer, D.A.; Bloom, G.S. IQGAP1 interactome analysis by in vitro reconstitution and live cell 3-color FRET microscopy. Cytoskeleton 2013, 70, 819–836. [Google Scholar] [CrossRef][Green Version]
- Fábián, Á.; Horváth, G.; Vámosi, G.; Vereb, G.; Szöllősi, J. TripleFRET measurements in flow cytometry. Cytom. Part A J. Int. Soc. Anal. Cytol. 2013, 83, 375–385. [Google Scholar] [CrossRef]
- Fazekas, Z.; Petrás, M.; Fábián, Á.; Pályi-Krekk, Z.; Nagy, P.; Damjanovich, S.; Vereb, G.; Szöllősi, J. Two-sided fluorescence resonance energy transfer for assessing molecular interactions of up to three distinct species in confocal microscopy. Cytom. Part A J. Int. Soc. Anal. Cytol. 2008, 73, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Grefen, C.; Blatt, M.R. A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). BioTechniques 2012, 53, 311–314. [Google Scholar] [CrossRef]
- Ladwig, F.; Dahlke, R.I.; Stührwohldt, N.; Hartmann, J.; Harter, K.; Sauter, M. Phytosulfokine Regulates Growth in Arabidopsis through a Response Module at the Plasma Membrane That Includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 2015, 27, 1718–1729. [Google Scholar] [CrossRef] [PubMed]
- Mohrholz, A.; Sun, H.; Glöckner, N.; Hummel, S.; Kolukisaoglu, Ü.; Schneeberger, K.; Harter, K. The striking flower-in-flower phenotype of Arabidopsis thaliana Nossen (No-0) is caused by a novel LEAFY allele. Plants 2019, 8, 599. [Google Scholar] [CrossRef]
- Jmol: An Open-Source Java Viewer for Chemical Structures in 3D. Available online: http://www.jmol.org/ (accessed on 23 September 2022).
- MATLAB. 9.8.0.1417392 (R2020a); The MathWorks Inc.: Natick, MA, USA, 2020. [Google Scholar]
- Ohmi, Y.; Ise, W.; Harazono, A.; Takakura, D.; Fukuyama, H.; Baba, Y.; Narazaki, M.; Shoda, H.; Takahashi, N.; Ohkawa, Y.; et al. Sialylation converts arthritogenic IgG into inhibitors of collagen-induced arthritis. Nat. Commun. 2016, 7, 11205. [Google Scholar] [CrossRef] [PubMed]
- de Winter, J.C.F. Using the Student’s “t”-Test with Extremely Small Sample Sizes. Pract. Assess. Res. Eval. 2013, 18, 10. [Google Scholar]
- Balleza, E.; Kim, J.M.; Cluzel, P. Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 2018, 15, 47–51. [Google Scholar] [CrossRef]
- Campbell, R.E.; Tour, O.; Palmer, A.E.; Steinbach, P.A.; Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. A Monomeric Red Fluorescent Protein. Proceedings of the National Academy of Sciences of the United States of America. 2002, 99, 7877–7882. [Google Scholar] [CrossRef]
- Cranfill, P.J.; Sell, B.R.; Baird, M.A.; Allen, J.R.; Lavagnino, Z.; Gruiter, H.M. de; Piston, D.W. Quantitative assessment of fluorescent proteins. Nat. Methods 2016, 13, 557–562. [Google Scholar] [CrossRef]
- Koushik, S.V.; Blank, P.S.; Vogel, S.S. Anomalous surplus energy transfer observed with multiple FRET acceptors. PloS ONE 4. 2009, e8031. [Google Scholar] [CrossRef]
- Kremers, G.-J.; Goedhart, J.; van Munster, E.B.; Gadella, T W. J.; JR. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry. 2006, 45, 6570–6580. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. FRET study of a trifluorophore-labeled DNAzyme. J. Am. Chem. Soc. 2002, 124, 15208–15216. [Google Scholar] [CrossRef]
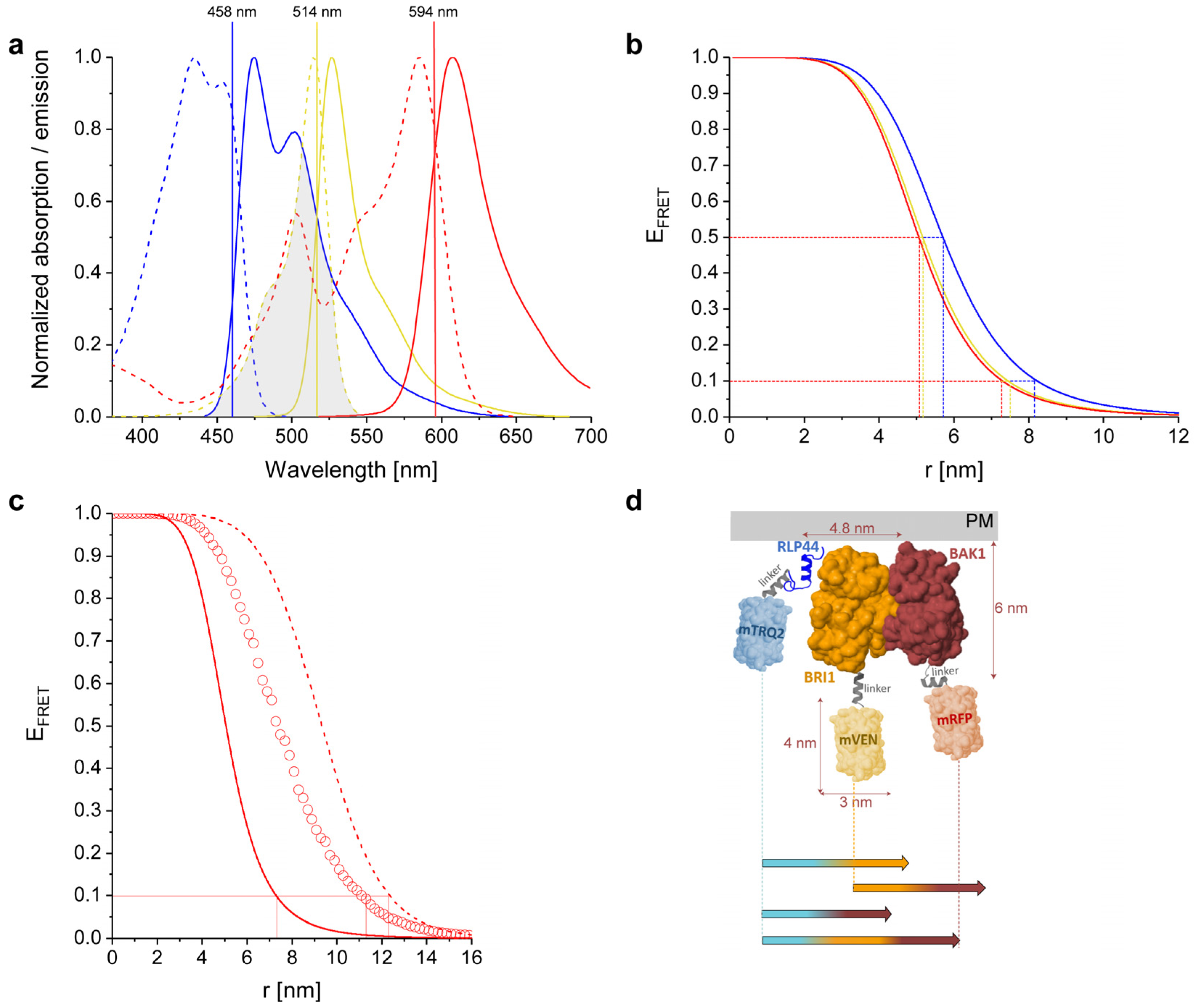
| FRET Combinations | R0 [nm] | r10% [nm], D:A = 1:1 |
|---|---|---|
| mTRQ2-mVEN | 5.7 | 8.2 |
| mVEN-mRFP | 5.2 | 7.4 |
| mTRQ2-mRFP | 5.1 | 7.3 |
| mTRQ2-mVEN-mRFP (middle position) | - | 12.4 |
| mTRQ2-mVEN-mRFP (random position) | - | 11.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glöckner, N.; zur Oven-Krockhaus, S.; Rohr, L.; Wackenhut, F.; Burmeister, M.; Wanke, F.; Holzwart, E.; Meixner, A.J.; Wolf, S.; Harter, K. Three-Fluorophore FRET Enables the Analysis of Ternary Protein Association in Living Plant Cells. Plants 2022, 11, 2630. https://doi.org/10.3390/plants11192630
Glöckner N, zur Oven-Krockhaus S, Rohr L, Wackenhut F, Burmeister M, Wanke F, Holzwart E, Meixner AJ, Wolf S, Harter K. Three-Fluorophore FRET Enables the Analysis of Ternary Protein Association in Living Plant Cells. Plants. 2022; 11(19):2630. https://doi.org/10.3390/plants11192630
Chicago/Turabian StyleGlöckner, Nina, Sven zur Oven-Krockhaus, Leander Rohr, Frank Wackenhut, Moritz Burmeister, Friederike Wanke, Eleonore Holzwart, Alfred J. Meixner, Sebastian Wolf, and Klaus Harter. 2022. "Three-Fluorophore FRET Enables the Analysis of Ternary Protein Association in Living Plant Cells" Plants 11, no. 19: 2630. https://doi.org/10.3390/plants11192630
APA StyleGlöckner, N., zur Oven-Krockhaus, S., Rohr, L., Wackenhut, F., Burmeister, M., Wanke, F., Holzwart, E., Meixner, A. J., Wolf, S., & Harter, K. (2022). Three-Fluorophore FRET Enables the Analysis of Ternary Protein Association in Living Plant Cells. Plants, 11(19), 2630. https://doi.org/10.3390/plants11192630







