Comparative Transcriptome and Anatomic Characteristics of Stems in Two Alfalfa Genotypes
Abstract
1. Introduction
2. Results
2.1. Nutritional Quality Analysis
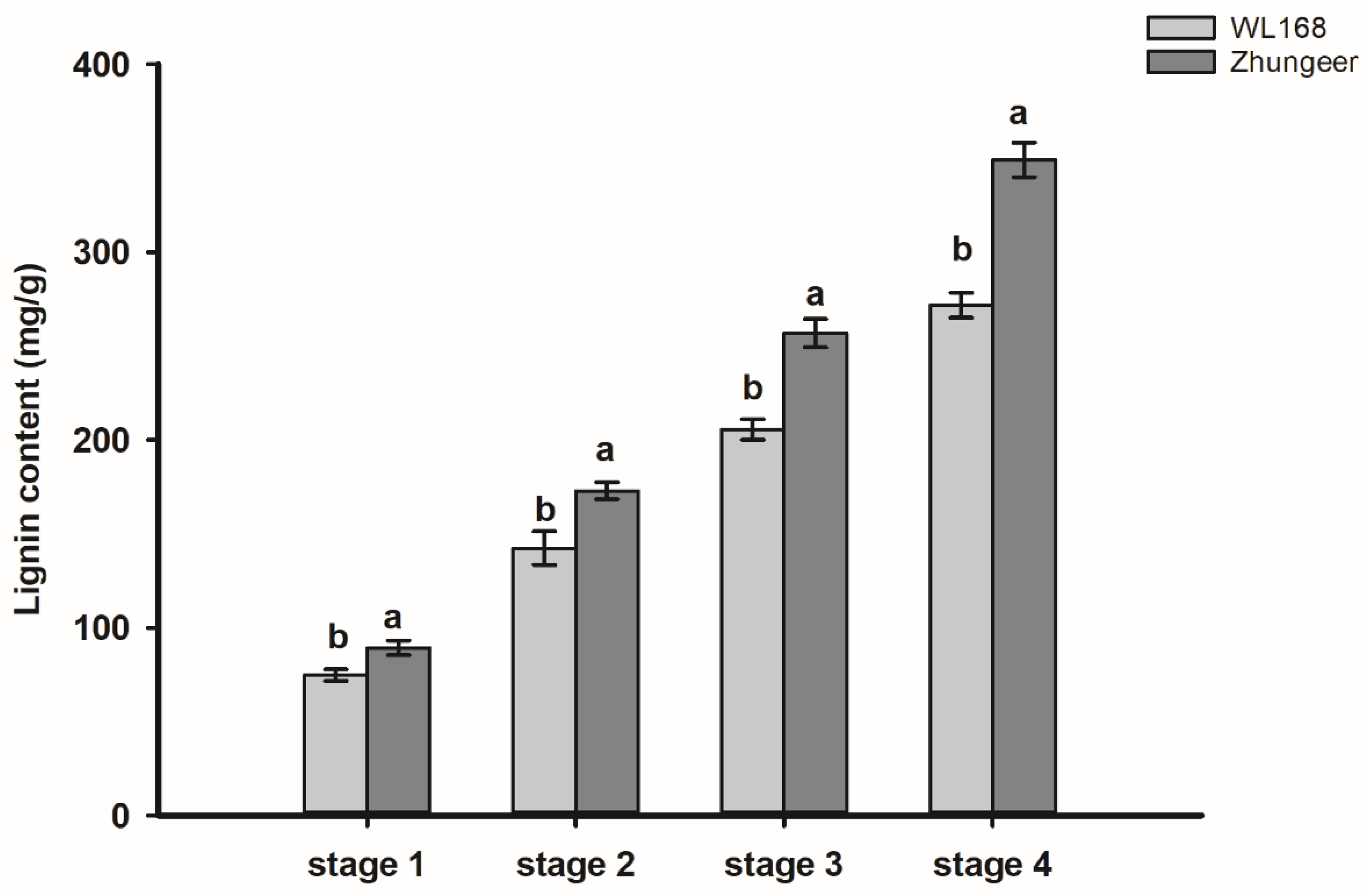
2.2. Lignin Content Analysis
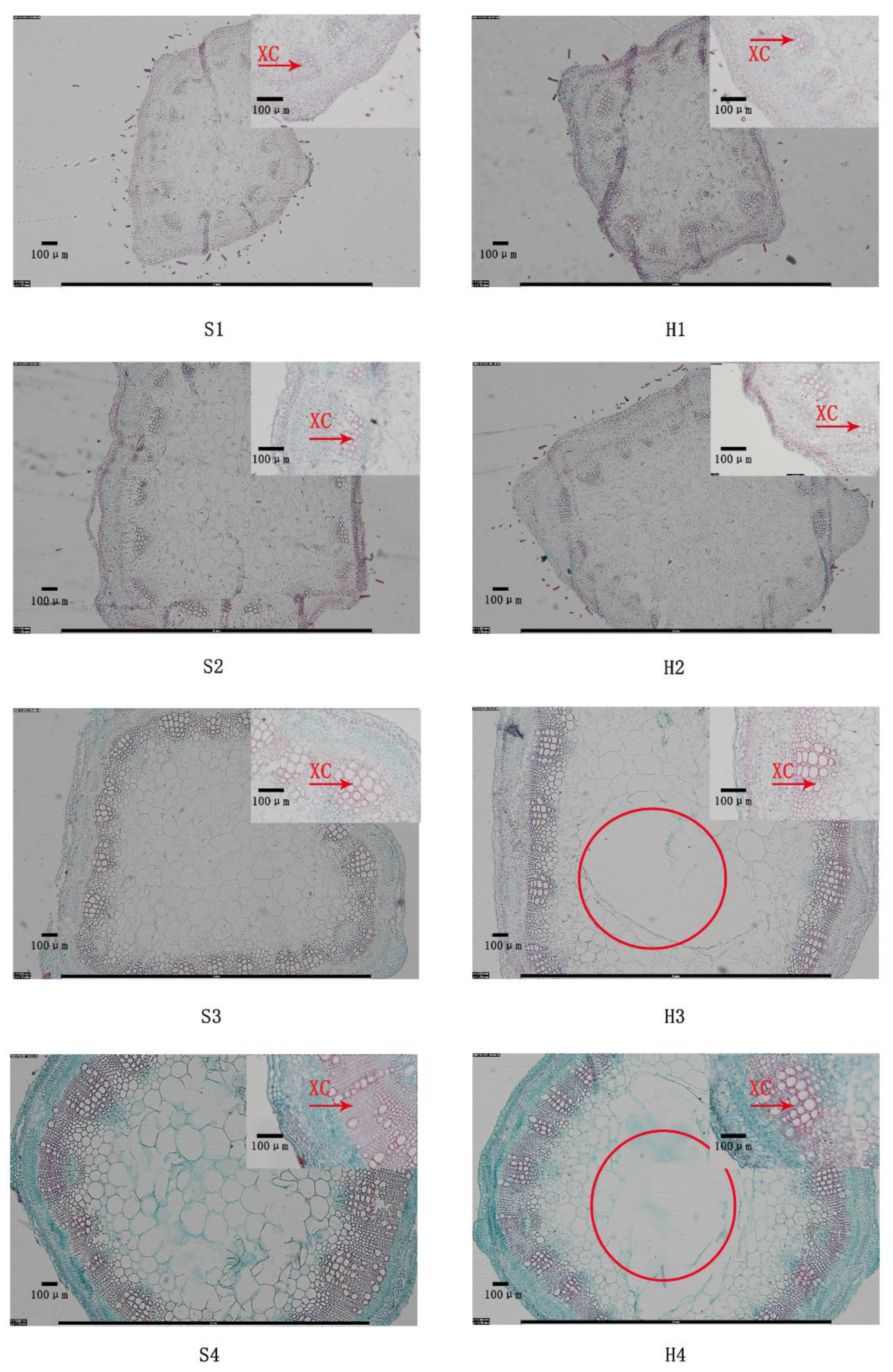
2.3. Stem Structure Analysis
2.4. Summary of Transcriptome Sequencing, Assembly and Annotation
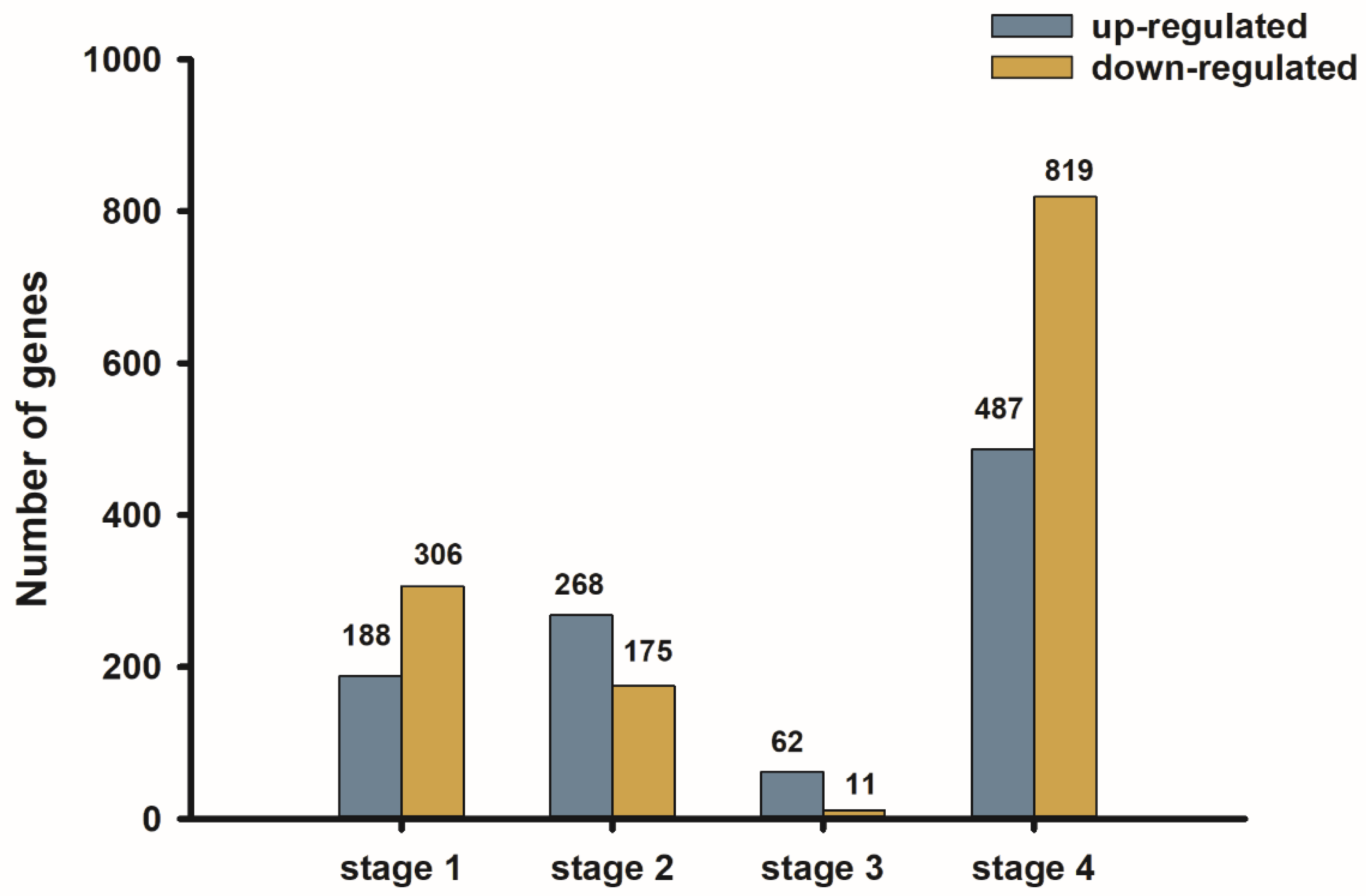
2.5. Differentially Expressed Genes and Functional Enrichment Analysis
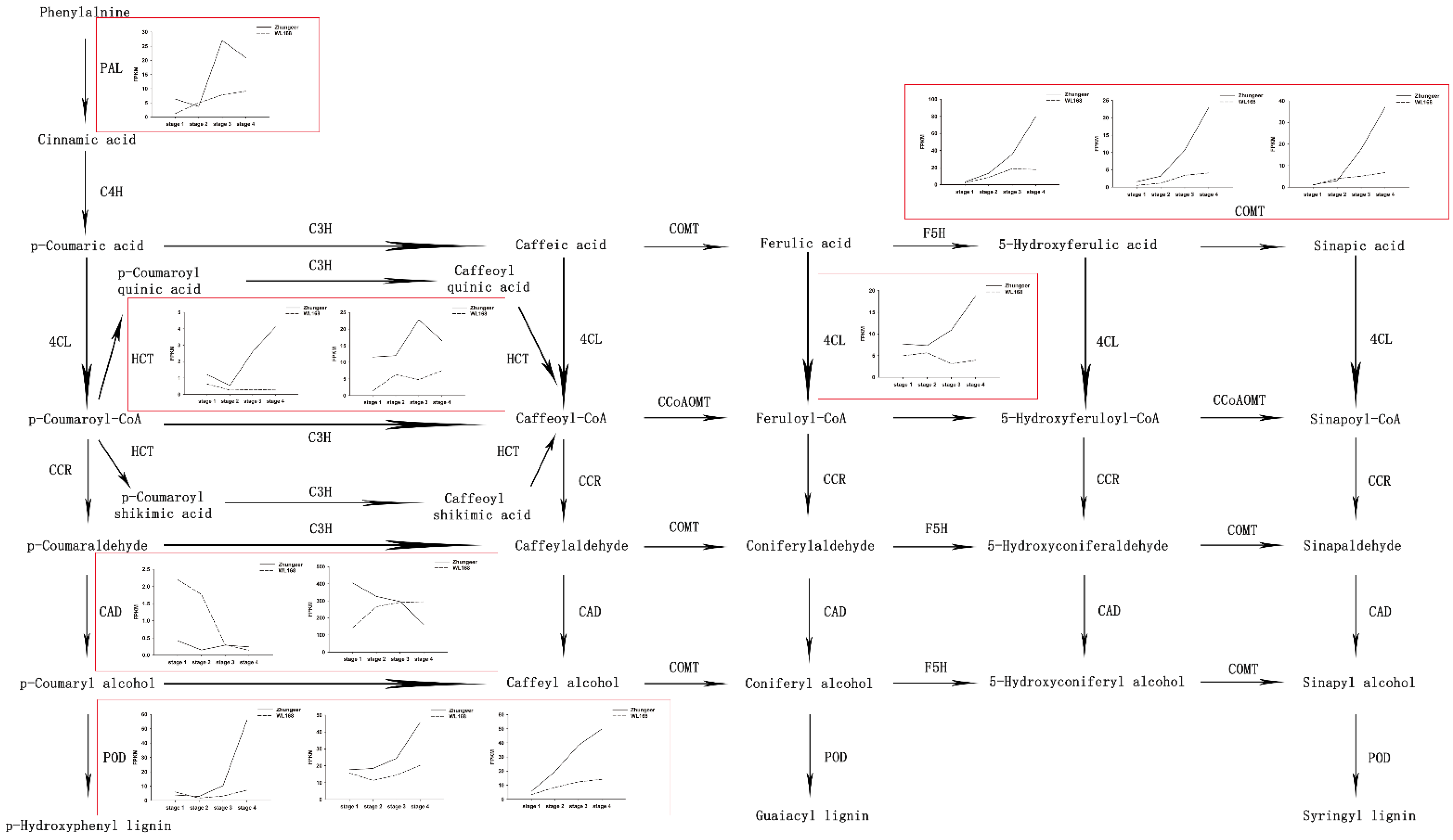
2.6. Expression Profiles of Candidate Genes Involved in Lignin Biosynthesis at Different Developmental Stages in Alfalfa
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Sample Collection
4.2. Nutritional Quality Analysis
4.3. Total Lignin Content Assay
4.4. Histological Analysis
4.5. cDNA Library Construction and Transcriptome Sequencing
4.6. Transcriptome Assembly
4.7. Annotation and Classification
4.8. Differentially Expressed Genes Analysis
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Engels, F.M.; Jung, H.G. Alfalfa stem tissues: Cell-wall development and lignification. Ann. Bot. 1998, 82, 561–568. [Google Scholar] [CrossRef]
- Doblin, M.S.; Pettolino, F.; Bacic, A. Plant cell walls: The skeleton of the plant world. Funct. Plant Biol. 2010, 37, 357–381. [Google Scholar] [CrossRef]
- Jung, H.G.; Engels, F.M. Alfalfa stem tissues: Cell wall deposition, composition, and degradability. Crop Sci. 2002, 42, 524–534. [Google Scholar] [CrossRef]
- Ding, S.Y.; Liu, Y.S.; Zeng, Y.; Himmel, M.E.; Baker, J.O.; Bayer, E.A. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 2012, 338, 1055–1060. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.S.; Chen, F.; Shadle, G.; Jackson, L.; Aljoe, H.; Dixon, R.A. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. USA 2005, 102, 16573–16578. [Google Scholar] [CrossRef] [PubMed]
- Dien, B.S.; Miller, D.J.; Hector, R.E.; Dixon, R.A.; Chen, F.; McCaslin, M.; Reisen, P.; Sarath, G.; Cotta, M.A. Enhancing alfalfa conversion efficiencies for sugar recovery and ethanol production by altering lignin composition. Bioresour. Technol. 2011, 102, 6479–6486. [Google Scholar] [CrossRef]
- Hu, J.Q.; Qi, Q.; Zhao, Y.L.; Tian, X.-M.; Lu, H.; Gai, Y.; Jiang, X.-N. Unraveling the impact of Pto4CL1 regulation on the cell wall components and wood properties of perennial transgenic Populus tomentosa. Plant Physiol. Biochem. 2019, 139, 672–680. [Google Scholar] [CrossRef]
- Piñera-Chavez, F.J.; Berry, P.M.; Foulkes, M.J.; Jesson, M.A.; Reynolds, M.P. Avoiding lodging in irrigated spring wheat. I. Stem and root structural requirements. Field Crops Res. 2016, 196, 325–336. [Google Scholar] [CrossRef]
- Huang, J.; Liu, W.; Zhou, F.; Peng, Y.; Wang, N. Mechanical properties of maize fibre bundles and their contribution to lodging resistance. Biosyst. Eng. 2016, 151, 298–307. [Google Scholar] [CrossRef]
- Jones, L.; Ennos, A.R.; Turner, S.R. Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 2001, 26, 205–216. [Google Scholar] [CrossRef]
- Wang, C.; Ruan, R.W.; Yuan, X.H.; Hu, D.; Yang, H.; Li, Y.; Yi, Z.L. Effects of nitrogen fertilizer and planting density on the lignin synthesis in the culm in relation to lodging resistance of buckwheat. Plant Prod. Sci. 2015, 18, 218–227. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, J.; Li, H.; Cao, X.; Xia, X.; He, Z. Lodging resistance and yield potential of winter wheat: Effect of planting density and genotype. Front. Agric. Sci. Eng. 2015, 2, 168–178. [Google Scholar] [CrossRef]
- You, T.T.; Mao, J.Z.; Yuan, T.Q.; Wen, J.L.; Xu, F. Structural elucidation of the lignins from stems and foliage of Arundo donax Linn. J. Agric. Food Chem. 2013, 61, 5361–5370. [Google Scholar] [CrossRef]
- Wang, Y.; Chantreau, M.; Sibout, R.; Hawkins, S. Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 2013, 4, 220. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.F.; Jung, H.J.G.; Sheaffer, C.C.; Samac, D.A. Alfalfa leaf protein and stem cell wall polysaccharide yields under hay and biomass management systems. Crop Sci. 2007, 47, 1407–1415. [Google Scholar] [CrossRef]
- Barros, J.; Temple, S.; Dixon, R.A. Development and commercialization of reduced lignin alfalfa. Curr. Opin. Biotechnol. 2019, 56, 48–54. [Google Scholar] [CrossRef]
- Sulc, R.M.; Arnold, A.M.; Cassida, K.A.; Albrecht, K.A.; Hall, M.H.; Min, D.; Xu, X.; Undersander, D.J.; van Santen, E. Changes in forage nutritive value of reduced-lignin alfalfa during regrowth. Crop Sci. 2021, 61, 1478–1487. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Ann. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Liu, C.J. Deciphering the enigma of lignification: Precursor transport, oxidation, and the topochemistry of lignin assembly. Mol. Plant 2012, 5, 304–317. [Google Scholar] [CrossRef]
- Ralph, J.; Lundquist, K.; Brunow, G.; Lu, F.; Kim, H.; Schatz, P.F.; Marita, J.M.; Hatfield, R.D.; Ralph, S.A.; Christensen, J.H.; et al. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem. Rev. 2004, 3, 29–60. [Google Scholar] [CrossRef]
- Shadle, G.; Chen, F.; Reddy, M.S.; Jackson, L.; Nakashima, J.; Dixon, R.A. Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 2007, 68, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Getachew, G.; Ibáñez, A.M.; Pittroff, W.; Dandekar, A.M.; McCaslin, M.; Goyal, S.; Reisen, P.; DePeters, E.J.; Putnam, D.H. A comparative study between lignin down regulated alfalfa lines and their respective unmodified controls on the nutritional characteristics of hay. Anim. Feed Sci. Technol. 2011, 170, 192–200. [Google Scholar] [CrossRef]
- Tong, Z.; Li, H.; Zhang, R.; Ma, L.; Dong, J.; Wang, T. Co-downregulation of the hydroxycinnamoyl-CoA: Shikimate hydroxycinnamoyl transferase and coumarate 3-hydroxylase significantly increases cellulose content in transgenic alfalfa (Medicago sativa L.). Plant Sci. 2015, 239, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Chapple, C. Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol. 2013, 24, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.F.; Chandra, R.; Berleth, T.; Beatson, R.P. Rapid, microscale, acetyl bromide-based method for high-throughput determination of lignin content in Arabidopsis thaliana. J. Agric. Food Chem. 2008, 56, 6825–6834. [Google Scholar] [CrossRef]
- Sheaffer, C.C.; Martin, N.P.; Lamb, J.F.; Cuomo, G.R.; Jewett, J.G.; Quering, S.R. Leaf and stem properties of alfalfa entries. Agron. J. 2000, 92, 733–739. [Google Scholar] [CrossRef]
- Lamb, J.F.; Hans-Joachim, G.J.; Riday, H. Harvest impacts on alfalfa stem neutral detergent fiber concentration and digestibility and cell wall concentration and composition. Crop Sci. 2012, 52, 2402–2412. [Google Scholar] [CrossRef]
- Albrecht, K.A.; Wedin, W.F.; Buxton, D.R. Cell-Wall Composition and Digestibility of Alfalfa Stems and Leaves 1. Crop Sci. 1987, 27, 735–741. [Google Scholar] [CrossRef]
- Akin, D.E.; Robinson, E.L.; Barton, F.E.; Himmelsbach, D.S. Changes with maturity in anatomy, histochemistry, chemistry, and tissue digestibility of bermudagrass plant parts. J. Agric. Food Chem. 1977, 25, 179–186. [Google Scholar] [CrossRef]
- Jung, H.G.; Vogel, K.P. Influence of lignin on digestibility of forage cell wall material. J. Anim. Sci. 1986, 62, 1703–1712. [Google Scholar] [CrossRef]
- Grev, A.M.; Wells, M.S.; Catalano, D.N.; Martinson, K.L.; Jungers, J.M.; Sheaffer, C.C. Stem and leaf forage nutritive value and morphology of reduced lignin alfalfa. Agron. J. 2020, 112, 406–417. [Google Scholar] [CrossRef]
- Liu, N.; Yu, P. Molecular clustering, interrelationships and carbohydrate conformation in hull and seeds among barley cultivars. J. Cereal Sci. 2011, 53, 379–383. [Google Scholar] [CrossRef]
- Jung, H.J.G.; Samac, D.A.; Sarath, G. Modifying crops to increase cell wall digestibility. Plant Sci. 2012, 185, 65–77. [Google Scholar] [CrossRef]
- Guo, D.; Chen, F.; Inoue, K.; Blount, J.W.; Dixon, R.A. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 2001, 13, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Grev, A.M.; Wells, M.S.; Samac, D.A.; Martinson, K.L.; Sheaffer, C.C. Forage accumulation and nutritive value of reduced lignin and reference alfalfa cultivars. Agron. J. 2017, 109, 2749–2761. [Google Scholar] [CrossRef]
- Lanning, S.P.; Fox, P.; Elser, J.; Martin, J.M.; Blake, N.K.; Talbert, L.E. Microsatellite markers associated with a secondary stem solidness locus in wheat. Crop Sci. 2006, 46, 1701–1703. [Google Scholar] [CrossRef]
- Kong, E.; Liu, D.; Guo, X.; Yang, W.; Sun, J.; Li, X.; Zhan, K.; Cui, D.; Lin, J.; Zhang, A. Anatomical and chemical characteristics associated with lodging resistance in wheat. Crop J. 2013, 1, 43–49. [Google Scholar] [CrossRef]
- Zhang, S.; Shi, Y.; Cheng, N.; Du, H.; Fan, W.; Wang, C. De novo characterization of fall dormant and nondormant alfalfa (Medicago sativa L.) leaf transcriptome and identification of candidate genes related to fall dormancy. PLoS ONE 2015, 10, e0122170. [Google Scholar] [CrossRef]
- Arshad, M.; Gruber, M.Y.; Hannoufa, A. Transcriptome analysis of microRNA156 overexpression alfalfa roots under drought stress. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, X.; Wang, L.; Liu, H.; Zhao, Y.; Yi, K.; Cui, G.; Yin, X. Transcriptome profiling unveils the mechanism of phenylpropane biosynthesis in rhizome development of Caucasian clover. PLoS ONE 2021, 16, e0254669. [Google Scholar] [CrossRef]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Hannoufa, A.; Prates, L.L.; Christensen, D.; Wang, Y.; Yu, P. Silencing TT8 and HB12 decreased protein degradation and digestion, microbial synthesis, and metabolic protein in relation to molecular structures of alfalfa (Medicago sativa). J. Agric. Food Chem. 2019, 67, 7898–7907. [Google Scholar] [CrossRef] [PubMed]
- Jeong, M.J.; Bo, S.C.; Dong, W.B.; Shin, S.C.; Bae, H. Differential expression of kenaf phenylalanine ammonia-lyase (pal) ortholog during developmental stages and in response to abiotic stresses. Plant Omics 2012, 3, 392–399. [Google Scholar] [CrossRef]
- Chowdhury, M.E.K.; Choi, B.; Cho, B.K.; Kim, J.B.; Park, S.U.; Natarajan, S.; Lim, H.-S.; Bae, H. Regulation of 4CL, encoding 4-coumarate: Coenzyme A ligase, expression in kenaf under diverse stress conditions. Plant Omics 2013, 6, 254–262. [Google Scholar]
- Hoffmann, L.; Maury, S.; Martz, F.; Geoffroy, P.; Legrand, M. Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Biol. Chem. 2003, 278, 95–103. [Google Scholar] [CrossRef]
- Pu, Y.; Chen, F.; Ziebell, A.; Davison, B.H.; Ragauskas, A.J. NMR characterization of C3H and HCT down-regulated alfalfa lignin. BioEnergy Res. 2009, 2, 198–208. [Google Scholar] [CrossRef]
- Xu, Y.; Thammannagowda, S.; Thomas, T.P.; Azadi, P.; Schlarbaum, S.E.; Liang, H. LtuCAD1 is a cinnamyl alcohol dehydrogenase ortholog involved in lignin biosynthesis in Liriodendron tulipifera L., a basal angiosperm timber species. Plant Mol. Biol. Rep. 2013, 31, 1089–1099. [Google Scholar] [CrossRef]
- Preisner, M.; Kulma, A.; Zebrowski, J.; Dymińska, L.; Hanuza, J.; Arendt, M.; Starzycki, M.; Szopa, J. Manipulating cinnamyl alcohol dehydrogenase (CAD) expression in flax affects fibre composition and properties. BMC Plant Biol. 2014, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; de Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.-L.; Wang, G.-L.; Xiong, F.; Yu, X.-R.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. De novo assembly, transcriptome characterization, lignin accumulation and anatomic characteristics: Novel insights into lignin biosynthesis during celery leaf development. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef]
- Vogel, K.P.; Pedersen, J.F.; Masterson, S.D.; Toy, J.J. Evaluation of a Filter Bag System for NDF, and IVDMD Forage Analysis. Crop Sci. 1999, 1, 276–279. [Google Scholar] [CrossRef]
- Bean, B.W.; Baumhardt, R.L.; McCollum, F.T., III; McCuistion, K.C. Comparison of sorghum classes for grain and forage yield and forage nutritive value. Field Crops Res. 2013, 142, 20–26. [Google Scholar] [CrossRef]
- Moreira-Vilar, F.C.; Siqueira-Soares, R.D.C.; Finger-Teixeira, A.; Oliveira, D.M.D.; Ferro, A.P.; da Rocha, G.J.; de Ferrarese, M.L.L.; dos Santos, W.D.; Ferrarese-Filho, O. The acetyl bromide method is faster, simpler and presents best recovery of lignin in different herbaceous tissues than Klason and thioglycolic acid methods. PLoS ONE 2014, 9, e110000. [Google Scholar] [CrossRef]
- Su, G.; An, Z.; Zhang, W.; Liu, Y. Light promotes the synthesis of lignin through the production of H2O2 mediated by diamine oxidases in soybean hypocotyls. J. Plant Physiol. 2005, 162, 1297–1303. [Google Scholar] [CrossRef]
- Tong, N.-N.; Peng, L.-P.; Liu, Z.-A.; Li, Y.; Zhou, X.-Y.; Wang, X.-R.; Shu, Q.-Y. Comparative transcriptomic analysis of genes involved in stem lignin biosynthesis in woody and herbaceous Paeonia species. Physiol. Plant. 2021, 173, 961–977. [Google Scholar] [CrossRef]
- Shi, R.; Jiao, W.; Yun, L.; Zhang, Z.; Zhang, X.; Wang, Q.; Li, Y.; Mi, F. Utilization of Transcriptome, Small RNA, and Degradome Sequencing to Provide Insights Into Drought Stress and Rewatering Treatment in Medicago ruthenica. Front. Plant Sci. 2021, 12, 675903. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, X.; Liu, W. De novo transcriptomic analysis during Lentinula edodes fruiting body growth. Gene 2018, 641, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Q.; Tan, Y.; Shuang, S.; Dai, R.; Jiang, X.; Temeur, B. Combined Transcriptome and Metabolome Analysis of Alfalfa Response to Thrips Infection. Genes 2021, 12, 1967. [Google Scholar] [CrossRef] [PubMed]


| Length Range | Transcript | Unigene |
|---|---|---|
| 300–500 | 26,086 (20.85%) | 16,511 (29.46%) |
| 500–1000 | 36,879 (29.48%) | 16,535 (29.50%) |
| 1000–2000 | 39,075 (31.24%) | 14,339 (25.58%) |
| >2000 | 23,053 (18.43%) | 8669 (15.47%) |
| Total Number | 125,093 | 56,054 |
| Total Length | 161,338,984 | 64,388,875 |
| N50 Length | 1761 | 1668 |
| Mean Length | 1290 | 1148.69 |
| Database | Number Annotated | Percentage (%) |
|---|---|---|
| COG | 15,281 | 27.26 |
| GO | 25,913 | 46.23 |
| KEGG | 17,045 | 30.41 |
| KOG | 23,860 | 42.57 |
| Pfam | 30,672 | 54.72 |
| Swissprot | 25,608 | 45.68 |
| eggNOG | 40,075 | 71.49 |
| Nr | 44,610 | 79.58 |
| Overall | 45,398 | 80.99 |
| GO Terms | H1 vs. S1 | H2 vs. S2 | H3 vs. S3 | H4 vs. S4 |
|---|---|---|---|---|
| Biological Process | ||||
| metabolic process | 137 | 95 | 10 | 390 |
| cellular process | 115 | 93 | 11 | 398 |
| single-organism process | 101 | 75 | 10 | 272 |
| biological regulation | 34 | 38 | 2 | 137 |
| localization | 16 | 13 | 1 | 76 |
| response to stimulus | 42 | 33 | 4 | 109 |
| cellular component organization or biogenesis | 12 | 21 | 1 | 74 |
| signaling | 14 | 16 | 2 | 51 |
| developmental process | 4 | 11 | 2 | 24 |
| multicellular organismal process | 11 | 11 | 2 | 24 |
| reproductive process | 8 | 3 | 0 | 11 |
| multi-organism process | 15 | 3 | 0 | 5 |
| Cellular Component | ||||
| cell | 71 | 61 | 6 | 322 |
| cell part | 71 | 61 | 6 | 321 |
| membrane | 101 | 68 | 9 | 242 |
| organelle | 34 | 40 | 4 | 222 |
| membrane part | 86 | 54 | 8 | 207 |
| macromolecular complex | 7 | 14 | 1 | 78 |
| organelle part | 15 | 18 | 2 | 109 |
| membrane-enclosed lumen | 1 | 4 | 0 | 25 |
| extracellular region | 13 | 7 | 0 | 8 |
| cell junction | 9 | 4 | 0 | 7 |
| symplast | 9 | 4 | 0 | 7 |
| Molecular Function | ||||
| catalytic activity | 142 | 101 | 11 | 362 |
| binding | 110 | 94 | 13 | 396 |
| transporter activity | 8 | 5 | 1 | 42 |
| structural molecule activity | 0 | 2 | 0 | 18 |
| nucleic acid binding transcription factor activity | 4 | 1 | 0 | 9 |
| molecular transducer activity | 3 | 2 | 1 | 5 |
| signal transducer activity | 3 | 2 | 1 | 5 |
| antioxidant activity | 8 | 3 | 0 | 3 |
| electron carrier activity | 0 | 1 | 1 | 5 |
| transcription factor activity, protein binding | 2 | 0 | 0 | 4 |
| nutrient reservoir activity | 2 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Wang, X.; Zhang, R.; Fu, Q.; Tang, F.; Shi, F.; Temuer, B.; Zhang, Z. Comparative Transcriptome and Anatomic Characteristics of Stems in Two Alfalfa Genotypes. Plants 2022, 11, 2601. https://doi.org/10.3390/plants11192601
Wu J, Wang X, Zhang R, Fu Q, Tang F, Shi F, Temuer B, Zhang Z. Comparative Transcriptome and Anatomic Characteristics of Stems in Two Alfalfa Genotypes. Plants. 2022; 11(19):2601. https://doi.org/10.3390/plants11192601
Chicago/Turabian StyleWu, Jierui, Xiaoyu Wang, Ruxue Zhang, Qingwen Fu, Fang Tang, Fengling Shi, Buhe Temuer, and Zhiqiang Zhang. 2022. "Comparative Transcriptome and Anatomic Characteristics of Stems in Two Alfalfa Genotypes" Plants 11, no. 19: 2601. https://doi.org/10.3390/plants11192601
APA StyleWu, J., Wang, X., Zhang, R., Fu, Q., Tang, F., Shi, F., Temuer, B., & Zhang, Z. (2022). Comparative Transcriptome and Anatomic Characteristics of Stems in Two Alfalfa Genotypes. Plants, 11(19), 2601. https://doi.org/10.3390/plants11192601





