Expression Profiling of Four Mango FT/TFL1-Encoding Genes under Different Fruit Load Conditions, and Their Involvement in Flowering Regulation
Abstract
1. Introduction
2. Results
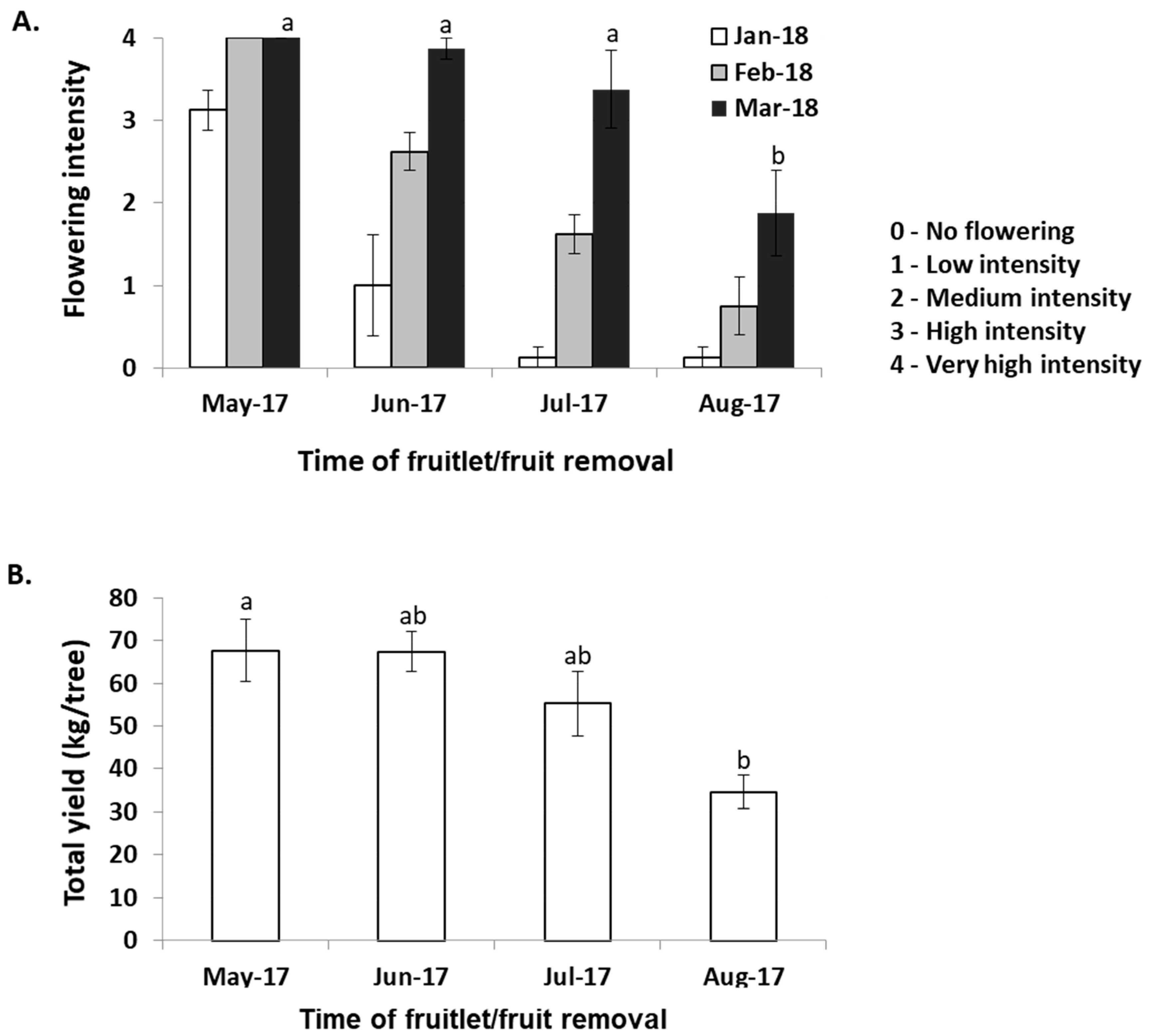
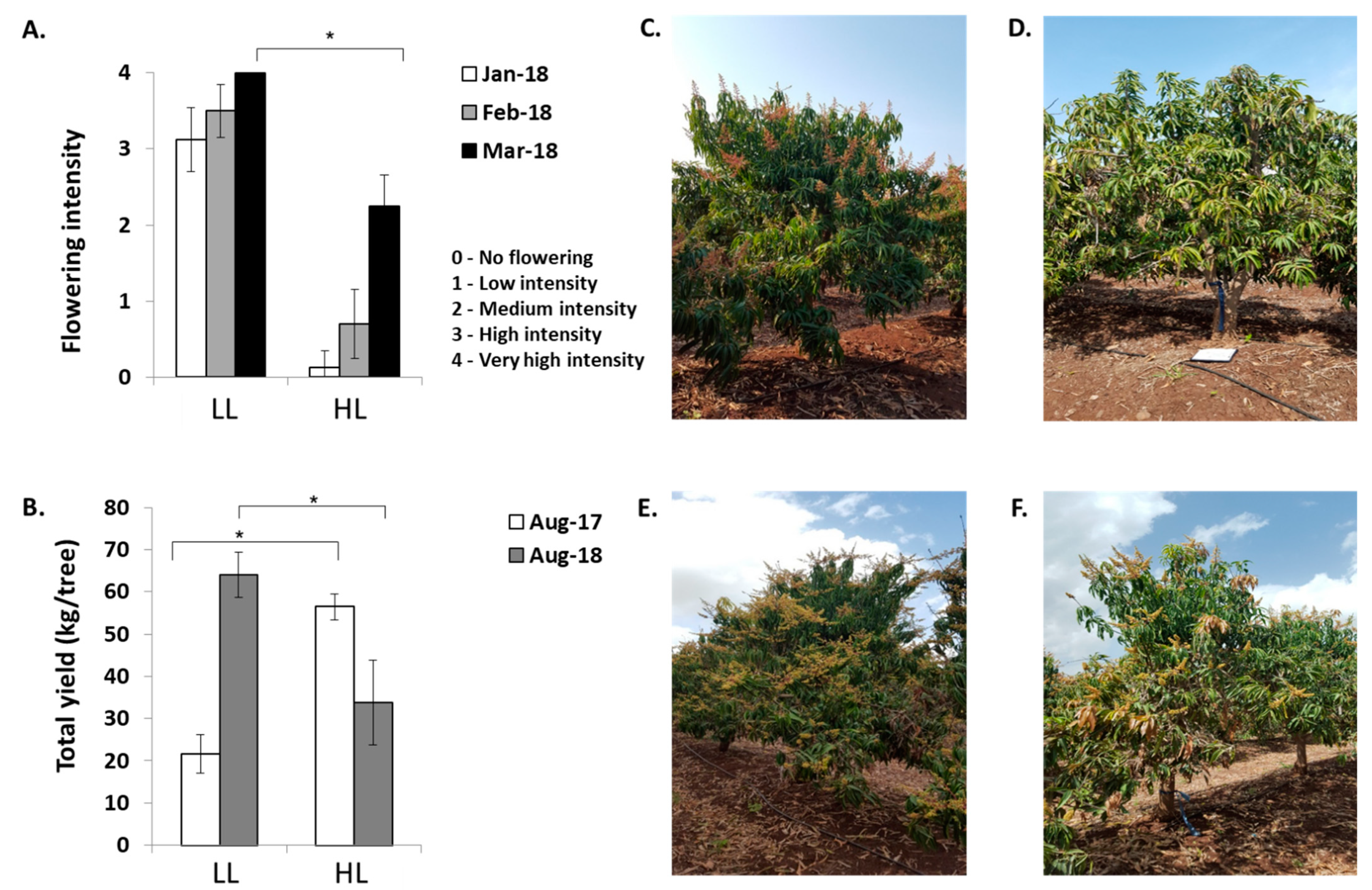
2.1. Exploring the Effects of Crop Load on Return Flowering in Mango ‘Shelly’ Cultivar
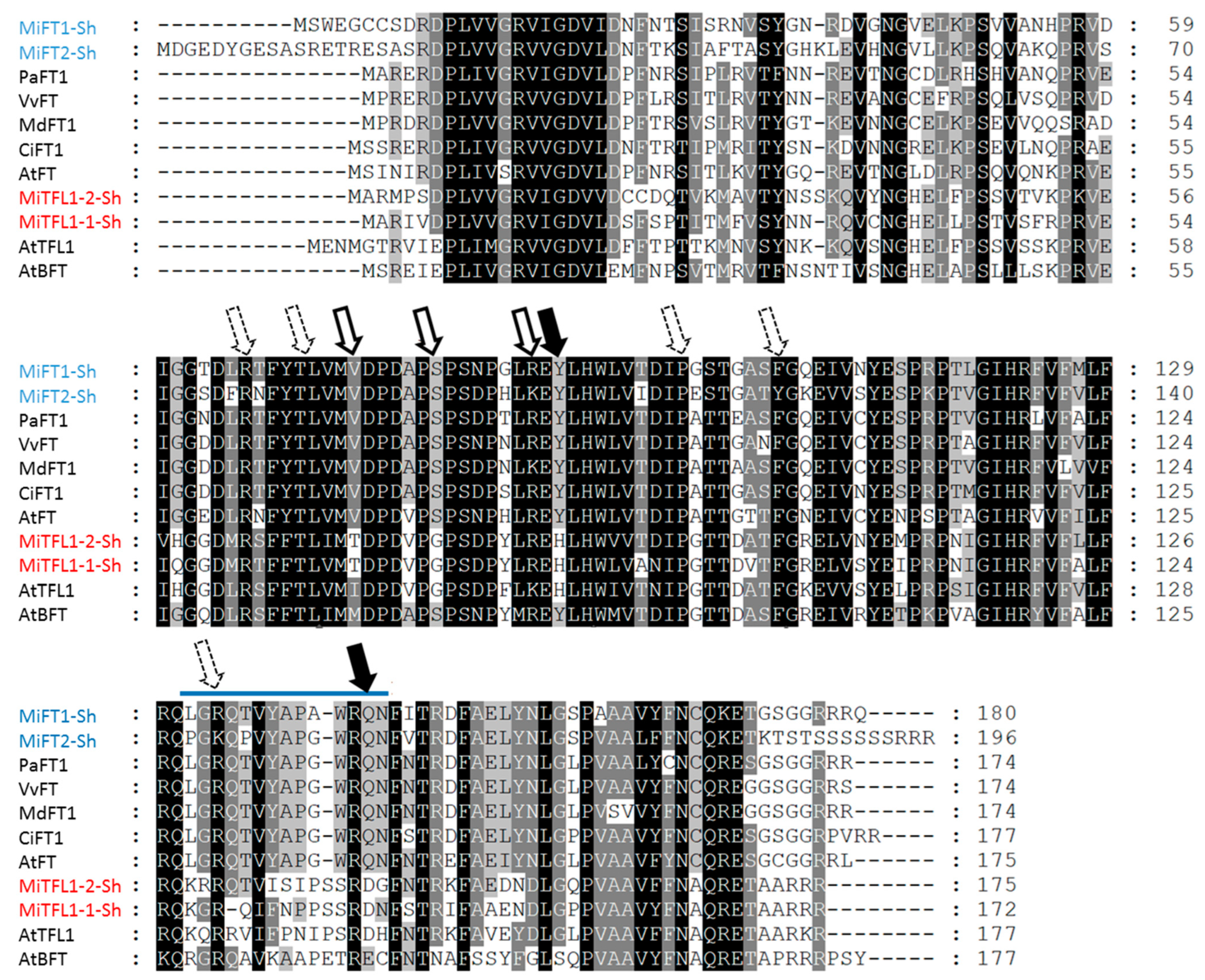
2.2. Identification and Characterization of MiFT/MiTFL1 Transcripts from Mango ‘Shelly’ Cultivar
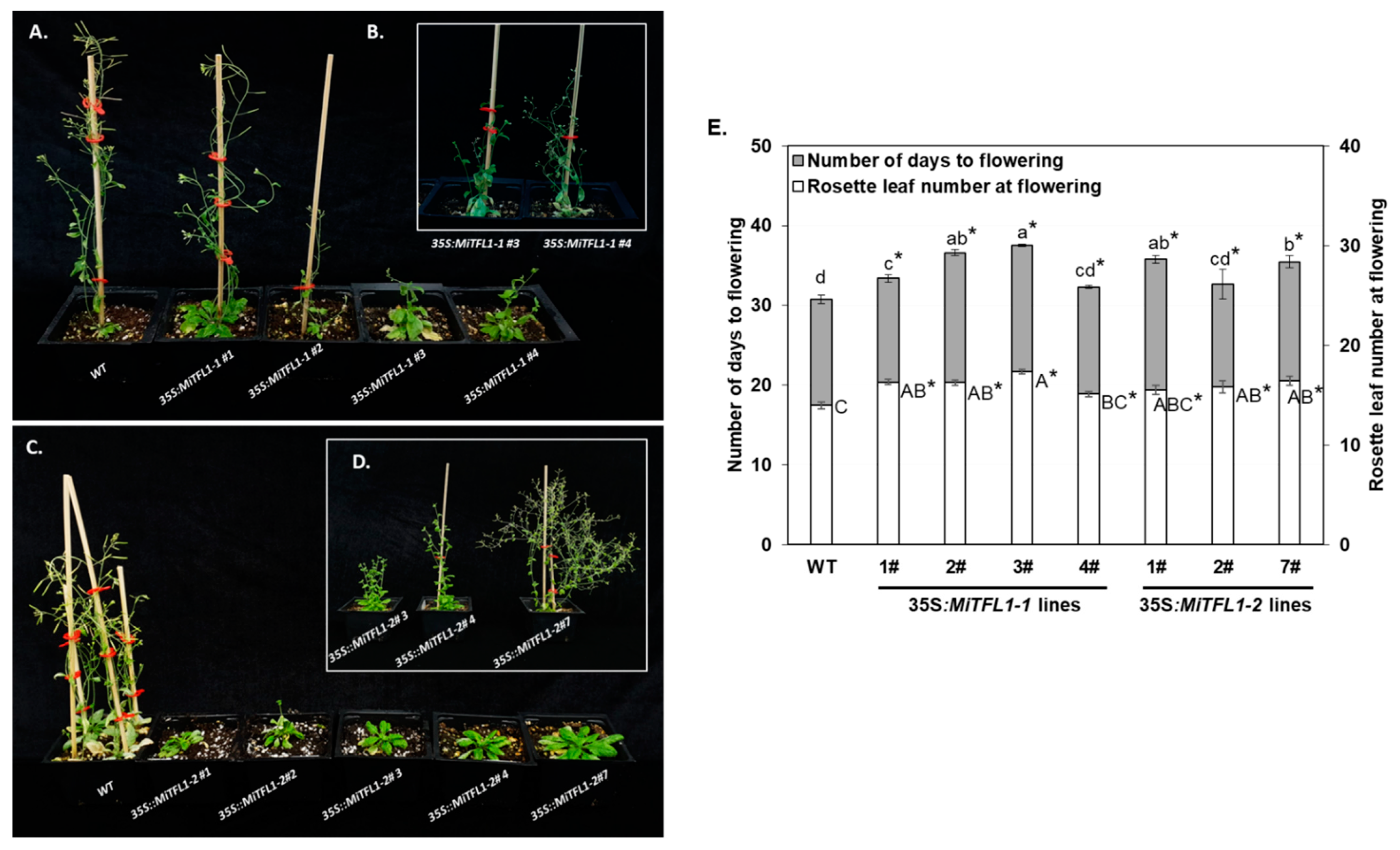
2.3. Functional Analysis of Mango cDNA Encoding FT/TFL1 Proteins in Transgenic Arabidopsis Plants
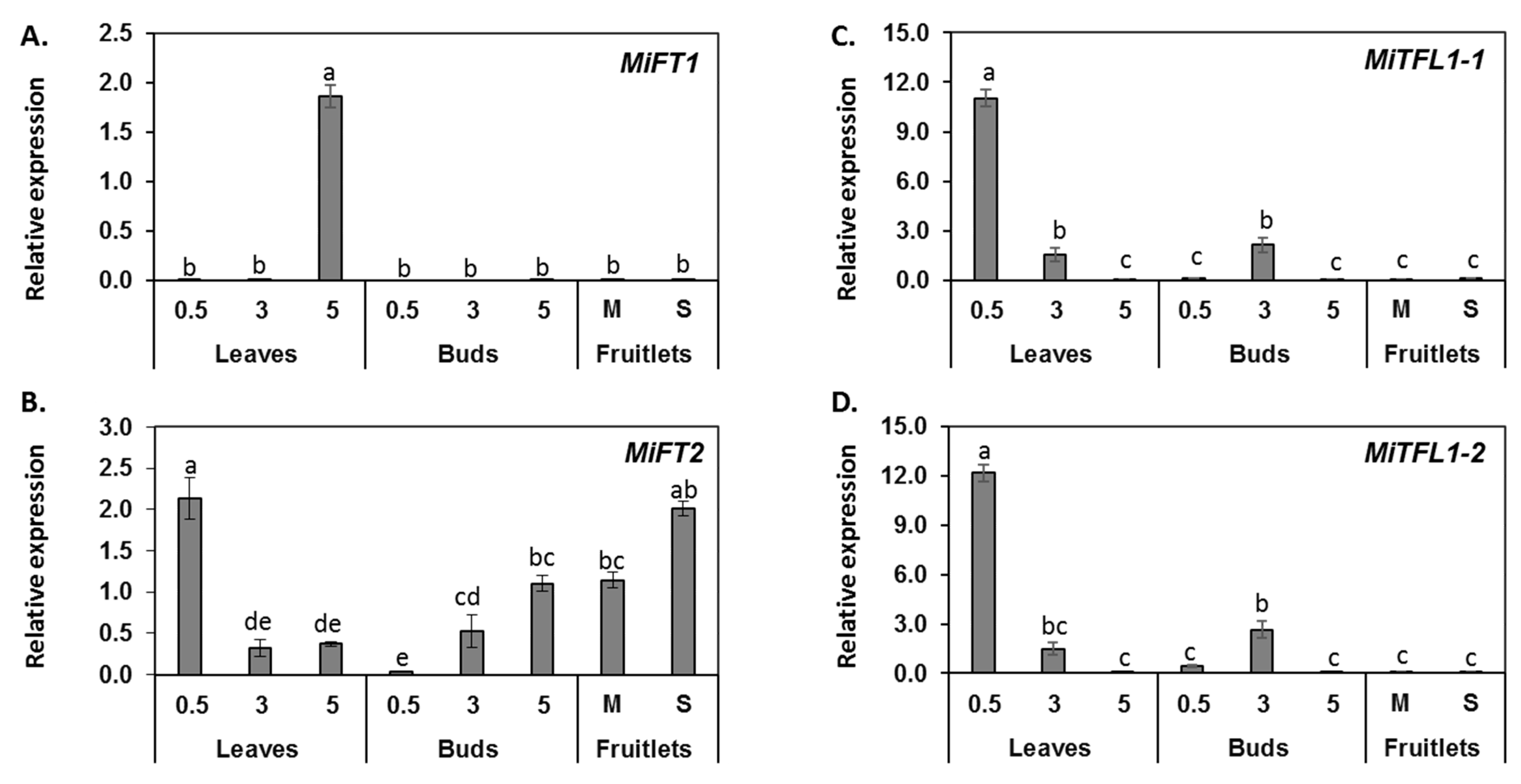
2.4. Tissue-Specific Expression Patterns of MiFT/TFL1 Genes
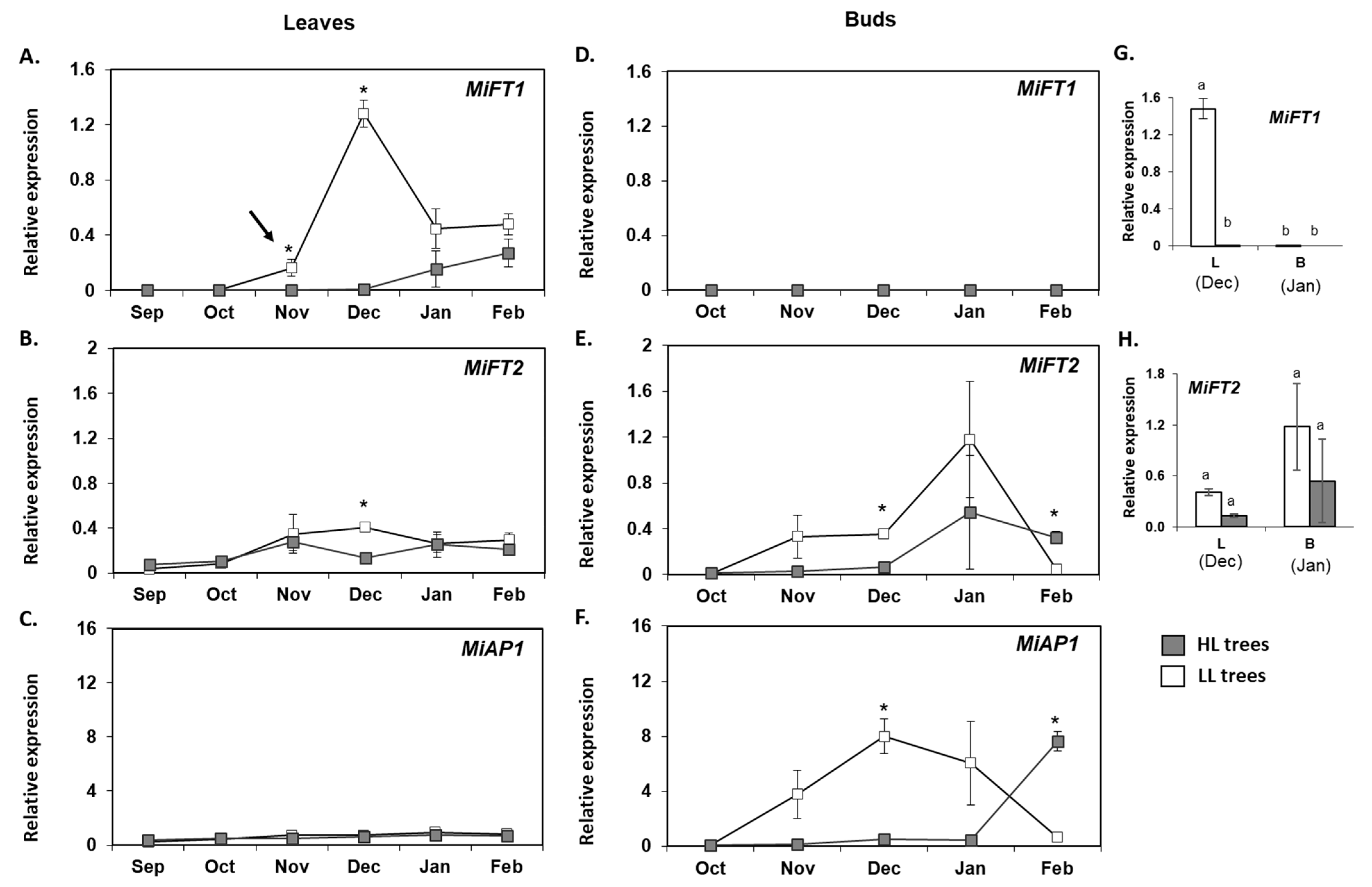
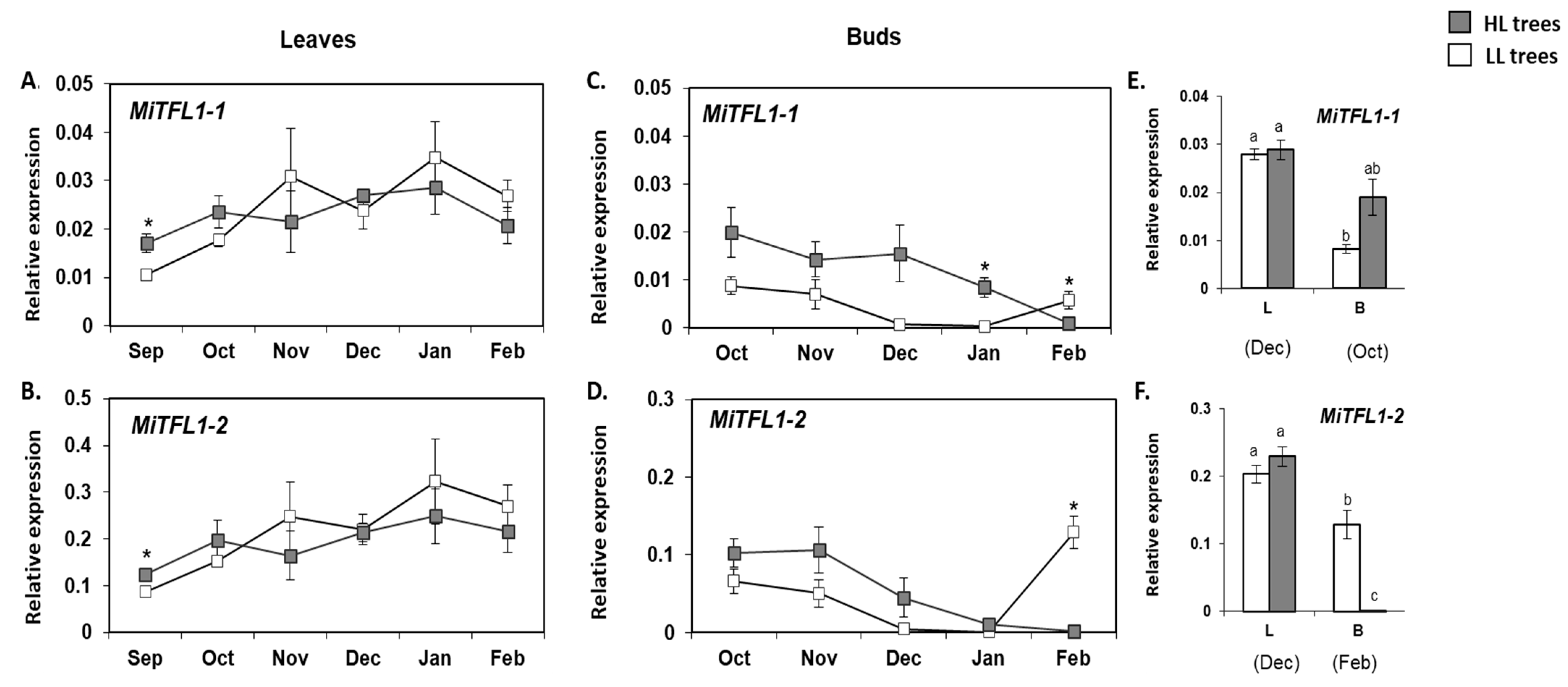
2.5. Monitoring the Expression of MiFT/TFL1 Genes under Different Fruit Load Conditions
3. Discussion
4. Materials and Methods
4.1. Plant Material and De-Fruiting Treatments
4.2. Cloning of MiFT/MiTFLs from Mango ‘Shelly’ Cultivar
4.3. Arabidopsis Transformation and Phenotypic Analysis
4.4. Plant Material and Tissue Collection from Juvenile and Adult Trees
4.5. Tissue Collection from Fruit Trees with Heavy and Low Fruit Loads
4.6. RNA Isolation and cDNA Synthesis
4.7. Real-Time qPCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mukherjee, S.K.; Litz, R.E. Introduction: Botany and importance. In The Mango: Botany, Production and Uses, 2nd ed.; Litz, R.E., Ed.; CAB International: Wallingford, UK, 2009; pp. 1–18. [Google Scholar]
- Saúco, V.G. Trends in world mango production and marketing. Acta Hortic. 2017, 1183, 351–363. [Google Scholar] [CrossRef]
- FAOSTAT Food and Agriculture Data 2016; Food and Agriculture Organization: Rome, Italy, 2016.
- Nakagawa, M.; Honsho, C.; Kanzaki, S.; Shimizu, K.; Utsunomiya, N. Isolation and Expression analysis of FLOWERING LOCUS T-like and gibberellin metabolism genes in biennial-bearing mango trees. Sci. Hortic. 2012, 139, 108–117. [Google Scholar] [CrossRef]
- Ramírez, F.; Davenport, T.L. Mango (Mangifera indica L.) flowering physiology. Sci. Hortic. 2010, 126, 65–72. [Google Scholar] [CrossRef]
- Monselise, S.P.; Goldschmidt, E.E. Alternate bearing in fruit trees. In Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 1982; Volume 4, pp. 128–173. [Google Scholar]
- Goldschmidt, E.E.; Sadka, A. Yield alternation: Horticulture, physiology, molecular biology, and evolution. In Horticultural Reviews; Warrington, I., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2005; pp. 363–418. [Google Scholar]
- Samach, A.; Smith, H.M. Plant science constraints to obtaining consistent annual yields in perennials. II: Environment and fruit load affect induction of flowering. Plant Sci. 2013, 207, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.H.; Sheen, T.F. Floral induction in axillary buds of mango (Mangifera indica L.) as affected by temperature. Sci. Hortic. 1987, 31, 81–87. [Google Scholar] [CrossRef]
- Núñez-Elisea, R.; Davenport, T.L. Flowering of mango trees in containers as influenced by seasonal temperature and water stress. Sci. Hortic. 1994, 58, 57–66. [Google Scholar] [CrossRef]
- Núñez-Elisea, R.; Davenport, T.L. Effect of leaf age, duration of cool temperature treatment, and photoperiod on bud dormancy release and floral initiation in mango. Sci. Hortic. 1995, 62, 63–73. [Google Scholar] [CrossRef]
- Tomer, E. Inhibition of flowering in mango by gibberellic acid. Sci. Hortic. 1984, 24, 299–303. [Google Scholar] [CrossRef]
- Núñez-Elisea, R.; Davenport, T.L. Gibberellin and temperature effects on dormancy release and shoot morphogenesis of mango (Mangifera indica L.). Sci. Hortic. 1998, 77, 11–21. [Google Scholar] [CrossRef]
- Protacio, C.M.; Quinto, J.E.; Serrano, E.P.; Marquez, I.P.; Rodriguez, F.M. Unravelling the mechanism of mango flowering. Acta Hortic. 2009, 820, 259–270. [Google Scholar] [CrossRef]
- Nunez-Elisea, R.; Davenport, T.L.; Caldeira, M.L. Bud initiation and morphogenesis in “Tommy Atkins” mango as affected by temperature and triazole growth retardants. Acta Hortic. 1993, 341, 192–198. [Google Scholar] [CrossRef]
- Upreti, K.K.; Reddy, Y.T.N.; Prasad, S.R.S.; Bindu, G.V.; Jayaram, H.L.; Rajan, S. Hormonal changes in response to paclobutrazol induced early flowering in mango cv. Totapuri. Sci. Hortic. 2013, 150, 414–418. [Google Scholar] [CrossRef]
- Pin, P.A.; Nilsson, O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012, 35, 1742–1755. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Varkonyi-Gasic, E. FT and florigen long-distance flowering control in plants. Curr. Opin. Plant Biol. 2016, 33, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Shannon, S.; Meeks-Wagner, D.R. Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell 1993, 5, 639–655. [Google Scholar] [CrossRef]
- Ratcliffe, O.J.; Amaya, I.; Vincent, C.A.; Rothstein, S.; Carpenter, R.; Coen, E.S.; Bradley, D.J. A common mechanism controls the life cycle and architecture of plants. Development 1998, 1615, 1609–1615. [Google Scholar] [CrossRef]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation tagging of the floral inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Kobayashi, Y.; Goto, K.; Abe, M.; Araki, T. TWIN SISTER of FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005, 46, 1175–1189. [Google Scholar] [CrossRef]
- Yoo, S.Y.; Kardailsky, I.; Lee, J.S.; Weigel, D.; Ahn, J.H. Acceleration of flowering by overexpression of MFT (MOTHER OF FT and TFL1). Mol. Cells 2004, 17, 95–101. [Google Scholar]
- Mimida, N.; Goto, K.; Kobayashi, Y.; Araki, T.; Ahn, J.H.; Weigel, D.; Murata, M.; Motoyoshi, F.; Sakamoto, W. Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 2001, 6, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.J.; Chung, K.S.; Jung, S.H.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 2010, 63, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Taoka, K.I.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S.; et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Bäurle, I.; Dean, C. The timing of developmental transitions in plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Périlleux, C.; Bouché, F.; Randoux, M.; Orman-Ligeza, B. Turning meristems into fortresses. Trends Plant Sci. 2019, 24, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Moraes, T.S.; Dornelas, M.C.; Martinelli, A.P. FT/TFL1: Calibrating plant architecture. Front. Plant Sci. 2019, 10, 97. [Google Scholar] [CrossRef]
- Zhu, Y.; Klasfeld, S.; Jeong, C.W.; Jin, R.; Goto, K.; Yamaguchi, N.; Wagner, D. TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 2020, 11, 5118. [Google Scholar] [CrossRef]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef]
- Ahn, J.H.; Miller, D.; Winter, J.; Banfield, M.J.; Hwan, J.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef]
- Endo, M.; Yoshida, M.; Sasaki, Y.; Negishi, K.; Horikawa, K.; Daimon, Y.; Kurotani, K.; Notaguchi, M.; Abe, M.; Araki, T. Re-evaluation of florigen transport kinetics with separation of functions by mutations that uncouple flowering initiation and long-distance transport. Plant J. 2018, 59, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, P.; Gao, Y.; Li, Y.; Wu, L.; Gao, J.; Zhao, M.; Xia, Q. Isolation and functional characterization of a novel FLOWERING LOCUS T homolog (NtFT5) in Nicotiana tabacum. J. Plant Physiol. 2018, 231, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Harig, L.; Beinecke, F.A.; Oltmanns, J.; Muth, J.; Müller, O.; Rüping, B.; Twyman, R.M.; Fischer, R.; Prüfer, D.; Noll, G.A. Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 2012, 72, 908–921. [Google Scholar] [CrossRef]
- Endo, T.; Shimada, T.; Fujii, H.; Kobayashi, Y.; Araki, T.; Omura, M. Ectopic expression of an FT homolog from Citrus confers an early flowering phenotype on trifoliate orange (Poncirus trifoliata L. Raf.). Transgenic Res. 2005, 1, 703–712. [Google Scholar] [CrossRef]
- Tränkner, C.; Lehmann, S.; Hoenicka, H.; Hanke, M.-V.; Fladung, M.; Lenhardt, D.; Dunemann, F.; Gau, A.; Schlangen, K.; Malnoy, M.; et al. Over-expression of an FT-homologous gene of apple induces early flowering in annual and perennial plants. Planta 2010, 232, 1309–1324. [Google Scholar] [CrossRef]
- Haberman, A.; Bakhshian, O.; Cerezo-Medina, S.; Paltiel, J.; Adler, C.; Ben-Ari, G.; Mercado, J.A.; Pliego-Alfaro, F.; Lavee, S.; Samach, A. A possible role for flowering locus T-encoding genes in interpreting environmental and internal cues affecting olive (Olea europaea L.) flower induction. Plant Cell Environ. 2017, 40, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Moss, S.M.A.; Wang, T.; Voogd, C.; Brian, L.A.; Wu, R.; Hellens, R.P.; Allan, A.C.; Putterill, J.; Varkonyi-Gasic, E. AcFT promotes kiwifruit in vitro flowering when overexpressed and Arabidopsis flowering when expressed in the vasculature under its own promoter. Plant Direct 2018, 2, e00068. [Google Scholar] [CrossRef]
- Freiman, A.; Shlizerman, L.; Golobovitch, S.; Yablovitz, Z.; Korchinsky, R.; Cohen, Y.; Samach, A.; Chevreau, E.; Le Roux, P.M.; Patocchi, A.; et al. Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2. Planta 2012, 235, 1239–1251. [Google Scholar] [CrossRef]
- Flachowsky, H.; Szankowski, I.; Waidmann, S.; Peil, A.; Tränkner, C.; Hanke, M.V. The MdTFL1 gene of apple (Malus × domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 2012, 32, 1288–1301. [Google Scholar] [CrossRef]
- Muñoz-Fambuena, N.; Mesejo, C.; Carmen González-Mas, M.; Primo-Millo, E.; Agustí, M.; Iglesias, D.J. Fruit regulates seasonal expression of flowering genes in alternate-bearing ‘Moncada’ mandarin. Ann. Bot. 2011, 108, 511–519. [Google Scholar] [CrossRef]
- Muñoz-Fambuena, N.; Mesejo, C.; González-Mas, M.C.; Primo-Millo, E.; Agustí, M.; Iglesias, D.J. Fruit load modulates flowering-related gene expression in buds of alternate-bearing ‘Moncada’ mandarin. Ann. Bot. 2012, 110, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Shalom, L.; Samuels, S.; Zur, N.; Shlizerman, L.; Zemach, H.; Weissberg, M. Alternate bearing in citrus: Changes in the expression of flowering control genes and in global gene expression in ON- versus OFF-crop trees. PLoS ONE 2012, 7, e46930. [Google Scholar] [CrossRef] [PubMed]
- Ziv, D.; Zviran, T.; Zezak, O.; Samach, A.; Irihimovitch, V. Expression profiling of FLOWERING LOCUS T-Like gene in alternate bearing ‘Hass’ avocado trees suggests a role for PaFT in avocado flower induction. PLoS ONE 2014, 9, e110613. [Google Scholar] [CrossRef] [PubMed]
- Haberman, A.; Ackerman, M.; Crane, O.; Kelner, J.; Costes, E.; Samach, A. Different flowering response to various fruit loads in apple cultivars correlates with degree of transcript reaccumulation of a TFL1-encoding gene. Plant J. 2016, 87, 161–173. [Google Scholar] [CrossRef]
- Vyavahare, S.N.; Krishna, B.; Joshi, S.S.; Chaudhari, R.S.; Subramaniam, V.R.; Sane, P.V. Characterization of mango Flowering Locus T (FT) and Terminal Flower 1 (TFL1) genes. Acta Hortic. 2017, 1, 113–124. [Google Scholar] [CrossRef]
- Fan, Z.-Y.; He, X.-H.; Fan, Y.; Yu, H.-X.; Wang, Y.-H.; Xie, X.-J.; Liu, Y.; Mo, X.; Wang, J.-Y.; Luo, C. Isolation and functional characterization of three MiFTs genes from mango. Plant Physiol. Biochem. 2020, 155, 169–176. [Google Scholar] [CrossRef]
- Wang, Y.-H.; He, X.-H.; Yu, H.-X.; Mo, X.; Fan, Y.; Fan, Z.-Y.; Xie, X.-J.; Liu, Y.; Luo, C. Overexpression of four MiTFL1 genes from mango delays the flowering time in transgenic Arabidopsis. BMC Plant Biol. 2021, 21, 407. [Google Scholar] [CrossRef]
- Mango Genome Consortium; Bally, I.S.E.; Bombarely, A.; Chambers, A.H.; Cohen, Y.; Dillon, N.L.; Innes, D.J.; Islas-Osuna, M.A.; Kuhn, D.N.; Mueller, L.A.; et al. The ‘Tommy Atkins’ mango genome reveals candidate genes for fruit quality. BMC Plant Biol. 2021, 21, 108. [Google Scholar] [CrossRef]
- Wang, P.; Luo, Y.; Huang, J.; Gao, S.; Zhu, G.; Dang, Z.; Gai, J.; Yang, M.; Zhu, M.; Zhang, H.; et al. The genome evolution and domestication of tropical fruit mango. Genome Biol. 2020, 21, 60. [Google Scholar] [CrossRef]
- Lavi, U.; Kaufman, D.; Sharon, D.; Gazit, S.; Tomer, E. ‘Shelly’: A new mango cultivar. HortScience 1997, 32, 138. [Google Scholar] [CrossRef]
- Khezri, M.; Heerema, R.; Brar, G.; Ferguson, L. Alternate bearing in pistachio (Pistacia vera L.): A review. Trees 2020, 34, 855–868. [Google Scholar] [CrossRef]
- Dag, A.; Bustan, A.; Avni, A.; Tzipori, I.; Lavee, S.; Riov, J. Timing of fruit removal affects concurrent vegetative growth and subsequent return bloom and yield in olive (Olea europaea L.). Sci. Hortic. 2010, 123, 469–472. [Google Scholar] [CrossRef]
- Kotoda, N.; Hayashi, H.; Suzuki, M.; Igarashi, M.; Hatsuyama, Y.; Kidou, S.-I.; Igasaki, T.; Nishiguchi, M.; Yano, K.; Shimizu, T.; et al. Molecular characterization of Flowering LOCUS T-like genes of apple (Malus × domestica Borkh.). Plant Cell Physiol. 2010, 51, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, F.; Endo, T.; Shimada, T.; Fujii, H.; Shimizu, T.; Omura, M.; Ikoma, Y. Increased CiFT abundance in the stem correlates with floral induction by low temperature in Satsuma mandarin (Citrus unshiu Marc.). J. Exp. Bot. 2007, 58, 3915–3927. [Google Scholar] [CrossRef] [PubMed]
- Pillitteri, L.J.; Lovatt, C.J.; Walling, L.L. Isolation and characterization of a TERMINAL FLOWER homolog and its correlation with juvenility in Citrus. Plant Physiol. 2004, 135, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Moraes, T.S.; Immink, R.G.H.; Martinelli, A.P.; Angenent, G.C.; van Esse, W.; Dornelas, M.C. Passiflora organensis FT/TFL1 gene family and their putative roles in phase transition and floral initiation. Plant Reprod. 2021, 35, 105–126. [Google Scholar] [CrossRef]
- Esumi, T.; Kitamura, Y.; Hagihara, C.; Yamane, H.; Tao, R. Identification of a TFL1 ortholog in Japanese apricot (Prunus mume Sieb. et Zucc.). Sci. Hortic. 2010, 125, 608–616. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, Y.; Zhang, L.; Su, W.; Peng, J.; Yang, X.; Song, H.; Gao, Y.; Lin, S. EjTFL1 genes promote growth but inhibit flower bud differentiation in loquat. Front. Plant Sci. 2020, 11, 576. [Google Scholar] [CrossRef]
- Jean Finnegan, E.; Kovac, K.A.; Jaligot, E.; Sheldon, C.C.; James Peacock, W.; Dennis, E.S. The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J. 2005, 44, 420–432. [Google Scholar] [CrossRef]
- Agustí, M.; Mesejo, C.; Muñoz-Fambuena, N.; Vera-Sirera, F.; de Lucas, M.; Martínez-Fuentes, A.; Reig, C.; Iglesias, D.J.; Primo-Millo, E.; Blázquez, M.A. Fruit-dependent epigenetic regulation of flowering in Citrus. New Phytol. 2020, 225, 376–384. [Google Scholar] [CrossRef]
- Rantanen, M.; Kurokura, T.; Jiang, P.; Mouhu, K.; Hytönen, T. Strawberry Homologue of TERMINAL FLOWER1 Integrates Photoperiod and Temperature Signals to Inhibit Flowering. Plant J. 2015, 82, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Koskela, E.A.; Kurokura, T.; Toivainen, T.; Sønsteby, A.; Heide, O.M.; Sargent, D.J.; Isobe, S.; Jaakola, L.; Hilmarsson, H.; Elomaa, P.; et al. Altered Regulation of TERMINAL FLOWER 1 Causes the Unique Vernalisation Response in an Arctic Woodland Strawberry Accession. New Phytol. 2017, 216, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J.; Gustafson-Brown, C.; Pinyopich, A.; Ditta, G.S.; Yanofsky, M.F. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 Specify Meristem Fate. Plant Cell 1999, 11, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Burondkar, M.M.; Upreti, K.K.; Ambavane, A.R. Hormonal Changes during Flowering in Response to Paclobutrazol Application in Mango Cv. Alphonso under Konkan Conditions. Indian J. Plant Physiol. 2016, 21, 306–311. [Google Scholar] [CrossRef]
- Abdel Rahim, A.O.S.; Elamin, O.M.; Bangerth, F.K. Effects of Paclobutrazol (PBZ) on Floral Induction and Associated Hormonal and Metabolic Changes of Biennially Bearing Mango (Mangifera Indica L.) Cultivars during off Year. ARPN J. Agric. Biol. Sci. 2011, 6, 55–67. [Google Scholar]
- Endo, T.; Shimada, T.; Nakata, Y.; Fujii, H.; Matsumoto, H.; Nakajima, N.; Ikoma, Y.; Omura, M. Abscisic Acid Affects Expression of Citrus FT Homologs upon Floral Induction by Low Temperature in Satsuma mandarin (Citrus unshiu Marc.). Tree Physiol. 2018, 38, 755–771. [Google Scholar] [CrossRef]
- Sherman, A.; Rubinstein, M.; Eshed, R.; Benita, M.; Ish-Shalom, M.; Sharabi-Schwager, M.; Rozen, A.; Saada, D.; Cohen, Y.; Ophir, R. Mango (Mangifera indica L.) Germplasm Diversity Based on Single Nucleotide Polymorphisms Derived from the Transcriptome. BMC Plant Biol. 2015, 15, 277. [Google Scholar] [CrossRef]
- Harrison, S.J.; Mott, E.K.; Parsley, K.; Aspinall, S.; Gray, J.C.; Cottage, A. A Rapid and Robust Method of Identifying Transformed Arabidopsis thaliana Seedlings Following Floral Dip Transformation. Plant Methods 2006, 2, 19. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gafni, I.; Rai, A.C.; Halon, E.; Zviran, T.; Sisai, I.; Samach, A.; Irihimovitch, V. Expression Profiling of Four Mango FT/TFL1-Encoding Genes under Different Fruit Load Conditions, and Their Involvement in Flowering Regulation. Plants 2022, 11, 2409. https://doi.org/10.3390/plants11182409
Gafni I, Rai AC, Halon E, Zviran T, Sisai I, Samach A, Irihimovitch V. Expression Profiling of Four Mango FT/TFL1-Encoding Genes under Different Fruit Load Conditions, and Their Involvement in Flowering Regulation. Plants. 2022; 11(18):2409. https://doi.org/10.3390/plants11182409
Chicago/Turabian StyleGafni, Itamar, Avinash Chandra Rai, Eyal Halon, Tali Zviran, Isaac Sisai, Alon Samach, and Vered Irihimovitch. 2022. "Expression Profiling of Four Mango FT/TFL1-Encoding Genes under Different Fruit Load Conditions, and Their Involvement in Flowering Regulation" Plants 11, no. 18: 2409. https://doi.org/10.3390/plants11182409
APA StyleGafni, I., Rai, A. C., Halon, E., Zviran, T., Sisai, I., Samach, A., & Irihimovitch, V. (2022). Expression Profiling of Four Mango FT/TFL1-Encoding Genes under Different Fruit Load Conditions, and Their Involvement in Flowering Regulation. Plants, 11(18), 2409. https://doi.org/10.3390/plants11182409








