Assessment of GGE, AMMI, Regression, and Its Deviation Model to Identify Stable Rice Hybrids in Bangladesh
Abstract
1. Introduction
2. Results and Discussion
2.1. Box Plot Sketch Depends on Descriptive Statistics
2.2. Frequency Distribution-Based Histogram of Some Hybrid Combinations
2.3. Analysis of Variance
2.4. General Genotypic Adaptation
2.5. Evaluation of Environment through the Which-Won-Where Pattern
2.6. Genotype Discrimination by GGE Biplot in Mean versus Stability Views
2.7. Discriminative vs. Representative
2.8. Ranking Genotypes
2.9. Grain Type
2.10. Stability Analysis for Different Characters of 26 Promising Hybrid Rice Grains
3. Materials and Methods
3.1. Materials of the Study
3.2. Layout and Design
3.3. Intercultural Practices
3.4. Data Collection
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yuan, L.P.; Virmani, S.S. Status of Hybrid Rice Research and Development. In Proceedings of the International Symposium on Hybrid Rice, Changsha, China, 6–10 October 1986; Hybrid Rice. International Rice Research Institute: Manila, Philippines, 1988. [Google Scholar]
- Shrestha, S.; Asch, F.; Dusserre, J.; Amanantsoanirina, A.; Brueck, H. Climate Effects on Yield Components as Affected by Genotypic Responses to Variable Environmental Conditions in Upland Rice Systems a Different Altitude. Field Crop. Res. 2012, 134, 216–228. [Google Scholar] [CrossRef]
- Islam, M.R.; Sarker, M.R.A.; Sharma, N.; Rahman, M.A.; Collard, B.C.Y.; Gregorio, G.B.; Ismail, A.M. Assessment of Adaptability of Recently Released Salt Tolerant Rice Varieties in Coastal Regions of South Bangladesh. Field Crop. Res. 2015, 190, 34–43. [Google Scholar] [CrossRef]
- Sarker, U.; Ferdous, N. Genotype × Seedling Age Interaction in Rice (Oryza sativa L.). Pak. J. Biol. Sci. 2002, 5, 275–277. [Google Scholar] [CrossRef][Green Version]
- Islam, M.A.; Sarker, U.; Hasan, M.R.; Islam, M.R.; Mahmud, M.N.H. Genotype x Environment (Fertilizer Dose) Interaction and Stability Analysis of Hybrid Seed Production of Rice (Oryza sativa L.). Eco-Friendly Agril. J. 2012, 5, 183–185. [Google Scholar]
- Islam, M.A.; Johora, F.T.; Sarker, U.; Mian, M.A.K. Adaptation of Chinese CMS Lines Interaction with Seedling Age and Row Ratio on Hybrid Seed Production of Rice (Oryza sativa L.). Bangladesh J. Agron. 2012, 25, 178–183. [Google Scholar]
- Islam, M.A.; Sarker, U.; Mian, M.A.K.; Ahmed, J.U. Genotype Seedling Age Interaction for Hybrid Seed Yield of Rice (Oryza sativa L.). Bangladesh J. Plant Breed. Genet. 2011, 24, 23–26. [Google Scholar] [CrossRef]
- Islam, M.A.; Sarker, U.; Mian, M.A.K.; Ahmed, J.U. Genotype Fertilizer Doses Interaction for Hybrid Seed Yield of Rice (Oryza sativa L.). Bangladesh J. Plant Breed. Genet. 2011, 24, 41–44. [Google Scholar] [CrossRef]
- Islam, M.A.; Mian, M.A.K.; Rasul, G.; Johora, F.T.; Sarker, U. Interaction Effect Between Genotypes, Row Ratio and Fertilizer Dose on Hybrid Seed Production of Rice (Oryza sativa. L.). In Proceedings of the 10th Conference proceeding of Bangladesh Society of Agronomy 4, Joydebpur, Gazipur, Bangladesh, 8 October 2011; pp. 134–141. [Google Scholar]
- Biswas, A.; Sarker, U.; Banik, B.R.; Rohman, M.M.; Talukder, M.Z.A. Genotype x Environment Interaction for Grain Yield of Maize (Zea mays L.) Inbreds under Salinity Stress. Bangladesh J. Agril. Res. 2014, 39, 293–301. [Google Scholar] [CrossRef]
- Sarker, U.; Biswas, P.S.; Prasad, B. Stability for Grain Yield and Yield Components in Rice (Oryza sativa L.). Bangladesh J. Agril. Res. 2007, 32, 559–564. [Google Scholar]
- Biswas, J.C.; Sarker, U.; Biswas, S.R. Influence of Seeding Time and Nitrogen Rates on Yield of Direct-Seeded Rice (Oryza sativa L.) in Wet Season. Agriculturists 2004, 2, 45–50. [Google Scholar]
- Sarker, U.; Rasul, M.G.; Mian, M.A.K. Effect of Sowing Time and Seedling Age on Performance of Two Boro Rice Varieties. J. Asiat. Soc. Bangladesh Sci. 2002, 28, 91–100. [Google Scholar]
- Sarker, U. Stability for Grain Yield under Different Planting Times in Rice. Bangladesh J. Agril. Res. 2002, 27, 425–430. [Google Scholar]
- Yan, W.; Hunt, L.A.; Sheng, Q.; Szlavnics, Z. Cultivar Evaluation and Mega-Environment Investigation Based on the GGE biplot. Crop. Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Magaji, U.; Abdullah, N.; Ramli, A.; Hussin, G. Assessing the Representative and Discriminative Ability of Test Environments for Rice Breeding in Malaysia Using GGE Biplot. Int. J. Sci. Tech. Res. 2017, 6, 8–16. [Google Scholar]
- Hasan-Ud-Daula, M.; Sarker, U. Variability, Heritability, Character Association, and Path Coefficient Analysis in Advanced Breeding Lines of Rice (Oryza sativa L.). Genetika 2020, 52, 711–726. [Google Scholar] [CrossRef]
- Hasan, M.J.; Kulsum, M.U.; Majumder, R.R.; Sarker, U. Genotypic Variability for Grain Quality Attributes in Restorer Lines of Hybrid Rice. Genetika 2020, 52, 973–989. [Google Scholar] [CrossRef]
- Azad, A.K.; Sarker, U.; Ercisli, S.; Assouguem, A.; Ullah, R.; Almeer, R.; Sayed, A.A.; Peluso, I. Evaluation of Combining Ability and Heterosis of Popular Restorer and Male Sterile Lines for the Development of Superior Rice Hybrids. Agronomy 2022, 12, 965. [Google Scholar] [CrossRef]
- Ganapati, R.K.; MG, R.; MAK, M.; Sarker, U. Genetic Variability and Character Association of T-Aman Rice (Oryza sativa L.). Intl. J. Plant Biol. Res. 2014, 2, 1–4. [Google Scholar]
- Sarker, U.; Mian, M.A.K. Genetic Variations and Correlations between Floral Traits in Rice. Bangladesh J. Agril. Res. 2004, 29, 553–558. [Google Scholar]
- Biswas, P.S.; Sarker, U.; Bhuiyan, M.A.R.; Khatun, S. Genetic Divergence in Cold Tolerant Irrigated Rice (Oryza sativa L.). Agriculturists 2006, 4, 15–20. [Google Scholar]
- Sarker, U.; Biswas, P.S.; Prasad, B.; Mian, M.A.K. Correlated Response, Relative Selection Efficiency and Path Analysis in Cold Tolerant Rice. Bangladesh J. Plant Breed. Genet. 2001, 14, 33–36. [Google Scholar]
- Sarker, U.; Mian, M.A.K. Genetic Variability, Character Association and Path Analysis for Yield and Its Components in Rice. J. Asiat. Soc. Bangladesh Sci. 2003, 29, 47–54. [Google Scholar]
- Ali, M.A.; Sarker, U.; MAK, M.; Islam, M.A.; Johora, F.T. Estimation of Genetic Divergence in Boro Rice (Oryza sativa L.). Intl. J. BioRes 2014, 16, 28–36. [Google Scholar]
- Karim, D.; Sarker, U.; Siddique, M.N.A.; Miah, M.A.K.; Hasnat, M.Z. Variability and Genetic Parameter Analysis in Aromatic Rice. Int. J. Sustain. Crop Prod. 2007, 2, 15–18. [Google Scholar]
- Karim, D.; Siddique, M.N.A.; Sarker, U.; Hasnat, Z.; Sultana, J. Phenotypic and Genotypic Correlation Co-Efficient of Quantitative Characters and Character Association of Aromatic Rice. J. Biosci. Agric. Res. 2014, 1, 34–46. [Google Scholar]
- Rai, P.K.; Sarker, U.; Roy, P.C.; Islam, A. Character Association in F4 Generation of Rice (Oryza sativa L.). Bangladesh J. Plant Breed. Genet. 2013, 26, 39–44. [Google Scholar]
- Hasan, M.R.; Sarker, U.; Hossain, M.A.; Huda, K.M.K.; Mian, M.A.K.; Hossain, T.; Zahan, M.S.; Mahmud, M.N.H. Genetic Diversity in Micronutrient Dense Rice and Its Implication in Breeding Program. Ecofriendly Agril. J. 2012, 5, 168–174. [Google Scholar]
- Hasan, M.R.; Sarker, U.; Mian, M.A.K.; Hossain, T.; Mahmud, M.N.H. Genetic Variation in Micronutrient Dense Rice and Its Implication in Breeding for Higher Yield. Eco-Friendly Agril. J. 2012, 5, 175–182. [Google Scholar]
- Siddique, M.N.A.; Sarker, U.; Mian, M.A.K. Genetic diversity in restorer line of rice. In Proceedings of the International Conference on Plant Breeding and Seed for Food Security, Dhaka, Bangladesh, 12 March 2009; Bhuiyan, M.S.R., Rahman, L., Eds.; Plant Breeding and Genetics Society of Bangladesh: Kampala, Uganda, 2009; pp. 137–142. [Google Scholar]
- Nath, J.K.; Sarker, U.; Mian, M.A.K.; Hossain, T. Genetic Divergence in T. Aman Rice. Ann. Bangladesh Agric. 2008, 12, 51–60. [Google Scholar]
- Rahman, M.H.; Sarker, U.; Main, M.A.K. Assessment of Variability of Floral and Yield Traits; I Restorer Lines of Rice. Ann. Bangladesh Agric. 2007, 11, 87–94. [Google Scholar]
- Rahman, M.H.; Sarker, U.; Main, M.A.K. Assessment of Variability of Floral and Yield Traits; II Maintainer Lines of Rice. Ann. Bangladesh Agric. 2007, 11, 95–102. [Google Scholar]
- Biswas, A.; Sarker, U.; Banik, B.R.; Rohman, M.M.; Mian, M.A.K. Genetic Divergence Study in Salinity Stress Tolerant Maize (Zea mays L.). Bangladesh J. Agric. Res. 2014, 39, 621–630. [Google Scholar]
- Azam, M.G.; Sarker, U.; Banik, B.R. Genetic Variability of Yield and Its Contributing Characters on CIMMYT Maize Inbreds under Drought Stress. Bangladesh J. Agric. Res. 2014, 39, 419–426. [Google Scholar]
- Azam, M.G.A.; Sarker, U.; Mian, M.A.K.; Banik, B.R.; Talukder, M.Z.A. Genetic Divergence on Quantitative Characters of Exotic Maize Inbreds (Zea mays L.). Bangladesh J. Plant Breed. Genet. 2013, 26, 9–14. [Google Scholar]
- Ashraf, A.T.M.; Rahman, M.M.; Hossain, M.M.; Sarker, U. Study of Correlation and Path Analysis in the Selected Okra Accessions. Asian Res. J. Agric. 2020, 12, 1–11. [Google Scholar] [CrossRef]
- Ashraf, A.T.M.; Rahman, M.M.; Hossain, M.M.; Sarker, U. Study of the Genetic Analysis of Some Selected Okra Accessions. Intl. J. Adv. Res. 2020, 8, 549–556. [Google Scholar] [CrossRef]
- Ashraf, A.T.M.; Rahman, M.M.; Hossain, M.M.; Sarker, U. Performance Evaluation of Some Selected Okra Accessions. Intl. J. Plant Soil Sci. 2020, 32, 13–20. [Google Scholar] [CrossRef]
- Talukder, M.Z.A.; Sarker, U.; Harun-Or-Rashid, M.; Zakaria, M. Genetic Diversity of Coconut (Cocos nucifera L.) in Barisal Region. Ann. Bangladesh Agric. 2015, 19, 13–21. [Google Scholar]
- Talukder, M.Z.A.; Sarker, U.; Khan ABM, M.M.; Moniruzzaman, M.; Zaman, M.M. Genetic Variability and Correlation Coefficient of Coconut (Cocos nucifera L.) in Barisal Region. Intl. J. BioRes 2011, 11, 15–21. [Google Scholar]
- Kayesh, E.; Sharker, M.S.; Roni, M.S.; Sarker, U. Integrated Nutrient Management for Growth, Yield and Profitability of Broccoli. Bangladesh J. Agric. Res. 2019, 44, 13–26. [Google Scholar] [CrossRef]
- Rashad, M.M.I.; Sarker, U. Genetic Variations in Yield and Yield Contributing Traits of Green Amaranth. Genetika 2020, 52, 393–407. [Google Scholar]
- Sarker, U.; Rabbani, M.G.; Oba, S.; Eldehna, W.M.; Al-Rashood, S.T.; Mostafa, N.M.; Eldahshan, O.A. Phytonutrients, Colorant Pigments, Phytochemicals, and Antioxidant Potential of Orphan Leafy Amaranthus Species. Molecules 2022, 27, 2899. [Google Scholar] [CrossRef] [PubMed]
- Sarker, U.; Lin, Y.P.; Oba, S.; Yoshioka, Y.; Ken, H. Prospects and potentials of underutilized leafy Amaranths as vegetable use for health promotion. Plant Physiol. Biochem. 2022, 182, 104–123. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S.; Ercisli, S.; Assouguem, A.; Alotaibi, A.; Ullah, R. Bioactive Phytochemicals and Quenching Activity of Radicals in Selected Drought-Resistant Amaranthus tricolor Vegetable Amaranth. Antioxidants 2022, 11, 578. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.N.; Sarker, U.; Raihan, M.S.; Al-Huqail, A.A.; Siddiqui, M.H.; Oba, S. Influence of Salinity Stress on Color Parameters, Leaf Pigmentation, Polyphenol and Flavonoid Contents, and Antioxidant Activity of Amaranthus lividus Leafy Vegetables. Molecules 2022, 27, 1821. [Google Scholar] [CrossRef] [PubMed]
- Sarker, U.; Oba, S.; Alsanie, W.F.; Gaber, A. Characterization of Phytochemicals, Nutrients, and Antiradical Potential in Slim Amaranth. Antioxidants 2022, 11, 1089. [Google Scholar] [CrossRef]
- Sarker, U.; Iqbal, M.A.; Hossain, M.N.; Oba, S.; Ercisli, S.; Muresan, C.C.; Marc, R.A. Colorant Pigments, Nutrients, Bioactive Components, and Antiradical Potential of Danta Leaves (Amaranthus lividus). Antioxidants 2022, 11, 1206. [Google Scholar] [CrossRef]
- Sarker, U.; Azam, M.G.; Talukder, M.Z.A. Genetic Variation in Mineral Profiles, Yield Contributing Agronomic Traits, and Foliage Yield of Stem Amaranth. Genetika 2022, 54, 91–108. [Google Scholar]
- Rahman, M.M.; Sarker, U.; Swapan, M.A.H.; Raihan, M.S.; Oba, S.; Alamri, S.; Siddiqui, M.H. Combining Ability Analysis and Marker-Based Prediction of Heterosis in Yield Reveal Prominent Heterotic Combinations from Diallel Population of Rice. Agronomy 2022, 12, 1797. [Google Scholar] [CrossRef]
- Azam, M.G.; Sarker, U.; Hossain, M.A.; Iqbal, M.S.; Islam, M.R.; Hossain, M.F.; Ercisli, S.; Kul, R.; Assouguem, A.; AL-Huqail, A.A.; et al. Genetic Analysis in Grain Legumes [Vigna radiata (L.) Wilczek] for Yield Improvement and Identifying Heterotic Hybrids. Plants 2022, 11, 1774. [Google Scholar] [CrossRef]
- Prodhan, M.M.; Sarker, U.; Hoque, M.A.; Biswas, M.S.; Ercisli, S.; Assouguem, A.; Ullah, R.; Almutairi, M.H.; Mohamed, H.R.H.; Najda, A. Foliar Application of GA3 Stimulates Seed Production in Cauliflower. Agronomy 2022, 12, 1394. [Google Scholar] [CrossRef]
- Das, K.R.; Imon, A.H.M.R. A Brief Review of Tests for Normality. Am. J. Theor. Appl. Stat. 2016, 5, 5–12. [Google Scholar] [CrossRef]
- Sahu, P.K.; Sharma, D.; Mondal, S.; Kumar, V.; Singh, S.; Baghel, S.; Tiwari, A.; Vishwakarma, G.; Das, B.K. Genetic Variability for Grain Quality Traits in Indigenous Rice Landraces of Chhattisgarh. Indian. J. Exp. Biol. Agric. Sci. 2017, 5, 439–455. [Google Scholar]
- Imon, A.H.M.R.; Das, K. Analyzing Length or Size Based Data: A Study on the Lengths of Peas Plants. Malays. J. Math. Sci. 2015, 9, 1–20. Available online: http://einspem.upm.edu.my/journal/fullpaper/vol9/1.%20imon%20&%20keya.pdf (accessed on 1 April 2022).
- Samak, A.N.R.; Hittalamani, S.; Shashidhar, N.; Hanumareddy, B. Exploratory Studies on Genetic Variability and Genetic Control for Protein and Micronutrient Content in F4 and F5 Generations of Rice (Oryza sativa L.). Asian J. Plant Sci. 2011, 10, 376–379. [Google Scholar] [CrossRef]
- Kamdar, J.H.; Jasani, M.D.; Ajay, B.C.; Bera, S.K.; George, J.J. Effect of Selection Response for Yield Related Traits in Early and Later Generations of Groundnut (Arachis hypogaea L.). Crop Breed. Appl. Biotechnol. 2020, 20, e317320215. [Google Scholar] [CrossRef]
- Chandrasekharan, N.; Chandran, S.; Sudha, M.; Ganesan, K.N.; Ravikesavan, R.; Senthil, N. Estimation of Variability, Heritability, Genetic Advance and Assessment of Frequency Distribution for Morphological Traits in Intercross Population of Maize. Electron. J. Plant Breed. 2020, 11, 574–580. [Google Scholar] [CrossRef]
- Raghavendra, P.; Hittalmani, S. Genetic Parameters of Two BC2F1 Populations for Development of Superior Male Sterile Lines Pertaining to Morpho-floral Traits for Aerobic Rice (Oryza sativa L.). SAARC J. Agric. 2015, 13, 198–213. [Google Scholar]
- Dinesh, H.B.; Viswanatha, K.P.; Lohithaswa, H.C.; Pavan, R.; Poonam, S. Genetic Association Estimates Using Third- and Fourth-Degree Statistics in Early Segregating Generations of Cowpea. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 867–873. [Google Scholar] [CrossRef]
- Nandini, C.; Sujata, B.; Krishnappa, M.; Chandrashekhar, A.; Prabhakar. Gene Action, Genetic Parameters and Association in Segregating Populations of Two Little Millet (Panicum miliare L.) Crosses. Green Farming 2017, 8, 523–528. [Google Scholar]
- Hasan, M.J.; Kulsum, M.U.; Hossain, M.S.; Billah, M.M.; Ansari, A. Genotype-Location Interaction of Indica Rice Using AMMI model. Bangladesh J. Plant Breed. Genet. 2011, 24, 9–18. [Google Scholar] [CrossRef]
- Kulsum, M.U.; Sarker, U.; Karim, M.A.; Mian, M.A.K. Additive Main Effects and Multiplicative Interaction (AMMI) Analysis for Yield of Hybrid Rice in Bangladesh. Trop. Agric. Develop. 2012, 56, 53–61. [Google Scholar]
- Matin, M.Q.I.; Rasul, M.G.; Islam, A.K.M.A.; Mian, M.A.K.; Ahmed, J.U.; Amiruzzaman, M. Stability Analysis for Yield and Yield Contributing Characters in Hybrid Maize (Zea mays L.). Afr. J. Agric. Res. 2017, 12, 2795–2806. [Google Scholar]
- Akter, A.; Hasan, M.J.; Kulsum, U.; Rahman, M.H.; Khatun, M.; Islam, M.R. GGE Biplot Analysis for Yield Stability in Multi-environment Trials of Promising Hybrid Rice (Oryza sativa L.). Bangladesh Rice J. 2015, 19, 1–8. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Magaji, U.; Miah, G.; Hussin, G.; Ramli, A. Genotype × Environment Interaction and Stability Analyses of Yield and Yield Components of Established and Mutant Rice Genotypes Tested in Multiple Locations in Malaysia. Acta Agric. Scand B Soil Plant Sci. 2017, 67, 590–606. [Google Scholar]
- Dia, M.; Wehner, T.C.; Hassell, R.; Price, D.S.; Boyhan, G.E.; Olson, S.; Juarez, B. Value of locations for Representing Mega- environments and for Discriminating Yield of Watermelon in the US. Crop Sci. 2016, 56, 1726–1735. [Google Scholar] [CrossRef]
- Rahman, N.M.F.; Mamun, M.A.A.; Ahmed, R.; Hossain, M.I.; Qayum, M.A.; Aziz, M.A.; Hossain, M.A.; Kabir, M.S. Stability and Adaptability Analysis of BRRI Developed Aus Varieties in Different Locations of Bangladesh. Bangladesh Rice J. 2018, 22, 65–72. [Google Scholar] [CrossRef]
- Matin, M.Q.I.; Rasul, M.G.; Islam, A.K.M.A.; Mian, M.A.K.; Ivy, N.A.; Ahmed, J.U. Genotype Environment Interaction in Maturity and Yield of Hybrid Maize (Zea mays L.). J. Agric. Sci. 2017, 2, 29–40. [Google Scholar]
- Balakrishnan, D.; Subrahmanyam, D.; Badri, J.; Raju, A.K.; Rao, Y.V.; Beerelli, K.; Mesapogu, S.; Surapaneni, M.; Ponnuswamy, R.; Padmavathi, G.; et al. Genotype × Environment Interactions of Yield Traits in Backcross Introgression Lines Derived from Oryza sativa cv. Swarna × Oryza nivara. Front. Plant Sci. 2016, 7, 1530. [Google Scholar] [CrossRef]
- Hashim, N.; Rafii, M.Y.; Oladosu, Y.; Ismail, M.R.; Ramli, A.; Arolu, F.; Chukwu, S. Integrating Multivariate and Univariate Statistical Models to Investigate Genotype–Environment Interaction of Advanced Fragrant Rice Genotypes under Rainfed Condition. Sustainability 2021, 13, 4555. [Google Scholar] [CrossRef]
- IRRI (Bangladesh Rice Research Institute). Standard Evaluation System (SES) for Rice, 5th ed.; IRRI: Manila, Philippines, 2013. [Google Scholar]
- Eberhart, S.A.; Russell, W.L. Stability parameters for comparing varieties. Crop Sci. 1966, 6, 36–40. [Google Scholar] [CrossRef]
- BRRI (Bangladesh Rice Research Institute). Adhunik Dhaner Chash; BRRI: Gazipur, Bangladesh, 2020. [Google Scholar]
- RStudio. RStudio: Integrated Development Environment for R (Computer Software v0.98.1074). 2014. Available online: http://www.rstudio.org/ (accessed on 19 March 2022).
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE Biplot vs. AMMI Analysis of Genotype-by-Environment data. Crop. Sci. 2007, 47, 643–653. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists, 1st ed.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Gauch, H.G.; Zobel, R.W. Identifying Mega-environments and Targeting Genotypes. Crop Sci. 1997, 37, 311–326. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists; CRC Press: New York, NY, USA, 2002; p. 71. [Google Scholar]
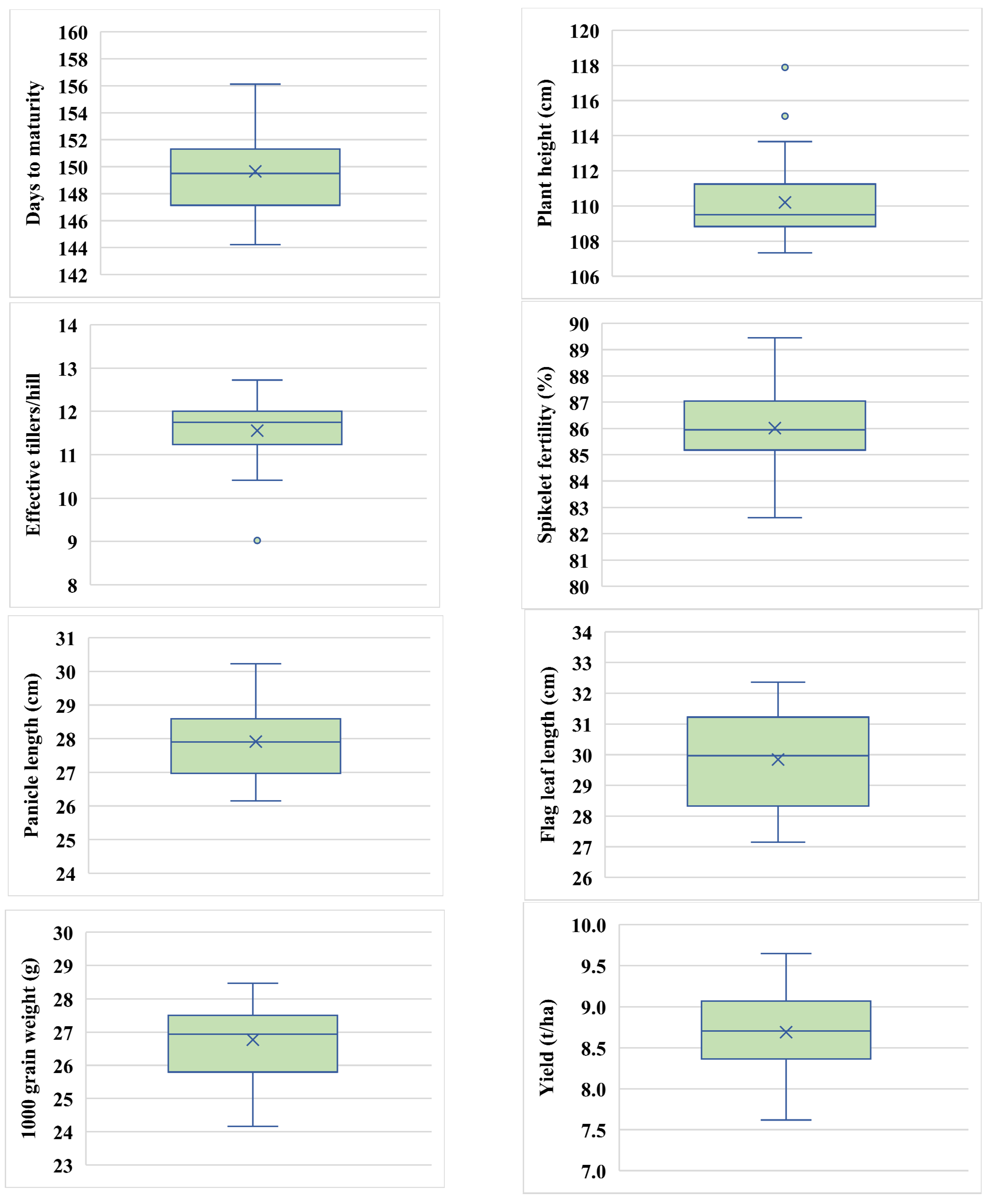

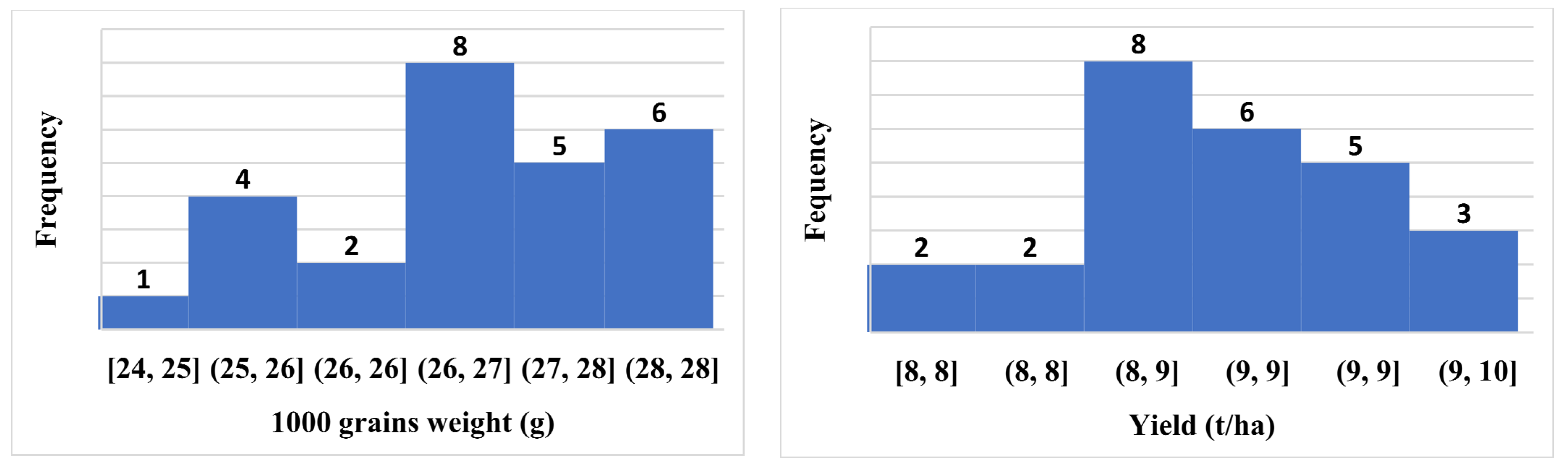





| Parameter | DTM | PH | ET/hill | SF% | PL | FLL | TGW | Grain Yield (t/ha) |
|---|---|---|---|---|---|---|---|---|
| Skewness | 0.44 | 1.71 | −1.69 | 0.09 | 0.25 | −0.19 | −0.41 | −0.08 |
| Kurtosis | −0.49 | 3.24 | 4.47 | 0.18 | −0.17 | −1.19 | −0.18 | −0.21 |
| Source of Variance | df | DTM | PH (cm) | ET/ hill | SF (%) | PL | FLL | TGW | Grain Yield (t/ha) |
|---|---|---|---|---|---|---|---|---|---|
| Location | 2 | 87.28 ** | 16.24 * | 0.23 ns | 8.87 * | 8.69 ** | 3.27 ** | 1.99 ** | 2.22 ** |
| Variety | 25 | 86.06 ** | 52.56 ** | 4.92 ** | 24.10 ** | 10.37 ** | 20.89 ** | 10.44 ** | 2.21 ** |
| Location × Rep | 6 | 2.91 ns | 4.03 ns | 1.59 * | 12.54 ** | 1.06 ** | 2.41 ** | 0.24 * | 0.30 ** |
| Location × Variety | 50 | 7.60 ** | 1.65 ns | 0.55 ns | 2.76 ns | 0.58 ** | 0.60 ns | 0.72 ** | 0.86 ** |
| Error | 150 | 2.73 | 3.66 | 0.45 | 2.19 | 0.22 | 0.55 | 0.12 | 0.08 |
| Total | 233 |
| Grain Type | Kernel Length (mm) | Kernel L × B Ratio | Hybrid Combination | Total |
|---|---|---|---|---|
| Medium slender | 5.0–6.0 | >3.0 | BRRI48A × BRRI38R, BRRI99A × BRRI38R, IR79156A × BRRI38R, IR79156A × BRRI45R, TejGold, Teea, SL8H | 7 |
| Long slender | >6.0 | >3.0 | BRRI99A × BRRI36R, IR79156A × BRRI49R, IR79156A × R line7, BRRI99A × BRRI31R, IR79156A × BRRI31R, H-2264, H-386 | 7 |
| Medium bold | 5.0–6.0 | 2.0–3.0 or <3.0 | BRRI35A × BRRI36R, BRRI35A × BRRI49R, BRRI99A × BRRI49R, BRRI35A × BRRI37R, BRRI35A × BRRI45R, BRRI99A × BRRI45R, BRRI35A × BRRI52R, BRRI hybrid dhan3, JhonokRaj, Heera-2, Gold | 11 |
| Long bold | >6.0 | 2.0–3.0 or <3.0 | BRRI hybrid dhan5 | 1 |
| Total | 26 |
| Sl No. | Combination | Environment | Pi | bi | S2di | |||
|---|---|---|---|---|---|---|---|---|
| Barisal (E1) | Gazipur (E2) | Ishwardi (E3) | Overall Mean | |||||
| 1 | BRRI35A × BRRI36R | 144.3 | 146.3 | 146.7 | 145.8 | −3.3 | 0.631 * | 0.00 |
| 2 | BRRI99A × BRRI36R | 144.0 | 148.7 | 149.3 | 147.3 | −1.8 | 1.452 | 0.01 |
| 3 | BRRI35A × BRRI49R | 146.3 | 158.3 | 157.7 | 154.1 | 5 | 3.308 | 3.37 |
| 4 | BRRI99A × BRRI49R | 102.7 | 146.7 | 148.0 | 132.4 | −16.7 | 12.802 | 18.61 |
| 5 | IR79156A × BRRI49R | 146.7 | 151.7 | 152.7 | 150.3 | 1.2 | 1.607 | 0.00 |
| 6 | BRRI35A × BRRI37R | 147.3 | 148.3 | 150.0 | 148.6 | −0.5 | 0.584 | 0.91 |
| 7 | BRRI48A × BRRI38R | 149.3 | 150.0 | 149.3 | 149.6 | 0.5 | 0.072 | 0.26 |
| 8 | BRRI99A × BRRI38R | 150.0 | 150.7 | 150.3 | 150.3 | 1.2 | 0.131 | 0.09 |
| 9 | IR79156A × BRRI38R | 149.7 | 149.3 | 152.3 | 150.4 | 1.3 | 0.442 | 3.85 |
| 10 | IR79156A × R line7 | 150.7 | 150.0 | 151.0 | 150.6 | 1.5 | −0.011 | 0.52 |
| 11 | BRRI35A × BRRI45R | 152.7 | 155.0 | 157.3 | 155.0 | 5.9 | 1.084 | 1.49 |
| 12 | BRRI99A × BRRI45R | 153.3 | 153.3 | 155.7 | 154.1 | 5 | 0.418 | 2.24 |
| 13 | IR79156A × BRRI45R | 154.0 | 153.7 | 152.7 | 153.4 | 4.3 | −0.274 | 0.37 |
| 14 | BRRI35A × BRRI52R | 147.3 | 147.0 | 147.3 | 147.2 | −1.9 | −0.035 | 0.07 |
| 15 | BRRI99A × BRRI31R | 147.3 | 150.3 | 149.0 | 148.9 | −0.2 | 0.619 | 1.46 |
| 16 | IR79156A × BRRI31R | 145.3 | 147.7 | 147.0 | 146.7 | −2.4 | 0.547 | 0.50 |
| 17 | H-2264 | 148.7 | 149.3 | 150.7 | 149.6 | 0.5 | 0.429 | 0.61 |
| 18 | H-386 | 149.7 | 152.0 | 151.0 | 150.9 | 1.8 | 0.488 | 0.84 |
| 19 | BRRI hybrid dhan3 | 146.0 | 146.0 | 147.7 | 146.6 | −2.5 | 0.298 | 1.15 |
| 20 | BRRI hybrid dhan5 | 147.7 | 147.0 | 148.0 | 147.6 | −1.5 | −0.011 | 0.52 |
| 21 | TejGold | 145.3 | 145.7 | 147.0 | 146.0 | −3.1 | 0.334 | 0.67 |
| 22 | JhonokRaj | 151.0 | 152.3 | 154.3 | 152.6 | 3.5 | 0.739 | 1.27 |
| 23 | Heera-2 | 155.3 | 156.7 | 156.3 | 156.1 | 7 | 0.322 | 0.14 |
| 24 | Gold | 148.3 | 148.7 | 149.3 | 148.8 | −0.3 | 0.215 | 0.16 |
| 25 | Teea | 144.0 | 144.0 | 144.7 | 144.2 | −4.9 | 0.120 | 0.19 |
| 26 | SL8H | 150.0 | 150.0 | 148.3 | 149.4 | 0.3 | −0.298 | 1.15 |
| Mean | 149.9 | 146.8 | 150.5 | 149.1 | ||||
| Ij | 0.8 | −2.3 | 1.4 | |||||
| CV% | 9.9 | 1.1 | 1.0 | |||||
| LSD (0.05) | 23.7 | 2.76 | 2.50 | |||||
| Sl No. | Combination | Environment | Pi | bi | S2di | |||
|---|---|---|---|---|---|---|---|---|
| Barisal (E1) | Gazipur (E2) | Ishwardi (E3) | Overall Mean | |||||
| 1 | BRRI35A × BRRI36R | 107.3 | 107.3 | 107.7 | 107.4 | −2.8 | 0.377 | 0.01 |
| 2 | BRRI99A × BRRI36R | 105.7 | 107.0 | 109.3 | 107.3 | −2.9 | 2.778 | 3.67 |
| 3 | BRRI35A × BRRI49R | 108.3 | 109.0 | 109.3 | 108.9 | −1.3 | 0.439 | 0.44 |
| 4 | BRRI99A × BRRI49R | 108.3 | 107.3 | 109.7 | 108.4 | −1.8 | 2.563 * | 0.00 |
| 5 | IR79156A × BRRI49R | 109.7 | 110.3 | 113.3 | 111.1 | 0.9 | 3.476 | 2.60 |
| 6 | BRRI35A × BRRI37R | 108.7 | 107.3 | 108.3 | 108.1 | −2.1 | 1.013 | 0.53 |
| 7 | BRRI48A × BRRI38R | 109.0 | 109.3 | 109.0 | 109.1 | −1.1 | −0.351 | 0.02 |
| 8 | BRRI99A × BRRI38R | 110.3 | 108.3 | 108.7 | 109.1 | −1.1 | 0.193 | 2.28 |
| 9 | IR79156A × BRRI38R | 111.3 | 111.3 | 112.7 | 111.8 | 1.6 | 1.516 | 0.23 |
| 10 | IR79156A × R line7 | 113.3 | 111.3 | 114.3 | 113.7 | 3.5 | 1.137 | 0.13 |
| 11 | BRRI35A × BRRI45R | 110.3 | 109.7 | 110.0 | 110.0 | −0.2 | 0.316 | 0.18 |
| 12 | BRRI99A × BRRI45R | 108.7 | 18.3 | 110.3 | 109.1 | −1.1 | 2.245 | 0.20 |
| 13 | IR79156A × BRRI45R | 111.0 | 112.3 | 111.7 | 111.7 | 1.5 | −0.638 | 0.72 |
| 14 | BRRI35A × BRRI52R | 109.3 | 109.3 | 110.3 | 109.7 | −0.5 | 1.137 | 0.13 |
| 15 | BRRI99A × BRRI31R | 109.0 | 107.3 | 109.7 | 108.7 | −1.5 | 2.501 | 0.28 |
| 16 | IR79156A × BRRI31R | 112.7 | 112.7 | 113.0 | 112.8 | 2.6 | 0.377 | 0.01 |
| 17 | H-2264 | 115.3 | 114.0 | 116.0 | 115.1 | 4.9 | 2.152 | 0.14 |
| 18 | H-386 | 117.7 | 118.0 | 118.0 | 117.9 | 7.7 | 0.029 | 0.07 |
| 19 | BRRI hybrid dhan3 | 110.0 | 109.7 | 110.0 | 109.9 | −0.3 | 0.347 | 0.02 |
| 20 | BRRI hybrid dhan5 | 110.0 | 109.3 | 110.3 | 109.9 | −0.3 | 1.075 | 0.04 |
| 21 | TejGold | 110.0 | 108.3 | 108.7 | 109.0 | −1.2 | 0.223 | 1.53 |
| 22 | JhonokRaj | 109.0 | 109.0 | 110.0 | 109.3 | −0.9 | 1.137 | 0.13 |
| 23 | Heera-2 | 111.0 | 109.7 | 109.7 | 110.1 | −0.1 | −0.125 | 1.18 |
| 24 | Gold | 110.7 | 109.3 | 109.7 | 109.9 | −0.3 | 0.254 | 0.93 |
| 25 | Teea | 109.3 | 108.7 | 109.0 | 109.0 | −1.2 | 0.316 | 0.18 |
| 26 | SL8H | 108.3 | 107.7 | 109.0 | 108.3 | −1.9 | 1.455 | 0.01 |
| Mean | 110.2 | 109.8 | 110.7 | 110.2 | ||||
| Ij | 0 | −0.4 | 0.5 | |||||
| CV% | 1.8 | 1.9 | 1.5 | |||||
| LSD (0.05) | 3.19 | 3.40 | 2.78 | |||||
| Sl No. | Combination | Environment | Pi | bi | S2di | |||
|---|---|---|---|---|---|---|---|---|
| Barisal (E1) | Gazipur (E2) | Ishwardi (E3) | Overall Mean | |||||
| 1 | BRRI35A × BRRI36R | 11.37 | 10.67 | 10.67 | 10.90 | −0.65 | 7.418 * | 0.00 |
| 2 | BRRI99A × BRRI36R | 11.77 | 11.63 | 10.90 | 11.43 | −0.12 | 5.061 | 0.28 |
| 3 | BRRI35A × BRRI49R | 8.800 | 8.800 | 9.467 | 9.022 | −2.528 | −3.318 | 0.23 |
| 4 | BRRI99A × BRRI49R | 12.47 | 12.20 | 11.80 | 12.16 | 0.61 | 4.815 | 0.09 |
| 5 | IR79156A × BRRI49R | 11.00 | 11.10 | 11.77 | 11.29 | −0.26 | −4.377 | 0.23 |
| 6 | BRRI35A × BRRI37R | 9.833 | 10.70 | 10.70 | 10.41 | −1.14 | −9.186 * | 0.00 |
| 7 | BRRI48A × BRRI38R | 12.17 | 11.87 | 11.30 | 11.78 | 0.23 | 5.998 | 0.17 |
| 8 | BRRI99A × BRRI38R | 10.63 | 10.80 | 10.73 | 10.72 | −0.83 | −1.436 | 0.00 |
| 9 | IR79156A × BRRI38R | 10.13 | 11.80 | 12.27 | 11.40 | −0.15 | −19.987 | 0.15 |
| 10 | IR79156A × R line7 | 10.27 | 11.00 | 11.93 | 11.07 | −0.48 | −12.417 | 0.48 |
| 11 | BRRI35A × BRRI45R | 11.57 | 11.67 | 11.40 | 11.54 | −0.01 | 0.266 | 0.04 |
| 12 | BRRI99A × BRRI45R | 12.10 | 11.93 | 11.83 | 11.96 | 0.41 | 2.263 | 0.01 |
| 13 | IR79156A × BRRI45R | 12.20 | 12.47 | 11.97 | 12.21 | 0.66 | −0.340 | 0.12 |
| 14 | BRRI35A × BRRI52R | 12.23 | 11.97 | 11.77 | 11.99 | 0.44 | 3.820 | 0.02 |
| 15 | BRRI99A × BRRI31R | 12.50 | 11.87 | 11.17 | 11.84 | 0.29 | 10.194 | 0.27 |
| 16 | IR79156A × BRRI31R | 11.43 | 10.93 | 11.57 | 11.31 | −0.24 | 2.148 | 0.20 |
| 17 | H-2264 | 11.93 | 11.73 | 11.97 | 11.88 | 0.33 | 0.958 | 0.03 |
| 18 | H-386 | 12.93 | 11.80 | 11.97 | 12.23 | 0.68 | 11.182 | 0.01 |
| 19 | BRRI hybrid dhan3 | 12.17 | 12.27 | 11.87 | 12.10 | 0.55 | 0.929 | 0.08 |
| 20 | BRRI hybrid dhan5 | 12.00 | 11.93 | 11.97 | 11.97 | 0.42 | 0.540 | 0.00 |
| 21 | TejGold | 12.80 | 12.77 | 12.60 | 12.72 | 1.17 | 1.182 | 0.01 |
| 22 | JhonokRaj | 12.07 | 12.00 | 12.07 | 12.04 | 0.49 | 0.374 | 0.00 |
| 23 | Heera-2 | 12.00 | 11.47 | 11.60 | 11.69 | 0.14 | 4.988 | 0.01 |
| 24 | Gold | 12.33 | 11.77 | 11.87 | 11.99 | 0.44 | 5.508 | 0.00 |
| 25 | Teea | 11.33 | 10.87 | 10.87 | 11.02 | −0.53 | 4.945 * | 0.00 |
| 26 | SL8H | 12.00 | 11.53 | 11.63 | 11.72 | 0.17 | 4.448 | 0.00 |
| Mean | 11.62 | 11.52 | 11.52 | 11.55 | ||||
| Ij | 0.07 | −0.03 | −0.03 | |||||
| CV% | 6.0 | 5.6 | 5.8 | |||||
| LSD (0.05) | 1.14 | 1.07 | 1.09 | |||||
| Sl No. | Combination | Environment | Pi | bi | S2di | |||
|---|---|---|---|---|---|---|---|---|
| Barisal (E1) | Gazipur (E2) | Ishwardi (E3) | Overall Mean | |||||
| 1 | BRRI35A × BRRI36R | 86.17 | 85.60 | 85.10 | 85.62 | −0.39 | −1.559 | 0.02 |
| 2 | BRRI99A × BRRI36R | 86.67 | 89.63 | 90.97 | 89.09 | 3.08 | 6.152 | 1.08 |
| 3 | BRRI35A × BRRI49R | 85.30 | 84.93 | 85.60 | 85.28 | −0.73 | 0.554 | 0.15 |
| 4 | BRRI99A × BRRI49R | 87.07 | 86.50 | 88.30 | 87.29 | 1.28 | 2.070 | 0.72 |
| 5 | IR79156A × BRRI49R | 80.73 | 83.50 | 85.17 | 83.13 | −2.88 | 6.405 | 0.70 |
| 6 | BRRI35A × BRRI37R | 85.33 | 87.33 | 87.33 | 86.67 | 0.66 | 2.730 | 0.97 |
| 7 | BRRI48A × BRRI38R | 89.50 | 86.50 | 86.10 | 87.37 | 1.36 | −4.720 | 1.84 |
| 8 | BRRI99A × BRRI38R | 85.13 | 85.43 | 87.50 | 86.02 | 0.01 | 3.673 | 0.25 |
| 9 | IR79156A × BRRI38R | 86.43 | 87.10 | 86.83 | 86.79 | 0.78 | 0.491 | 0.17 |
| 10 | IR79156A × R line7 | 83.37 | 86.40 | 86.40 | 85.39 | −0.62 | 4.139 | 2.24 |
| 11 | BRRI35A × BRRI45R | 85.73 | 84.73 | 84.73 | 85.07 | −0.94 | −1.361 | 0.25 |
| 12 | BRRI99A × BRRI45R | 87.27 | 87.77 | 87.77 | 87.60 | 1.59 | 0.684 | 0.06 |
| 13 | IR79156A × BRRI45R | 86.30 | 85.73 | 85.73 | 85.92 | −0.09 | −0.770 | 0.08 |
| 14 | BRRI35A × BRRI52R | 85.80 | 85.23 | 85.23 | 85.42 | −0.59 | −0.770 | 0.08 |
| 15 | BRRI99A × BRRI31R | 86.90 | 86.90 | 87.07 | 86.96 | 0.95 | 0.265 | 0.00 |
| 16 | IR79156A × BRRI31R | 85.80 | 85.40 | 85.40 | 85.53 | −0.48 | −0.543 | 0.04 |
| 17 | H-2264 | 83.17 | 81.83 | 82.83 | 82.61 | −3.4 | −0.238 | 0.95 |
| 18 | H-386 | 83.47 | 83.37 | 84.83 | 83.89 | −2.12 | 2.180 | 0.26 |
| 19 | BRRI hybrid dhan3 | 88.53 | 88.47 | 87.80 | 88.27 | 2.26 | −1.141 | 0.03 |
| 20 | BRRI hybrid dhan5 | 89.40 | 89.30 | 89.63 | 89.44 | 3.43 | 0.392 | 0.02 |
| 21 | TejGold | 85.60 | 86.60 | 86.70 | 86.30 | 0.29 | 1.524 | 0.21 |
| 22 | JhonokRaj | 85.53 | 86.53 | 87.50 | 86.52 | 0.51 | 2.891 | 0.03 |
| 23 | Heera-2 | 84.67 | 86.60 | 86.67 | 85.98 | −0.03 | 2.744 | 0.87 |
| 24 | Gold | 85.83 | 84.93 | 84.87 | 85.21 | −0.8 | −1.330 | 0.18 |
| 25 | Teea | 84.03 | 84.03 | 84.90 | 84.32 | −1.69 | 1.370 | 0.07 |
| 26 | SL8H | 84.50 | 84.67 | 84.67 | 84.61 | −1.4 | 0.230 | 0.01 |
| Mean | 85.70 | 85.96 | 86.37 | 86.01 | ||||
| Ij | −0.31 | −0.05 | 0.36 | |||||
| CV% | 1.5 | 1.9 | 1.8 | |||||
| LSD (0.05) | 2.09 | 2.65 | 2.50 | |||||
| Sl No. | Combination | Environment | Pi | bi | S2di | |||
|---|---|---|---|---|---|---|---|---|
| Barisal (E1) | Gazipur (E2) | Ishwardi (E3) | Overall Mean | |||||
| 1 | BRRI35A × BRRI36R | 26.20 | 27.17 | 27.23 | 26.87 | −1.04 | 1.707 | 0.02 |
| 2 | BRRI99A × BRRI36R | 27.43 | 29.17 | 30.43 | 29.01 | 1.1 | 4.414 | 0.19 |
| 3 | BRRI35A × BRRI49R | 25.43 | 27.80 | 27.80 | 27.01 | −0.9 | 3.988 | 0.19 |
| 4 | BRRI99A × BRRI49R | 27.90 | 28.57 | 28.90 | 28.46 | 0.55 | 1.516 | 0.01 |
| 5 | IR79156A × BRRI49R | 25.50 | 26.23 | 26.73 | 26.16 | −1.75 | 1.825 | 0.03 |
| 6 | BRRI35A × BRRI37R | 25.13 | 27.13 | 27.13 | 26.47 | −1.44 | 3.370 | 0.13 |
| 7 | BRRI48A × BRRI38R | 28.77 | 28.50 | 28.40 | 28.56 | 0.65 | −0.567 * | 0.00 |
| 8 | BRRI99A × BRRI38R | 30.17 | 30.17 | 30.33 | 30.22 | 2.31 | 0.196 | 0.01 |
| 9 | IR79156A × BRRI38R | 27.60 | 27.60 | 27.90 | 27.70 | −0.21 | 0.354 | 0.03 |
| 10 | IR79156A × R line7 | 26.87 | 28.20 | 28.20 | 27.76 | −0.15 | 2.247 | 0.06 |
| 11 | BRRI35A × BRRI45R | 26.70 | 27.03 | 27.03 | 26.92 | −0.99 | 0.562 | 0.00 |
| 12 | BRRI99A × BRRI45R | 28.20 | 28.93 | 28.93 | 28.69 | 0.78 | 1.236 | 0.02 |
| 13 | IR79156A × BRRI45R | 28.77 | 29.07 | 29.07 | 28.97 | 1.06 | 0.505 | 0.00 |
| 14 | BRRI35A × BRRI52R | 26.80 | 27.07 | 27.07 | 26.98 | −0.93 | 0.449 | 0.00 |
| 15 | BRRI99A × BRRI31R | 28.30 | 28.40 | 28.33 | 28.34 | 0.43 | 0.090 | 0.00 |
| 16 | IR79156A × BRRI31R | 27.73 | 27.77 | 27.77 | 27.76 | −0.15 | 0.056 | 0.00 |
| 17 | H-2264 | 27.73 | 27.80 | 27.87 | 27.80 | −0.11 | 0.191 * | 0.00 |
| 18 | H-386 | 28.17 | 28.53 | 28.50 | 28.40 | 0.49 | 0.578 | 0.01 |
| 19 | BRRI hybrid dhan3 | 28.07 | 28.13 | 28.47 | 28.22 | 0.31 | 0.505 | 0.03 |
| 20 | BRRI hybrid dhan5 | 30.03 | 30.03 | 30.17 | 30.08 | 2.17 | 0.157 | 0.01 |
| 21 | TejGold | 26.07 | 26.07 | 26.33 | 26.16 | −1.75 | 0.314 | 0.03 |
| 22 | JhonokRaj | 28.73 | 28.73 | 29.00 | 28.82 | 0.91 | 0.314 | 0.03 |
| 23 | Heera-2 | 27.83 | 28.07 | 28.13 | 28.01 | 0.1 | 0.472 | 0.00 |
| 24 | Gold | 27.90 | 27.90 | 28.10 | 27.97 | 0.06 | 0.236 | 0.01 |
| 25 | Teea | 26.13 | 26.80 | 26.93 | 26.62 | −1.29 | 1.281 | 0.00 |
| 26 | SL8H | 27.83 | 27.83 | 27.83 | 27.83 | −0.08 | 0.000 | 0.00 |
| Mean | 27.54 | 28.03 | 28.18 | 27.91 | ||||
| Ij | −0.37 | 0.12 | 0.27 | |||||
| CV% | 1.5 | 1.8 | 1.8 | |||||
| LSD (0.05) | 0.67 | 0.81 | 0.83 | |||||
| Sl No. | Combination | Environment | Pi | bi | S2di | |||
|---|---|---|---|---|---|---|---|---|
| Barisal (E1) | Gazipur (E2) | Ishwardi (E3) | Overall Mean | |||||
| 1 | BRRI35A × BRRI36R | 29.43 | 30.60 | 30.43 | 30.16 | 0.32 | 2.656 | 0.20 |
| 2 | BRRI99A × BRRI36R | 32.00 | 31.33 | 31.33 | 31.56 | 1.72 | −1.732 | 0.04 |
| 3 | BRRI35A × BRRI49R | 27.00 | 27.23 | 27.23 | 27.16 | −2.68 | 0.606 | 0.01 |
| 4 | BRRI99A × BRRI49R | 30.83 | 30.50 | 30.50 | 30.61 | 0.77 | −0.866 | 0.01 |
| 5 | IR79156A × BRRI49R | 28.13 | 28.13 | 28.13 | 28.13 | −1.71 | 0.000 | 0.00 |
| 6 | BRRI35A × BRRI37R | 26.37 | 29.40 | 29.40 | 28.39 | −1.45 | 7.881 | 0.92 |
| 7 | BRRI48A × BRRI38R | 30.30 | 30.30 | 30.30 | 30.30 | 0.46 | 0.000 | 0.00 |
| 8 | BRRI99A × BRRI38R | 30.80 | 30.80 | 30.80 | 30.80 | 0.96 | 0.000 | 0.00 |
| 9 | IR79156A × BRRI38R | 31.23 | 31.23 | 31.23 | 31.23 | 1.39 | 0.000 | 0.00 |
| 10 | IR79156A × R line7 | 29.23 | 29.23 | 29.23 | 29.23 | −0.61 | 0.000 | 0.00 |
| 11 | BRRI35A × BRRI45R | 29.33 | 30.00 | 30.00 | 29.78 | −0.06 | 1.732 | 0.04 |
| 12 | BRRI99A × BRRI45R | 31.27 | 31.60 | 31.60 | 31.49 | 1.65 | 0.866 | 0.01 |
| 13 | IR79156A × BRRI45R | 31.40 | 31.13 | 31.13 | 31.22 | 1.38 | −0.693 | 0.01 |
| 14 | BRRI35A × BRRI52R | 27.37 | 28.43 | 28.43 | 28.08 | −1.76 | 2.771 | 0.11 |
| 15 | BRRI99A × BRRI31R | 32.37 | 32.33 | 32.37 | 32.36 | 2.52 | −0.012 | 0.00 |
| 16 | IR79156A × BRRI31R | 31.43 | 31.43 | 31.60 | 31.49 | 1.65 | 0.374 | 0.01 |
| 17 | H-2264 | 31.93 | 32.40 | 31.37 | 31.90 | 2.06 | −1.109 | 0.43 |
| 18 | H-386 | 29.47 | 29.67 | 30.13 | 29.76 | −0.08 | 1.568 | 0.03 |
| 19 | BRRI hybrid dhan3 | 28.53 | 28.60 | 30.03 | 29.06 | −0.78 | 3.393 | 0.47 |
| 20 | BRRI hybrid dhan5 | 31.07 | 31.07 | 31.33 | 31.16 | 1.32 | 0.599 | 0.02 |
| 21 | TejGold | 27.83 | 27.83 | 28.17 | 27.94 | −1.9 | 0.749 | 0.03 |
| 22 | JhonokRaj | 27.77 | 27.77 | 28.67 | 28.07 | −1.77 | 2.022 | 0.20 |
| 23 | Heera-2 | 28.47 | 28.80 | 28.87 | 28.71 | −1.13 | 1.015 | 0.01 |
| 24 | Gold | 30.50 | 30.50 | 31.03 | 30.68 | 0.84 | 1.198 | 0.07 |
| 25 | Teea | 29.10 | 29.27 | 29.67 | 29.34 | −0.5 | 1.331 | 0.02 |
| 26 | SL8H | 27.07 | 27.07 | 27.80 | 27.31 | −2.53 | 1.647 | 0.13 |
| Mean | 29.62 | 29.87 | 30.03 | 29.84 | ||||
| Ij | −0.22 | 0.03 | 0.19 | |||||
| CV% | 2.6 | 2.5 | 2.3 | |||||
| LSD (0.05) | 1.26 | 1.24 | 1.14 | |||||
| Sl No. | Combination | Environment | Pi | bi | S2di | |||
|---|---|---|---|---|---|---|---|---|
| Barisal (E1) | Gazipur (E2) | Ishwardi (E3) | Overall Mean | |||||
| 1 | BRRI35A × BRRI36R | 27.10 | 26.73 | 26.57 | 26.80 | 0.03 | −1.706 | 0.00 |
| 2 | BRRI99A × BRRI36R | 28.03 | 28.30 | 28.57 | 28.30 | 1.53 | 1.611 | 0.01 |
| 3 | BRRI35A × BRRI49R | 25.47 | 28.13 | 28.13 | 27.24 | 0.47 | 9.297 | 0.32 |
| 4 | BRRI99A × BRRI49R | 26.47 | 26.47 | 27.97 | 26.97 | 0.2 | 3.837 | 0.75 |
| 5 | IR79156A × BRRI49R | 23.40 | 24.30 | 24.80 | 24.17 | −2.6 | 4.416 | 0.01 |
| 6 | BRRI35A × BRRI37R | 24.60 | 26.10 | 26.17 | 25.62 | −1.15 | 5.400 | 0.08 |
| 7 | BRRI48A × BRRI38R | 25.57 | 25.57 | 25.57 | 25.57 | −1.2 | −0.001 | 0.00 |
| 8 | BRRI99A × BRRI38R | 26.63 | 27.47 | 27.47 | 27.19 | 0.42 | 2.904 | 0.03 |
| 9 | IR79156A × BRRI38R | 26.70 | 27.23 | 27.23 | 27.06 | 0.29 | 1.858 | 0.01 |
| 10 | IR79156A × R line7 | 25.30 | 25.30 | 25.53 | 25.38 | −1.39 | 0.596 | 0.02 |
| 11 | BRRI35A × BRRI45R | 25.70 | 25.93 | 25.93 | 25.86 | −0.91 | 0.812 | 0.00 |
| 12 | BRRI99A × BRRI45R | 27.77 | 27.83 | 27.83 | 27.81 | 1.04 | 0.231 * | 0.00 |
| 13 | IR79156A × BRRI45R | 28.77 | 26.40 | 26.40 | 27.02 | 0.25 | −6.510 | 0.16 |
| 14 | BRRI35A × BRRI52R | 26.33 | 26.53 | 26.17 | 26.34 | −0.43 | −0.242 | 0.06 |
| 15 | BRRI99A × BRRI31R | 28.40 | 28.50 | 28.50 | 28.47 | 1.7 | 0.348 | 0.00 |
| 16 | IR79156A × BRRI31R | 26.37 | 26.37 | 26.37 | 26.37 | −0.4 | −0.001 | 0.00 |
| 17 | H-2264 | 27.37 | 26.67 | 26.67 | 26.90 | 0.13 | −2.442 | 0.02 |
| 18 | H-386 | 26.50 | 26.33 | 26.33 | 26.39 | −0.38 | −0.582 | 0.00 |
| 19 | BRRI hybrid dhan3 | 27.27 | 27.47 | 27.47 | 27.40 | 0.63 | 0.696 | 0.00 |
| 20 | BRRI hybrid dhan5 | 28.27 | 28.27 | 28.13 | 28.22 | 1.45 | −0.342 | 0.01 |
| 21 | TejGold | 25.47 | 25.47 | 25.47 | 25.47 | −1.3 | −0.001 | 0.00 |
| 22 | JhonokRaj | 27.50 | 28.50 | 28.30 | 28.10 | 1.33 | 2.974 | 0.11 |
| 23 | Heera-2 | 27.80 | 28.00 | 28.10 | 27.97 | 1.2 | 0.952 | 0.00 |
| 24 | Gold | 26.93 | 27.20 | 27.30 | 27.14 | 0.37 | 1.184 | 0.00 |
| 25 | Teea | 26.83 | 26.83 | 26.73 | 26.80 | 0.03 | −0.257 | 0.00 |
| 26 | SL8H | 25.33 | 25.33 | 25.70 | 25.46 | −1.31 | 0.937 | 0.04 |
| Mean | 26.59 | 26.82 | 26.90 | 26.77 | ||||
| Ij | −0.18 | 0.05 | 0.13 | |||||
| CV% | 1.1 | 1.3 | 1.5 | |||||
| LSD (0.05) | 0.49 | 0.56 | 0.66 | |||||
| Sl No. | Combination | Environment | Pi | bi | S2di | |||
|---|---|---|---|---|---|---|---|---|
| Barisal (E1) | Gazipur (E2) | Ishwardi (E3) | Overall Mean | |||||
| 1 | BRRI35A × BRRI36R | 7.743 | 8.173 | 8.310 | 8.076 | −0.613 | 1.714 | 0.01 |
| 2 | BRRI99A × BRRI36R | 8.300 | 9.673 | 9.673 | 9.216 | 0.527 | 4.244 | 0.23 |
| 3 | BRRI35A × BRRI49R | 7.313 | 7.757 | 8.623 | 7.898 | −0.791 | 3.814 | 0.06 |
| 4 | BRRI99A × BRRI49R | 8.640 | 8.790 | 9.490 | 8.973 | 0.284 | 2.438 | 0.07 |
| 5 | IR79156A × BRRI49R | 6.297 | 7.630 | 8.930 | 7.619 | −1.07 | 7.787 | 0.02 |
| 6 | BRRI35A × BRRI37R | 6.253 | 8.853 | 8.963 | 8.023 | −0.666 | 8.345 | 0.74 |
| 7 | BRRI48A × BRRI38R | 9.370 | 8.487 | 8.460 | 8.772 | 0.083 | −2.805 | 0.09 |
| 8 | BRRI99A × BRRI38R | 9.927 | 8.927 | 9.583 | 9.479 | 0.79 | −1.238 | 0.43 |
| 9 | IR79156A × BRRI38R | 8.977 | 9.143 | 9.087 | 9.069 | 0.38 | 0.355 | 0.01 |
| 10 | IR79156A × R line7 | 7.650 | 9.063 | 8.997 | 8.570 | −0.119 | 4.179 | 0.28 |
| 11 | BRRI35A × BRRI45R | 8.420 | 8.520 | 8.523 | 8.488 | −0.201 | 0.319 | 0.00 |
| 12 | BRRI99A × BRRI45R | 9.510 | 9.417 | 9.233 | 9.387 | 0.698 | −0.805 | 0.00 |
| 13 | IR79156A × BRRI45R | 9.623 | 8.817 | 8.770 | 9.070 | 0.381 | −2.624 | 0.07 |
| 14 | BRRI35A × BRRI52R | 8.317 | 8.590 | 8.543 | 8.483 | −0.206 | 0.713 | 0.01 |
| 15 | BRRI99A × BRRI31R | 9.367 | 9.103 | 9.257 | 9.242 | 0.553 | −0.381 | 0.03 |
| 16 | IR79156A × BRRI31R | 8.380 | 8.980 | 9.150 | 8.837 | 0.148 | 2.334 | 0.02 |
| 17 | H-2264 | 8.143 | 8.687 | 8.617 | 8.482 | −0.207 | 1.482 | 0.05 |
| 18 | H-386 | 9.363 | 8.383 | 8.333 | 8.693 | 0.004 | −3.169 | 0.10 |
| 19 | BRRI hybrid dhan3 | 8.793 | 8.780 | 8.847 | 8.807 | 0.118 | 0.147 | 0.00 |
| 20 | BRRI hybrid dhan5 | 9.857 | 9.470 | 9.617 | 9.648 | 0.959 | −0.781 | 0.04 |
| 21 | TejGold | 8.287 | 8.320 | 8.313 | 8.307 | −0.382 | 0.084 * | 0.00 |
| 22 | JhonokRaj | 8.500 | 8.810 | 8.833 | 8.714 | 0.025 | 1.024 | 0.01 |
| 23 | Heera-2 | 8.557 | 8.873 | 8.890 | 8.773 | 0.084 | 1.026 | 0.01 |
| 24 | Gold | 8.713 | 8.553 | 8.467 | 8.578 | −0.111 | −0.739 * | 0.00 |
| 25 | Teea | 8.087 | 8.457 | 8.497 | 8.347 | −0.342 | 1.256 | 0.01 |
| 26 | SL8H | 8.960 | 8.060 | 8.083 | 8.368 | −0.321 | −2.715 | 0.11 |
| Mean | 8.513 | 8.704 | 8.850 | 8.689 | ||||
| Ij | −0.176 | 0.015 | 0.161 | 0 | ||||
| CV% | 2.7 | 3.2 | 3.7 | |||||
| LSD (0.05) | 0.37 | 0.45 | 0.53 | |||||
| Sl No. | Combination | Sl No. | Combination | Sl No. | Combination |
|---|---|---|---|---|---|
| 1 | BRRI35A × BRRI36R | 10 | IR79156A × R line7 | 19 | BRRI hybrid dhan3 |
| 2 | BRRI99A × BRRI36R | 11 | BRRI35A × BRRI45R | 20 | BRRI hybrid dhan5 |
| 3 | BRRI35A × BRRI49R | 12 | BRRI99A × BRRI45R | 21 | TejGold |
| 4 | BRRI99A × BRRI49R | 13 | IR79156A × BRRI45R | 22 | JhonokRaj |
| 5 | IR79156A × BRRI49R | 14 | BRRI35A × BRRI52R | 23 | Heera-2 |
| 6 | BRRI35A × BRRI37R | 15 | BRRI99A × BRRI31R | 24 | Gold |
| 7 | BRRI48A × BRRI38R | 16 | IR79156A × BRRI31R | 25 | Teea |
| 8 | BRRI99A × BRRI38R | 17 | H-2264 | 26 | SL8H |
| 9 | IR79156A × BRRI38R | 18 | H-386 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, M.J.; Kulsum, M.U.; Sarker, U.; Matin, M.Q.I.; Shahin, N.H.; Kabir, M.S.; Ercisli, S.; Marc, R.A. Assessment of GGE, AMMI, Regression, and Its Deviation Model to Identify Stable Rice Hybrids in Bangladesh. Plants 2022, 11, 2336. https://doi.org/10.3390/plants11182336
Hasan MJ, Kulsum MU, Sarker U, Matin MQI, Shahin NH, Kabir MS, Ercisli S, Marc RA. Assessment of GGE, AMMI, Regression, and Its Deviation Model to Identify Stable Rice Hybrids in Bangladesh. Plants. 2022; 11(18):2336. https://doi.org/10.3390/plants11182336
Chicago/Turabian StyleHasan, M Jamil, M Umma Kulsum, Umakanta Sarker, M Quamrul Islam Matin, Nazmul Hoque Shahin, M Shahjahan Kabir, Sezai Ercisli, and Romina Alina Marc. 2022. "Assessment of GGE, AMMI, Regression, and Its Deviation Model to Identify Stable Rice Hybrids in Bangladesh" Plants 11, no. 18: 2336. https://doi.org/10.3390/plants11182336
APA StyleHasan, M. J., Kulsum, M. U., Sarker, U., Matin, M. Q. I., Shahin, N. H., Kabir, M. S., Ercisli, S., & Marc, R. A. (2022). Assessment of GGE, AMMI, Regression, and Its Deviation Model to Identify Stable Rice Hybrids in Bangladesh. Plants, 11(18), 2336. https://doi.org/10.3390/plants11182336








.jpg)

