GmSWEET29 and Paralog GmSWEET34 Are Differentially Expressed between Soybeans Grown in Eastern and Western Canada
Abstract
1. Introduction
2. Results
2.1. Seed Protein, Seed Oil Content, and Yield
2.2. Quality Control
2.3. SWEET Family Analysis
2.4. GmSWEET29 Is Persistently Upregulated in Soybeans Grown in West Locations
2.5. GmSWEET20 and GmSWEET34 Show Some DE between East and West Locations
2.6. Glyma.09G119100, Glyma.19G066300, Glyma.06G200200 Are AtSWEET17 Homologs
2.7. Top DE SWEET Genes between East and West
2.8. Weather Report
3. Discussion
3.1. GmSWEET29 Is an AtSWEET2 Homolog and Is Upregulated in Soybeans Grown in the West
3.2. GmSWEET34 Is a GmSWEET29 Paralog and a AtSWEET2 Homolog
3.3. GmSWEET20, a AtSWEET12 Homolog, Is Downregulated in the West
3.4. Glyma.06G200200, Glyma.09G119100, and Glyma.19G066300 May Be SWEET-like Proteins
3.5. GmSWEET39 Is Not DE between Eastern and Western-Grown Soybeans in Canada
3.6. SWEET Genes Have Been Shown to Influence Yield
3.7. Abiotic and Biotic Considerations
4. Materials and Methods
4.1. G. max Lines
4.2. Planting and Growth
4.3. Sampling and Seed Content Measurements
4.4. RNA Extraction and cDNA Library Preparation
4.5. RNA-Sequencing
4.6. Differential Expression (DE) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef]
- Ehrlich, P.R.; Harte, J. Opinion: To feed the world in 2050 will require a global revolution. Proc. Natl. Acad. Sci. USA 2015, 112, 14743–14744. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.G.; Kumari, H.; Pati, P.; Naaz, S.; Prasad, T.; Kumari, R.; Qayyum, H.; Naaz, N.; Nitin, M. Recent approaches of systems biology and omics in plant research. J. Curr. Opin. Crop Sci. 2021, 2, 288–305. [Google Scholar]
- Ray, D.K.; Gerber, J.S.; MacDonald, G.K.; West, P.C. Climate variation explains a third of global crop yield variability. Nat. Commun. 2015, 6, 5989. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate trends and global crop production since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Liu, K. Chemistry and Nutritional Value of Soybean Components BT—Soybeans: Chemistry, Technology, and Utilization; Liu, K., Ed.; Springer: Boston, MA, USA, 1997; pp. 25–113. [Google Scholar] [CrossRef]
- Huang, S.; Yu, J.; Li, Y.; Wang, J.; Wang, X.; Qi, H.; Xu, M.; Qin, H.; Yin, Z.; Mei, H.; et al. Identification of Soybean Genes Related to Soybean Seed Protein Content Based on Quantitative Trait Loci Collinearity Analysis. J. Agric. Food Chem. 2019, 67, 258–274. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, S.; Luthria, D.; Bae, H.; Lakshman, D.; Mitra, A. Transgenic Soybeans and Soybean Protein Analysis: An Overview. J. Agric. Food Chem. 2013, 61, 11736–11743. [Google Scholar] [CrossRef]
- Ma, Y.; Kan, G.; Zhang, X.; Wang, Y.; Zhang, W.; Du, H.; Yu, D. Quantitative Trait Loci (QTL) Mapping for Glycinin and β-Conglycinin Contents in Soybean (Glycine max L. Merr.). J. Agric. Food Chem. 2016, 64, 3473–3483. [Google Scholar] [CrossRef]
- Yamada, T.; Mori, Y.; Yasue, K.; Maruyama, N.; Kitamura, K.; Abe, J. Knockdown of the 7S globulin subunits shifts distribution of nitrogen sources to the residual protein fraction in transgenic soybean seeds. Plant Cell Rep. 2014, 33, 1963–1976. [Google Scholar] [CrossRef] [PubMed]
- Breene, W.M.; Lin, S.; Hardman, L.; Orf, J. Protein and oil content of soybeans from different geographic locations. J. Am. Oil Chem. Soc. 1988, 65, 1927–1931. [Google Scholar] [CrossRef]
- Clemente, T.E.; Cahoon, E.B. Soybean Oil: Genetic Approaches for Modification of Functionality and Total Content. Plant Physiol. 2009, 151, 1030–1040. [Google Scholar] [CrossRef]
- Canadian Grain Commission, Quality of Canadian Oilseed-Type Soybeans, Winnipeg, Manitoba. 2020. Available online: https://www.grainscanada.gc.ca/en/grain-research/export-quality/oilseeds/soybean-oil/2020/pdf/Quality-Canadian-Soybean-oilseed-type-2020.pdf (accessed on 20 May 2021).
- Canadian Grain Commission, Quality of Canadian Oilseed-Type Soybeans, Winnipeg, Manitoba. 2019. Available online: https://www.grainscanada.gc.ca/en/grain-research/export-quality/oilseeds/soybean-oil/2019/pdf/report19.pdf (accessed on 5 January 2021).
- Canadian Grain Commission, Quality of Canadian OILSEED-Type Soybeans, Winnipeg, Manitoba. 2021. Available online: https://publications.gc.ca/collections/collection_2022/ccg-cgc/A92-40-2021-eng.pdf (accessed on 22 June 2022).
- Ort, N.; Morrison, M.; Cober, E.; McAndrew, D.; Lawley, Y. A comparison of soybean maturity groups for phenology, seed yield, and seed quality components between eastern Ontario and southern Manitoba. Can. J. Plant Sci. 2022, 1–11. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Yao, T.; Gai, X.T.; Pu, Z.J.; Gao, Y.; Xuan, Y.H. From Functional Characterization to the Application of SWEET Sugar Transporters in Plant Resistance Breeding. J. Agric. Food Chem. 2022, 70, 5273–5283. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.-Q.; Sosso, D.; Ducat, D.C.; Hou, B.-H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef]
- Tao, Y.; Cheung, L.S.; Li, S.; Eom, J.-S.; Chen, L.-Q.; Xu, Y.Y.; Perry, K.K.; Frommer, W.B.; Feng, L. Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 2015, 527, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Huh, J.-H.; Yu, Y.-C.; Ho, L.-H.; Chen, L.-Q.; Tholl, D.; Frommer, W.B.; Guo, W.-J. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 2015, 83, 1046–1058. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, S. Rice MtN3/Saliva/SWEET Family Genes and Their Homologs in Cellular Organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520–536. [Google Scholar] [CrossRef] [PubMed]
- Gautam, T.; Saripalli, G.; Gahlaut, V.; Kumar, A.; Sharma, P.K.; Balyan, H.S.; Gupta, P.K. Further studies on sugar transporter (SWEET) genes in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2019, 46, 2327–2353. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, X.; Xuan, Y.; Jiang, J.; Wei, Y.; Piao, Z. Genome Wide Identification and Expression Profiling of SWEET Genes Family Reveals Its Role during Plasmodiophora brassicae-Induced Formation of Clubroot in Brassica rapa. Front. Plant Sci. 2018, 9, 207. Available online: https://www.frontiersin.org/article/10.3389/fpls.2018.00207 (accessed on 21 May 2022). [CrossRef] [PubMed]
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.-C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.-Q.; et al. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]
- Duan, Z.; Zhang, M.; Zhang, Z.; Liang, S.; Fan, L.; Yang, X.; Yuan, Y.; Pan, Y.; Zhou, G.; Liu, S.; et al. Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol. J. 2022, 20, 1807–1818. [Google Scholar] [CrossRef]
- Zhang, H.; Goettel, W.; Song, Q.; Jiang, H.; Hu, Z.; Wang, M.L.; An, Y.-Q.C. Selection of GmSWEET39 for oil and protein improvement in soybean. PLoS Genet. 2020, 16, e1009114. [Google Scholar] [CrossRef]
- Rotundo, J.L.; Westgate, M.E. Meta-analysis of environmental effects on soybean seed composition. Field Crop. Res. 2009, 110, 147–156. [Google Scholar] [CrossRef]
- Cober, E.R.; Voldeng, H.D. Developing High-Protein, High-Yield Soybean Populations and Lines. Crop Sci. 2000, 40, 39–42. [Google Scholar] [CrossRef]
- Borisjuk, L.; Nguyen, T.H.; Neuberger, T.; Rutten, T.; Tschiersch, H.; Claus, B.; Feussner, I.; Webb, A.G.; Jakob, P.; Weber, H.; et al. Gradients of lipid storage, photosynthesis and plastid differentiation in developing soybean seeds. New Phytol. 2005, 167, 761–776. [Google Scholar] [CrossRef]
- Rawsthorne, S. Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 2001, 41, 182–196. [Google Scholar] [CrossRef]
- Eom, J.-S.; Chen, L.-Q.; Sosso, D.; Julius, B.T.; Lin, I.; Qu, X.-Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Yang, L.; Fang, Z.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y. Plant SWEET Family of Sugar Transporters: Structure, Evolution and Biological Functions. Biomolecules 2022, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.; Nelson, R.T.; Cannon, S.B.; Shoemaker, R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010, 38, D843–D846. [Google Scholar] [CrossRef]
- Lin, I.W.; Sosso, D.; Chen, L.-Q.; Gase, K.; Kim, S.-G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.-H.; Qu, X.-Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tao, Y.; Cheung, L.S.; Fan, C.; Chen, L.-Q.; Xu, S.; Perry, K.; Frommer, W.B.; Feng, L. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature 2014, 515, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.-J.; Nagy, R.; Chen, H.-Y.; Pfrunder, S.; Yu, Y.-C.; Santelia, D.; Frommer, W.B.; Martinoia, E. SWEET17, a Facilitative Transporter, Mediates Fructose Transport across the Tonoplast of Arabidopsis Roots and Leaves. Plant Physiol. 2014, 164, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Kiers, E.T.; Duhamel, M.; Beesetty, Y.; Mensah, J.A.; Franken, O.; Verbruggen, E.; Fellbaum, C.R.; Kowalchuk, G.A.; Hart, M.M.; Bago, A.; et al. Reciprocal Rewards Stabilize Cooperation in the Mycorrhizal Symbiosis. Science 2011, 333, 880–882. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete Transcriptome of the Soybean Root Hair Cell, a Single-Cell Model, and Its Alteration in Response to Bradyrhizobium japonicum Infection. Plant Physiol. 2010, 152, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.; Keim, P.; Vodkin, L.; Retzel, E.; Clifton, S.W.; Waterston, R.; Smoller, D.; Coryell, V.; Khanna, A.; Erpelding, J.; et al. A compilation of soybean ESTs: Generation and analysis. Genome 2002, 45, 329–338. [Google Scholar] [CrossRef]
- Shoemaker, R.C.; Polzin, K.; Labate, J.; Specht, J.; Brummer, E.C.; Olson, T.; Young, N.; Concibido, V.; Wilcox, J.; Tamulonis, J.; et al. Genome Duplication in Soybean (Glycine subgenus soja). Genetics 1996, 144, 329–338. [Google Scholar] [CrossRef]
- Tian, A.; Wang, J.; Cui, P.; Han, Y.-J.; Xu, H.; Cong, L.-J.; Huang, X.-G.; Wang, X.-L.; Jiao, Y.-Z.; Wang, B.-J.; et al. Characterization of soybean genomic features by analysis of its expressed sequence tags. Theor. Appl. Genet. 2004, 108, 903–913. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, R.; Spinner, L.; Klemens, P.A.W.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Géry, C.; et al. Disruption of the Sugar Transporters AtSWEET11 and AtSWEET12 Affects Vascular Development and Freezing Tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A Cascade of Sequentially Expressed Sucrose Transporters in the Seed Coat and Endosperm Provides Nutrition for the Arabidopsis Embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef]
- Andriotis, V.; Pike, M.J.; Schwarz, S.L.; Rawsthorne, S.; Wang, T.L.; Smith, A.M. Altered Starch Turnover in the Maternal Plant Has Major Effects on Arabidopsis Fruit Growth and Seed Composition. Plant Physiol. 2012, 160, 1175–1186. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, L.; Zhang, D.; Miao, Q.; Yang, J.; Xuan, Y.; Hu, Y. Essential Role of Sugar Transporter OsSWEET11 during the Early Stage of Rice Grain Filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef]
- Yang, J.; Luo, D.; Yang, B.; Frommer, W.B.; Eom, J. SWEET 11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.-L.; Ma, J.F.; Shou, H. The Soybean Sugar Transporter GmSWEET15 Mediates Sucrose Export from Endosperm to Early Embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Valifard, M.; Le Hir, R.; Müller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.H.; Bai, L.; Qin, Q.P.; Li, N.Y. Isolation and Comparison of Eight SWEET17 Genes from Six Loquat Cultivars. Russ. J. Plant Physiol. 2020, 67, 1063–1075. [Google Scholar] [CrossRef]
- Sonah, H.; O’Donoughue, L.; Cober, E.; Rajcan, I.; Belzile, F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol. J. 2015, 13, 211–221. [Google Scholar] [CrossRef]
- Liu, Z.; Fan, H.; Ma, Z. Comparison of SWEET gene family between maize and foxtail millet through genomic, transcriptomic, and proteomic analyses. Plant Genome 2022, 1–25. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.-B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; Mccarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Cober, E.R.; Daba, T.D.; Warkentin, A.K.; Tomasiewicz, D.J.; Mooleki, P.S.; Karppinen, E.M.; Frey, J.; Mohr, R.M.; Glenn, A.J.; Shaw, L.; et al. Soybean Seed Protein Content Is Lower but Protein Quality Is Higher in Western Canada Compared to Eastern Canada. Can. J. Plant Sci. 2022; accepted with revisions. [Google Scholar]
- Kuzyakov, Y.; Jones, D. Glucose uptake by maize roots and its transformation in the rhizosphere. Soil Biol. Biochem. 2006, 38, 851–860. [Google Scholar] [CrossRef]
- Tylka, G.L.; Marett, C.C. Known Distribution of the Soybean Cyst Nematode, Heterodera glycines, in the United States and Canada in 2020. Plant Health Prog. 2021, 22, 72–74. [Google Scholar] [CrossRef]
- Tylka, G.L.; Marett, C.C. Known Distribution of the Soybean Cyst Nematode, Heterodera glycines, in the United States and Canada, 1954 to 2017. Plant Health Prog. 2017, 18, 167–168. [Google Scholar] [CrossRef]
- Zhang, L.X.; Kyei-Boahen, S.; Zhang, J.; Zhang, M.H.; Freeland, T.B.; Watson, C.E.; Liu, X. Modifications of Optimum Adaptation Zones for Soybean Maturity Groups in the USA. Crop Manag. 2007, 6, 1–11. [Google Scholar] [CrossRef]
- Scott, W.; Aldrich, S.R. Modern Soybean Production; S & A Publications: Champaign, IL, USA, 1970. [Google Scholar]
- Voldeng, H.D.; Guillemette, R.J.D.; Leonard, D.A.; Cober, E.R. AC Harmony soybean. Can. J. Plant Sci. 1996, 76, 477–478. [Google Scholar] [CrossRef]
- Cober, E.R.; Bing, H.D.V.D.; Soper, R.J.D.G.J.; Sloan, A.; Hedges, B.R. 90A01 soybean. Can. J. Plant Sci. 2006, 86, 481–482. [Google Scholar] [CrossRef]
- Voldeng, H.D.; Guillemette, R.J.D.; Leonard, D.A.; Cober, E.R. AC Proteus soybean. Can. J. Plant Sci. 1996, 76, 153–154. [Google Scholar] [CrossRef]
- Pedersen, P.; Licht, M. Soybean Growth and Development, PM 1945; Iowa State University Extension: Ames, IA, USA, 2014. [Google Scholar]
- Sayols, S.; Scherzinger, D.; Klein, H. dupRadar: A Bioconductor package for the assessment of PCR artifacts in RNA-Seq data. BMC Bioinform. 2016, 17, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Daley, T.; Deng, C.; Li, T.; Smith, A. The Preseq Manual; The Smith Lab: Los Angeles, CA, USA, 2022; pp. 1–24. Available online: http://smithlabresearch.org/manuals/preseqmanual.pdf (accessed on 25 April 2022).
- Wang, L.; Nie, J.; Sicotte, H.; Li, Y.; Eckel-Passow, J.E.; Dasari, S.; Vedell, P.T.; Barman, P.; Wang, L.; Weinshiboum, R.; et al. Measure transcript integrity using RNA-seq data. BMC Bioinform. 2016, 17, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Bourgey, M.; Dali, R.; Eveleigh, R.; Chen, K.C.; Letourneau, L.; Fillon, J.; Michaud, M.; Caron, M.; Sandoval, J.; Lefebvre, F.; et al. GenPipes: An open-source framework for distributed and scalable genomic analyses. GigaScience 2019, 8, giz037. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 July 2021).
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–571. [Google Scholar] [CrossRef] [PubMed]

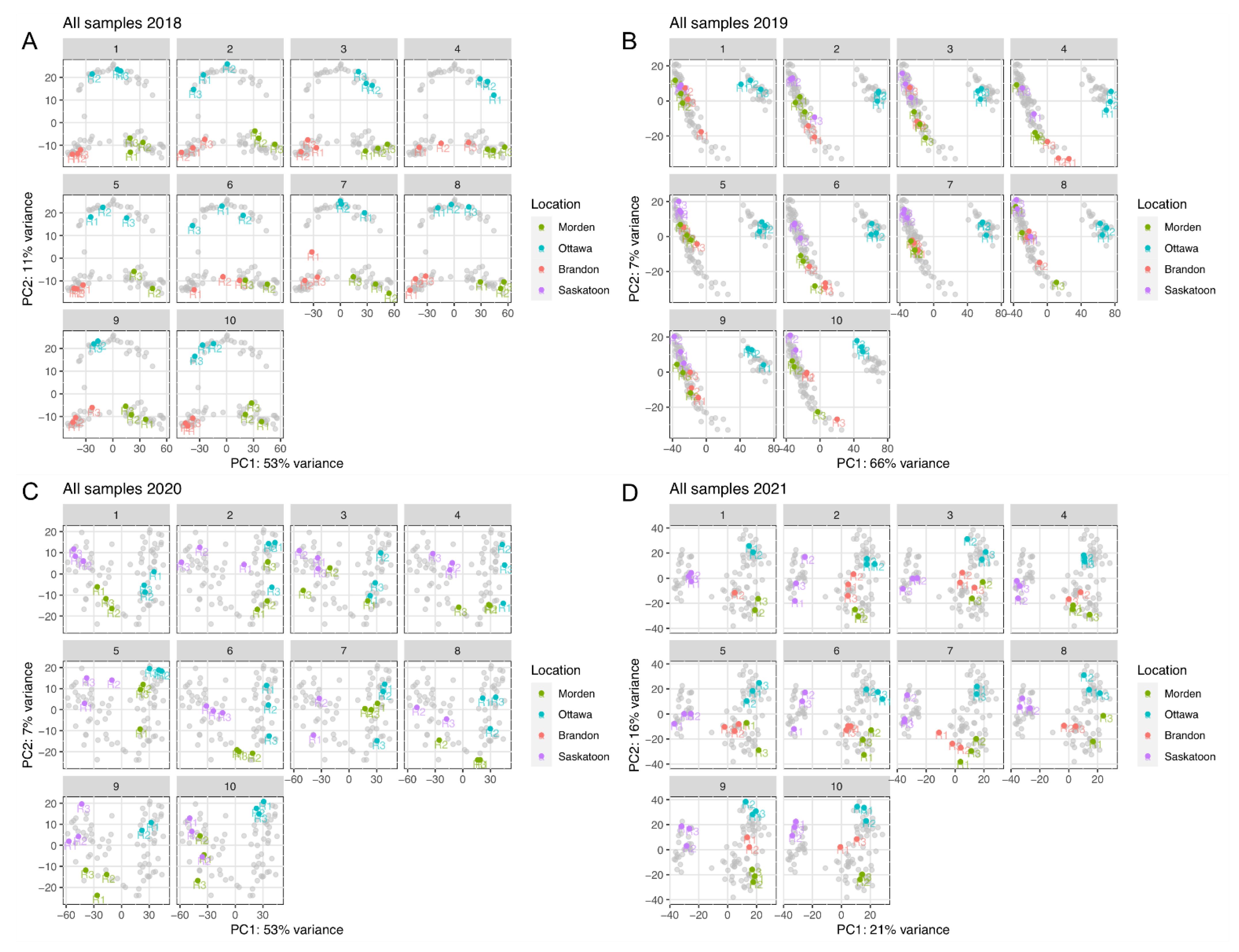
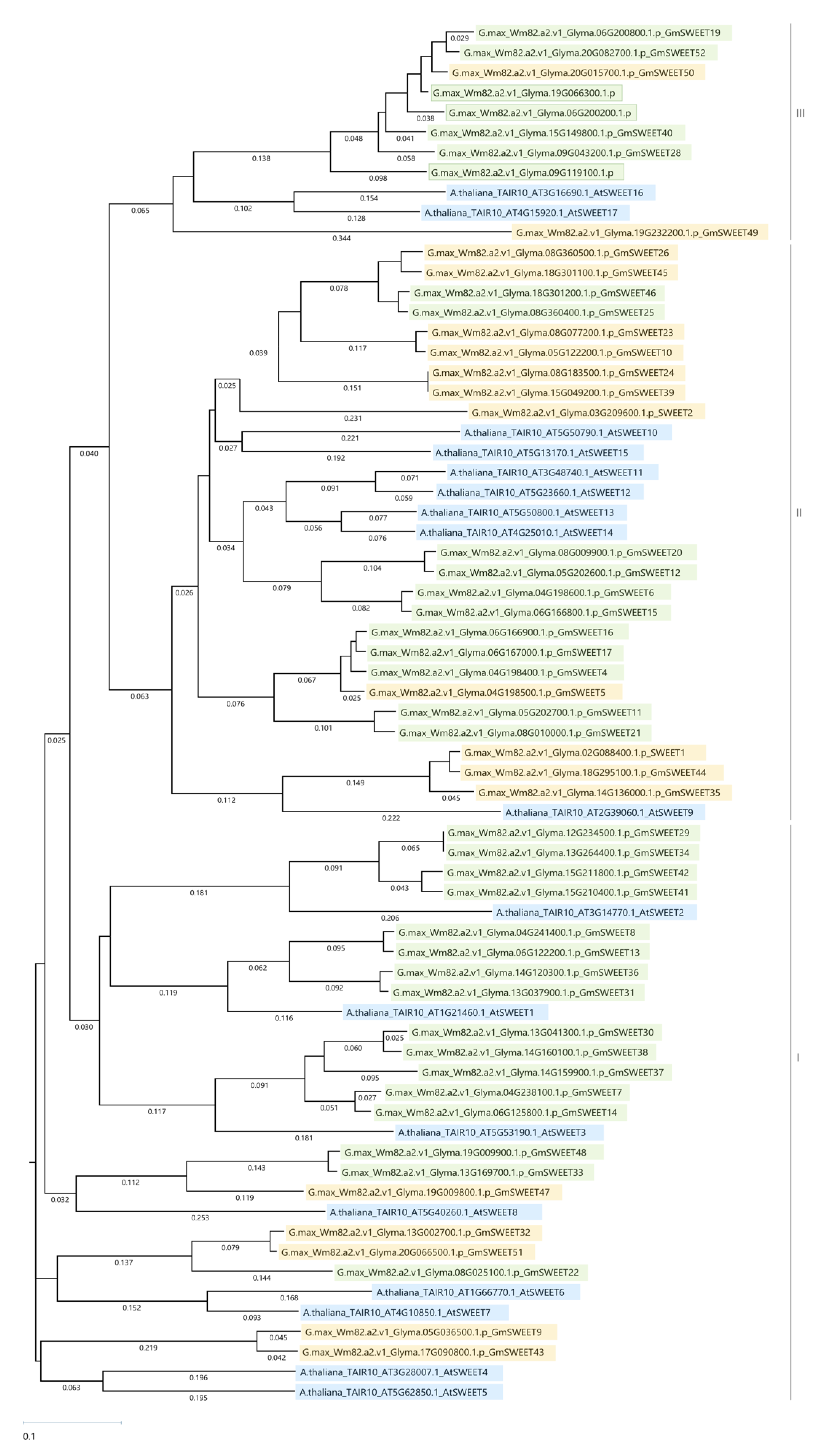
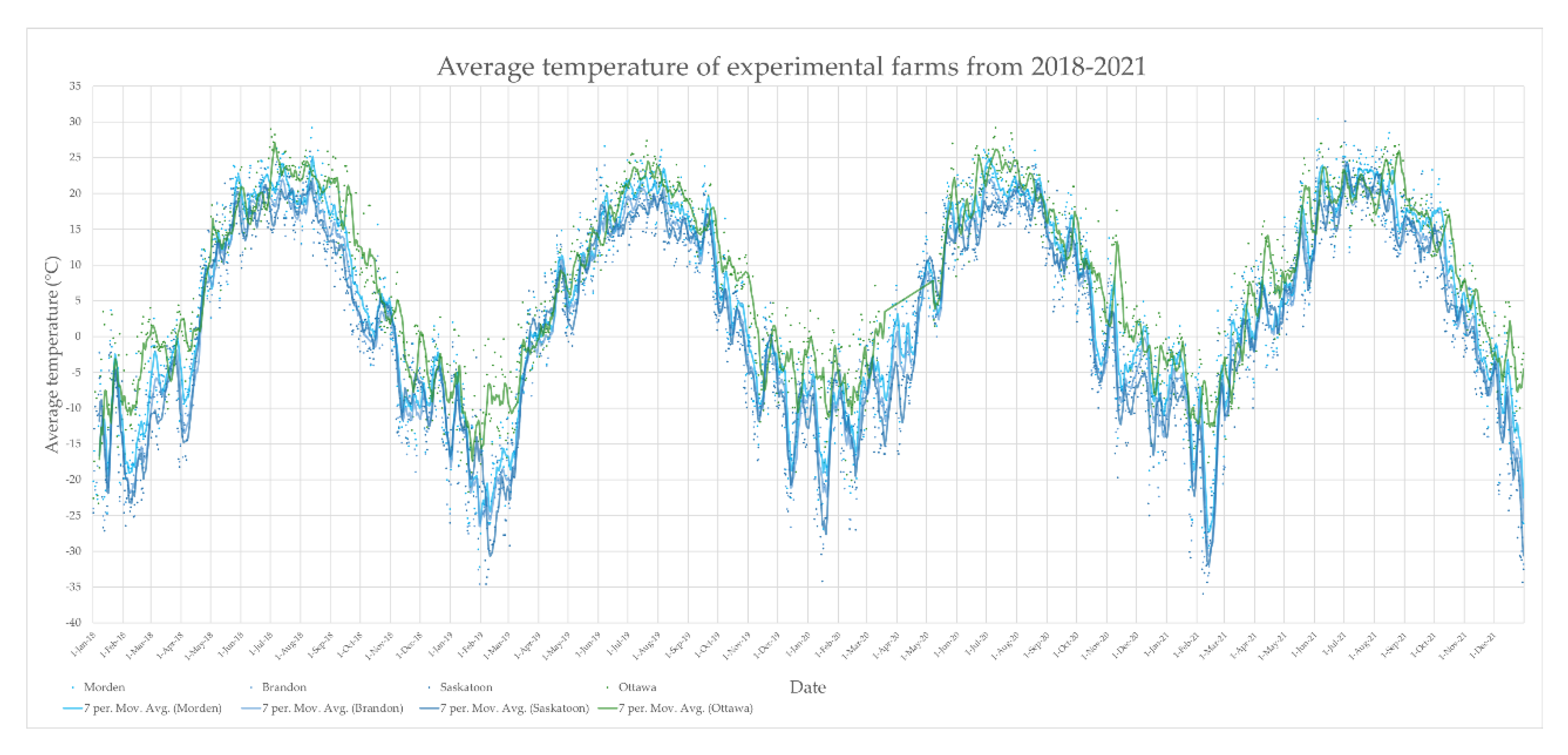
| Ottawa | Morden | Brandon | Saskatoon | West | East + West | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Line(s) | Genotype | Pro | Oil | Yield | Pro | Oil | Yield | Pro | Oil | Yield | Pro | Oil | Yield | Pro | Oil | Yield | Pro | Oil | Yield |
| 1 | X5583-1-041-5-5 | 40.4 | 22.8 | 2912.8 | 27.5 | 17.1 | 2214.3 | 35.6 | 21.9 | 2572.1 | 36.5 | 20.9 | 2447.2 | 33.2 | 20.0 | 2411.2 | 37.5 | 22.1 | 2715.2 |
| 2 | AC Harmony | 38.9 | 23.4 | 2589.7 | 27.2 | 17.4 | 1677.6 | 34.8 | 21.8 | 1866.0 | 36.2 | 21.5 | 2229.8 | 32.7 | 20.2 | 1924.5 | 36.7 | 22.4 | 2256.1 |
| 3 | AAC Halli | 41.6 | 21.7 | 2745.7 | 28.8 | 16.4 | 2110.9 | 38.1 | 20.5 | 2395.8 | 37.2 | 20.8 | 2253.3 | 34.7 | 19.2 | 2253.3 | 38.9 | 21.2 | 2544.8 |
| 4 | 90A01 | 42.3 | 21.4 | 2523.5 | 29.8 | 16.3 | 1803.5 | 39.4 | 20.8 | 2001.2 | 38.8 | 20.6 | 2244.0 | 36.0 | 19.2 | 2016.2 | 40.1 | 21.1 | 2306.2 |
| 5 | Maple Amber | 42.8 | 21.8 | 2516.9 | 29.8 | 16.5 | 1859.1 | 39.2 | 20.7 | 2222.3 | 38.5 | 20.5 | 1893.6 | 35.8 | 19.2 | 1991.7 | 40.1 | 21.2 | 2267.5 |
| 6 | OT13-08 | 43.8 | 21.7 | 2934.5 | 30.8 | 16.6 | 2265.5 | 40.3 | 21.4 | 2525.1 | 39.5 | 21.2 | 2214.6 | 36.9 | 19.7 | 2335.1 | 41.3 | 21.6 | 2659.6 |
| 7 | OT14-03 | 43.7 | 20.2 | 2742.1 | 31.1 | 15.4 | 2110.7 | 41.3 | 18.8 | 2090.4 | 40.0 | 19.5 | 2074.6 | 37.4 | 17.9 | 2091.9 | 41.6 | 19.8 | 2427.2 |
| 8 | AAC Springfield | 47.3 | 18.3 | 2365.2 | 32.6 | 14.5 | 1801.7 | 42.5 | 18.4 | 2000.1 | 44.4 | 18.0 | 1941.1 | 39.8 | 17.0 | 1914.3 | 44.6 | 18.5 | 2173.7 |
| 9 | Jari | 46.4 | 19.2 | 3054.2 | 31.9 | 14.7 | 2267.8 | 41.3 | 18.7 | 2087.6 | 41.3 | 18.5 | 1894.8 | 38.1 | 17.3 | 2083.4 | 43.0 | 19.0 | 2509.3 |
| 10 | AC Proteus | 49.4 | 16.6 | 2271.7 | 34.9 | 12.7 | 1824.2 | 45.9 | 16.1 | 1658.2 | 45.4 | 16.5 | 1475.0 | 42.1 | 15.1 | 1652.5 | 46.9 | 16.5 | 1947.0 |
| 1–10 | 43.6 | 20.7 | 2665.6 | 40.6 | 21.0 | 2658.0 | 39.9 | 19.9 | 2141.9 | 39.8 | 19.8 | 2066.8 | 40.0 | 20.2 | 2266.7 | 41.1 | 20.3 | 2380.7 | |
| Name | Gene ID (Wm82.a2.0) | Up | Down | Total |
|---|---|---|---|---|
| GmSWEET4 | Glyma.04G198400 | 4 | 1 | 5 |
| GmSWEET6 | Glyma.04G198600 | 32 | 3 | 35 |
| GmSWEET7 | Glyma.04G238100 | 0 | 27 | 27 |
| GmSWEET8 | Glyma.04G241400 | 7 | 2 | 9 |
| GmSWEET11 | Glyma.05G202700 | 6 | 16 | 22 |
| GmSWEET12 | Glyma.05G202600 | 7 | 19 | 26 |
| GmSWEET13 | Glyma.06G122200 | 10 | 5 | 15 |
| GmSWEET14 | Glyma.06G125800 | 5 | 31 | 36 |
| GmSWEET15 | Glyma.06G166800 | 26 | 3 | 29 |
| GmSWEET16 | Glyma.06G166900 | 1 | 0 | 1 |
| GmSWEET17 | Glyma.06G167000 | 2 | 28 | 30 |
| GmSWEET19 | Glyma.06G200800 | 1 | 1 | 2 |
| GmSWEET20 | Glyma.08G009900 | 4 | 47 | 51 |
| GmSWEET21 | Glyma.08G010000 | 6 | 25 | 31 |
| GmSWEET22 | Glyma.08G025100 | 2 | 6 | 8 |
| GmSWEET25 | Glyma.08G360400 | 25 | 0 | 25 |
| GmSWEET28 | Glyma.09G043200 | 24 | 12 | 36 |
| GmSWEET29 | Glyma.12G234500 | 62 | 9 | 71 |
| GmSWEET30 | Glyma.13G041300 | 5 | 34 | 39 |
| GmSWEET31 | Glyma.13G037900 | 19 | 4 | 23 |
| GmSWEET33 | Glyma.13G169700 | 23 | 0 | 23 |
| GmSWEET34 | Glyma.13G264400 | 55 | 1 | 56 |
| GmSWEET36 | Glyma.14G120300 | 14 | 1 | 15 |
| GmSWEET37 | Glyma.14G159900 | 4 | 0 | 4 |
| GmSWEET38 | Glyma.14G160100 | 2 | 38 | 40 |
| GmSWEET40 | Glyma.15G149800 | 8 | 26 | 34 |
| GmSWEET41 | Glyma.15G210400 | 7 | 13 | 20 |
| GmSWEET42 | Glyma.15G211800 | 17 | 5 | 22 |
| GmSWEET46 | Glyma.18G301200 | 17 | 0 | 17 |
| GmSWEET48 | Glyma.19G009900 | 3 | 0 | 3 |
| GmSWEET52 | Glyma.20G082700 | 9 | 1 | 10 |
| AtSWEET17 | Glyma.09G119100 | 1 | 2 | 3 |
| AtSWEET17 | Glyma.19G066300 | 3 | 2 | 5 |
| AtSWEET17 | Glyma.06G200200 | 0 | 1 | 1 |
| Total | 774 | 411 | 363 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hooker, J.C.; Nissan, N.; Luckert, D.; Zapata, G.; Hou, A.; Mohr, R.M.; Glenn, A.J.; Barlow, B.; Daba, K.A.; Warkentin, T.D.; et al. GmSWEET29 and Paralog GmSWEET34 Are Differentially Expressed between Soybeans Grown in Eastern and Western Canada. Plants 2022, 11, 2337. https://doi.org/10.3390/plants11182337
Hooker JC, Nissan N, Luckert D, Zapata G, Hou A, Mohr RM, Glenn AJ, Barlow B, Daba KA, Warkentin TD, et al. GmSWEET29 and Paralog GmSWEET34 Are Differentially Expressed between Soybeans Grown in Eastern and Western Canada. Plants. 2022; 11(18):2337. https://doi.org/10.3390/plants11182337
Chicago/Turabian StyleHooker, Julia C., Nour Nissan, Doris Luckert, Gerardo Zapata, Anfu Hou, Ramona M. Mohr, Aaron J. Glenn, Brent Barlow, Ketema A. Daba, Thomas D. Warkentin, and et al. 2022. "GmSWEET29 and Paralog GmSWEET34 Are Differentially Expressed between Soybeans Grown in Eastern and Western Canada" Plants 11, no. 18: 2337. https://doi.org/10.3390/plants11182337
APA StyleHooker, J. C., Nissan, N., Luckert, D., Zapata, G., Hou, A., Mohr, R. M., Glenn, A. J., Barlow, B., Daba, K. A., Warkentin, T. D., Lefebvre, F., Golshani, A., Cober, E. R., & Samanfar, B. (2022). GmSWEET29 and Paralog GmSWEET34 Are Differentially Expressed between Soybeans Grown in Eastern and Western Canada. Plants, 11(18), 2337. https://doi.org/10.3390/plants11182337







