Synthesis and Activity of 2-Acyl-cyclohexane-1,3-dione Congeners Derived from Peperomia Natural Products against the Plant p-Hydroxyphenylpyruvate Dioxygenase Herbicidal Molecular Target Site
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis of 2-Acyl-cyclohexane-1,3-diones and Analogs
2.2. Inhibition of HPPD
2.3. Herbicidal Activity of Tested Compounds
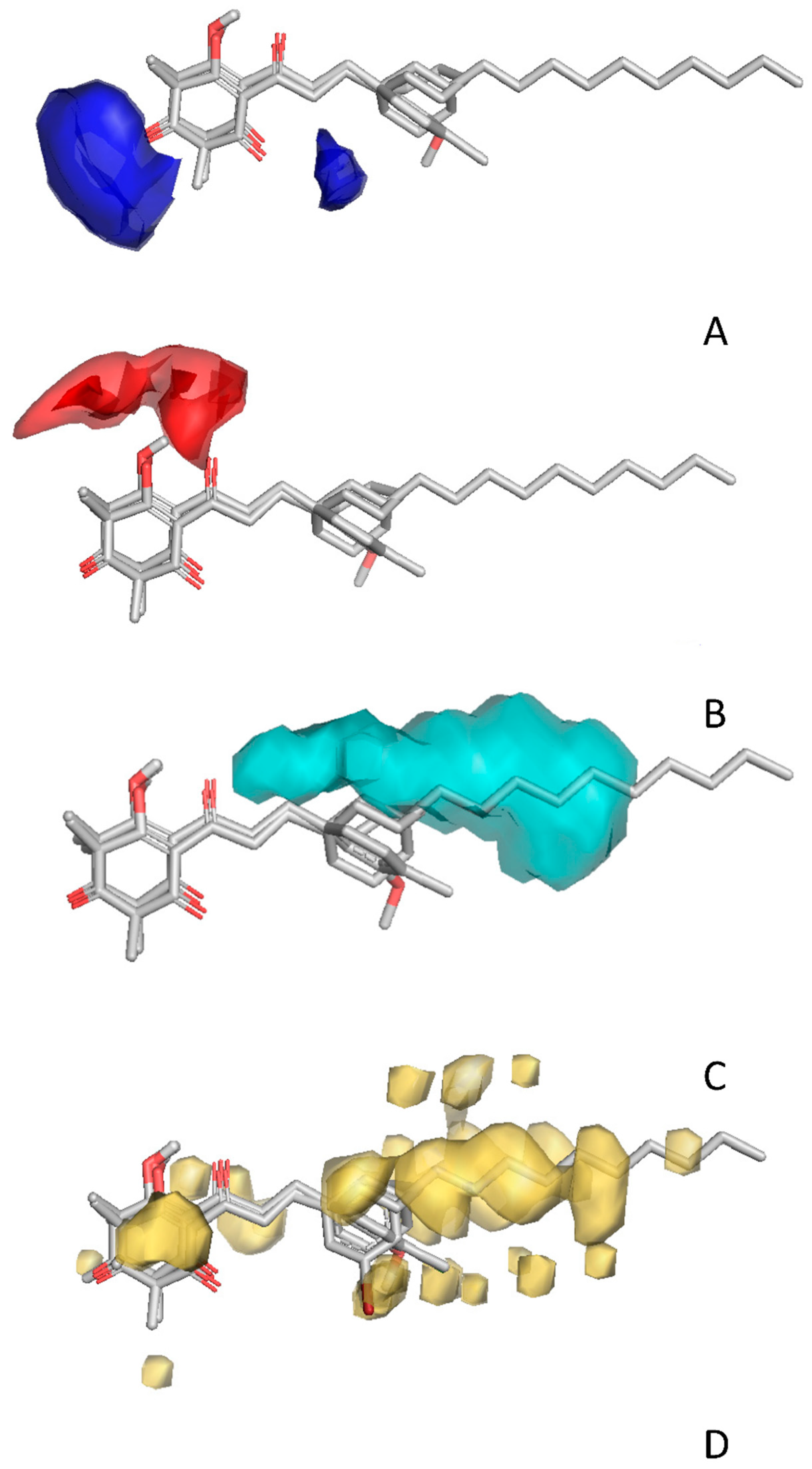
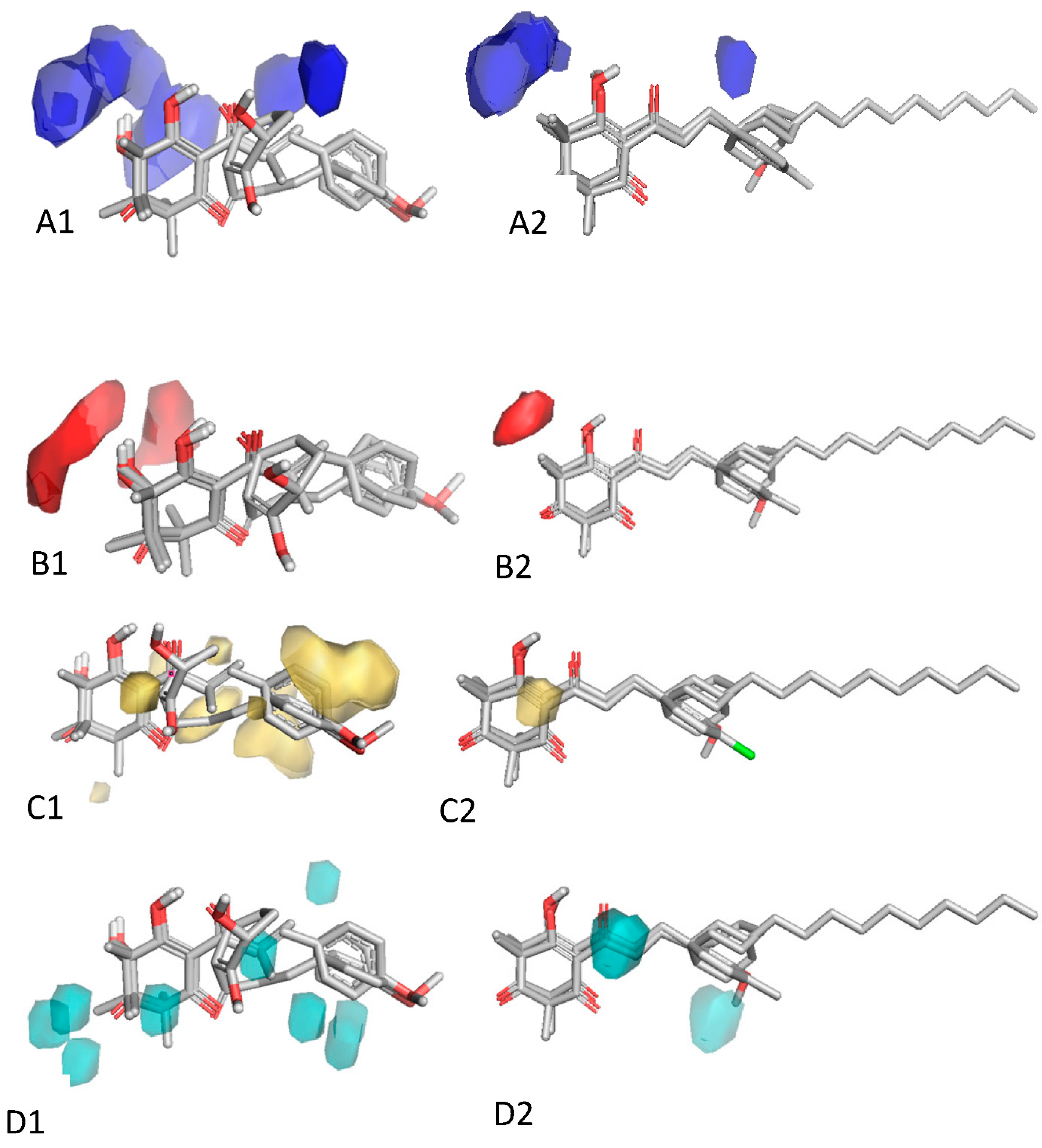
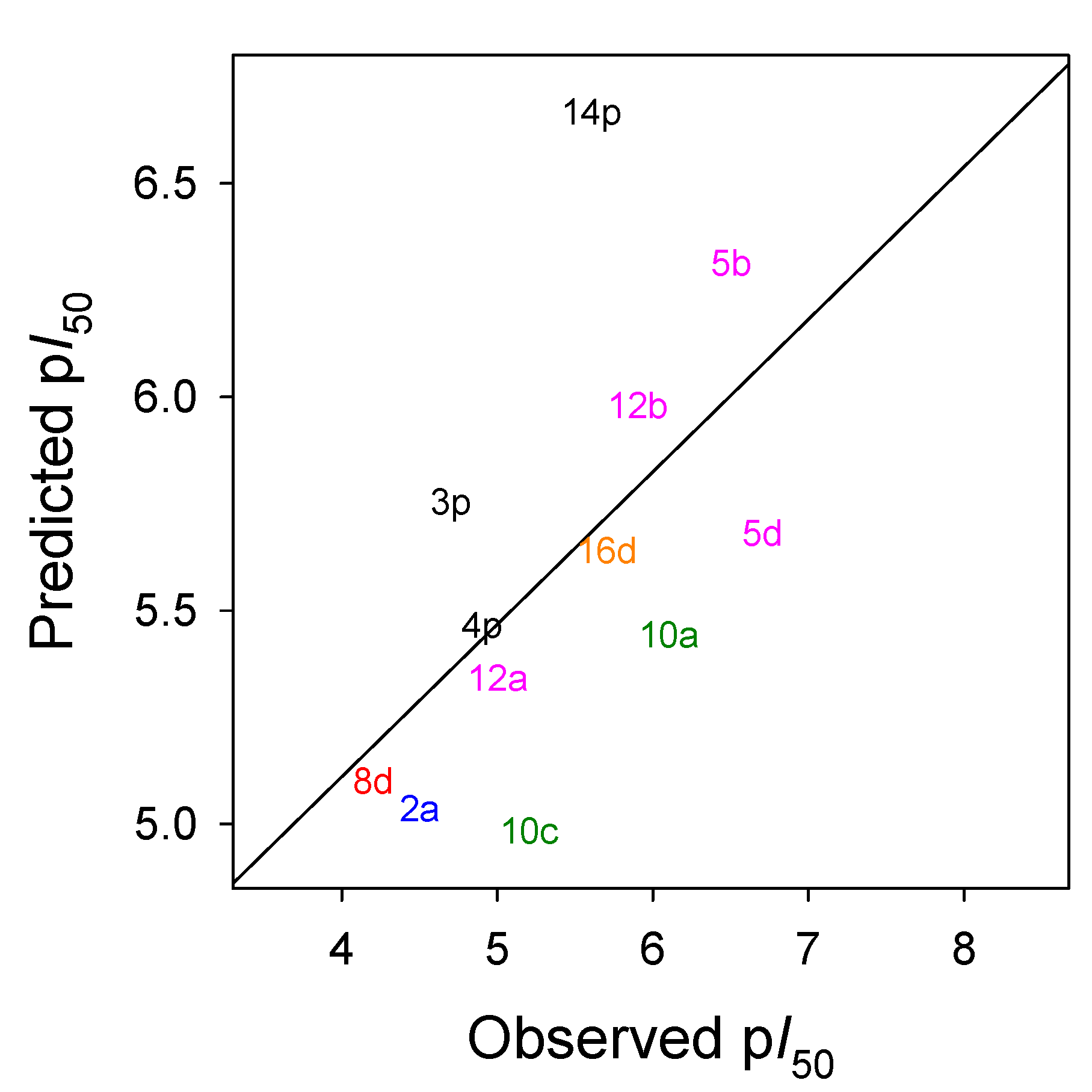
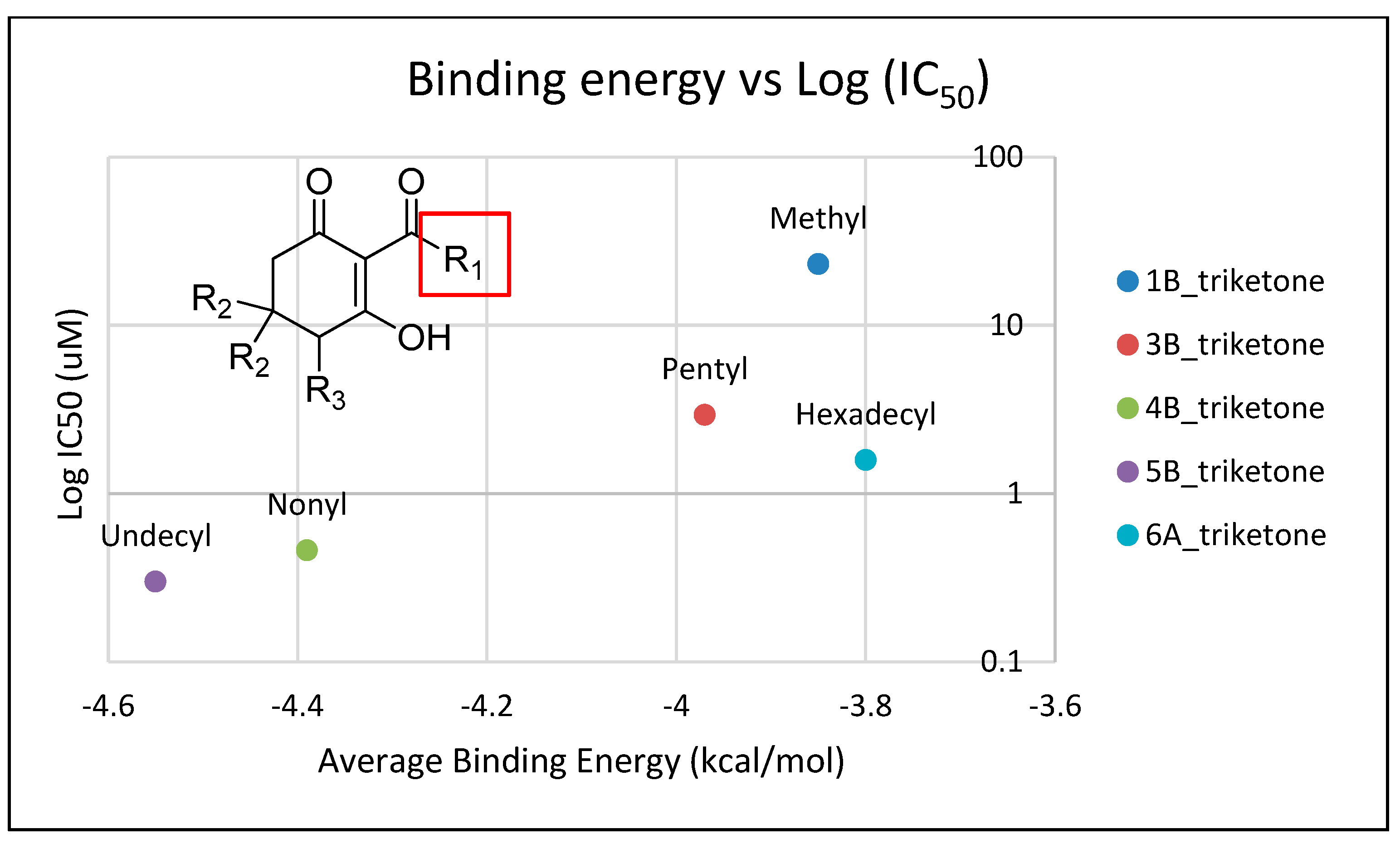
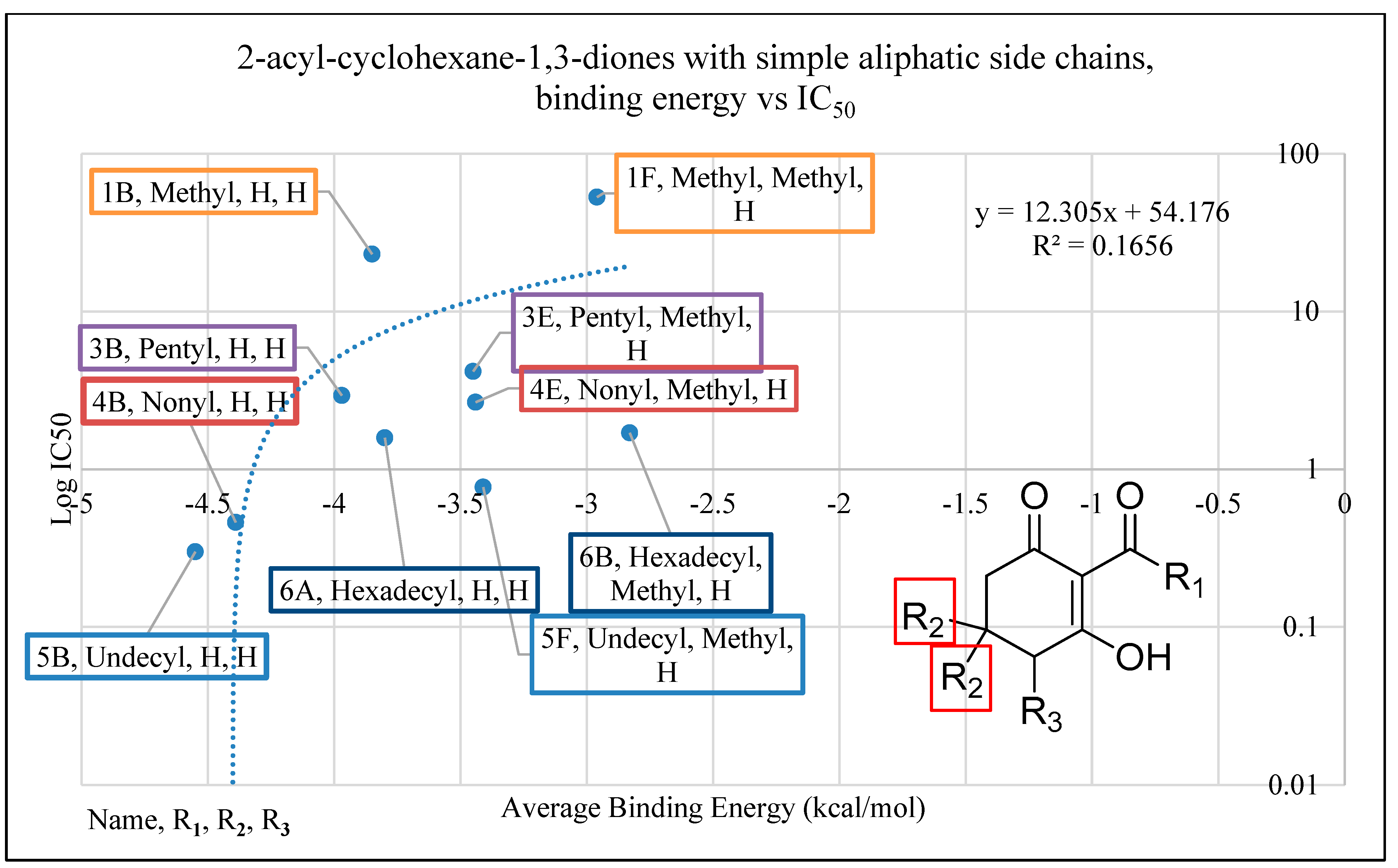
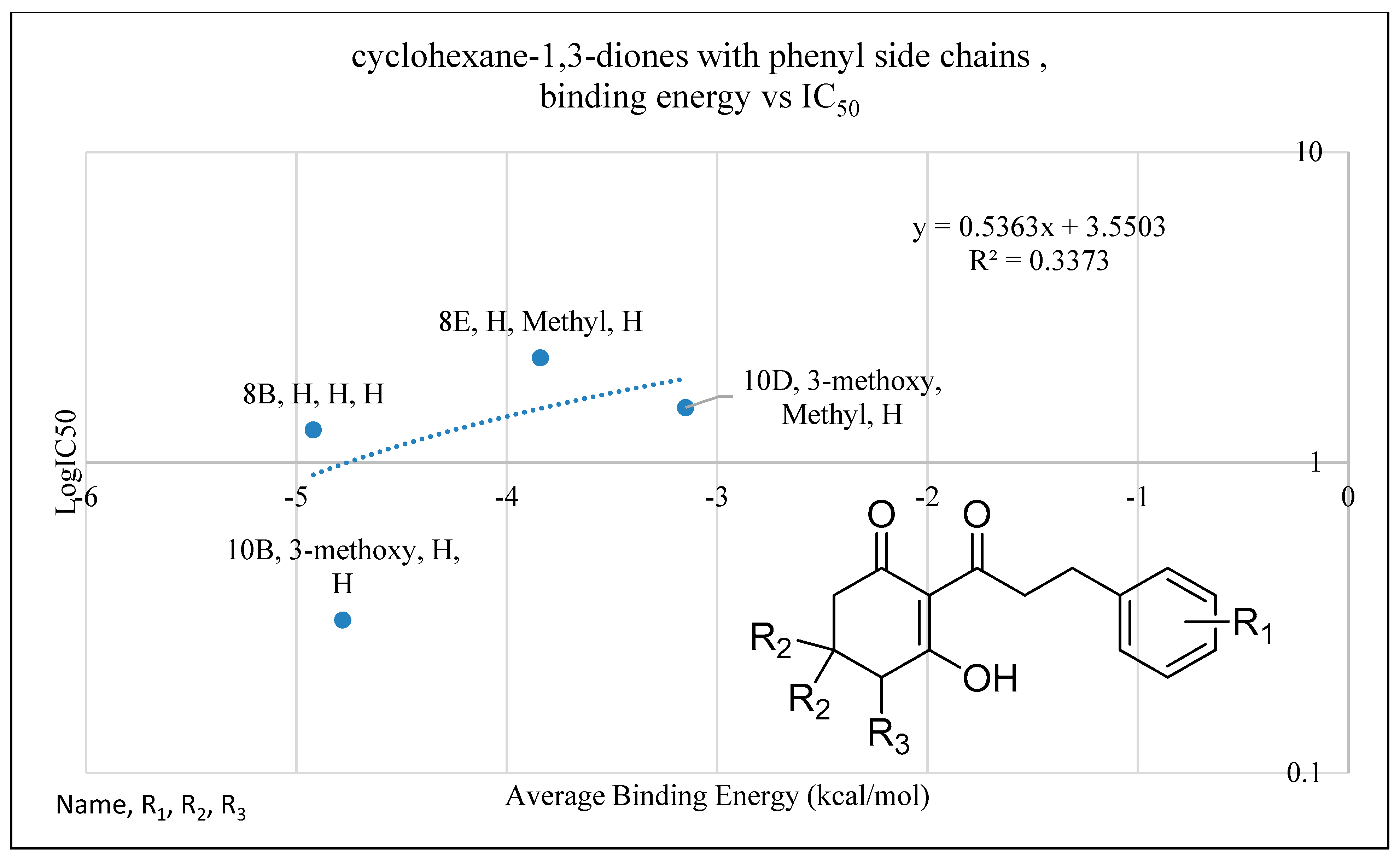
2.4. In Silico Analysis of Compound Interactions with HPPD
3. Conclusions
4. Materials and Methods
4.1. General Experimental Conditions
4.2. Synthesis of Saturated 2-Acyl-cyclohexane-1,3-diones
4.3. Synthesis of Unsaturated 2-Acyl-cyclohexane-1,3-diones (Supplemental Schemes S2 and S3)
4.4. Hydrogenation of Unsaturated 2-Acyl-cyclohexane-1,3-diones (Supplemental Scheme S4)
4.5. Synthesis of Hydroxylated 2-Acyl-Cyclohexane-1,3-diones (Supplemental Scheme S5)
4.5.1. Protection Step
4.5.2. Oxidation Step
4.5.3. Hydrogenation Step
4.5.4. Imine Hydrolysis Step
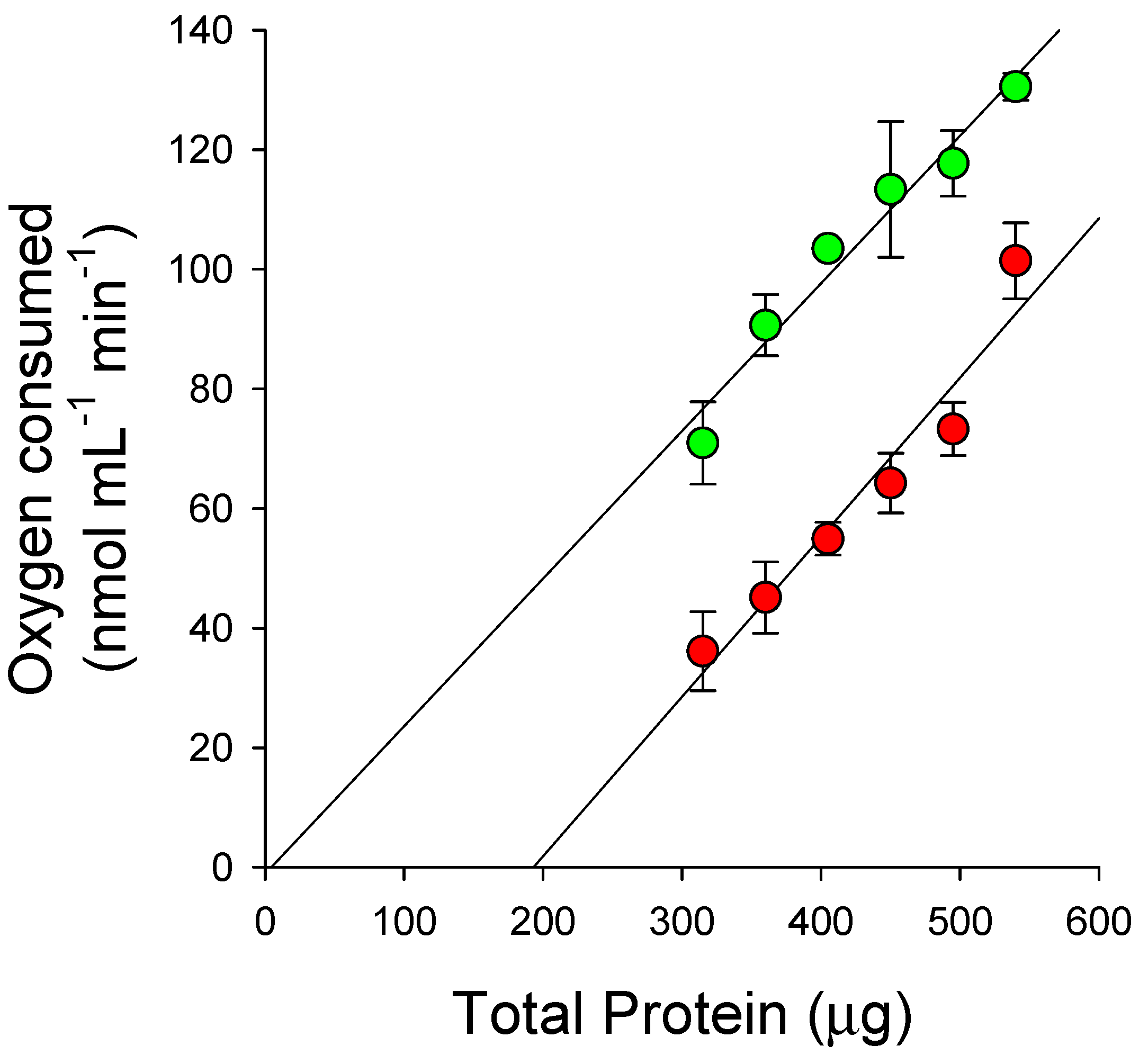
4.6. Recombinant Expression of HPPD and Enzyme Assays
4.7. Dose–Response Curve Analysis
4.8. Characterization of the Mechanism of HPPD Binding
4.9. Molecular Modeling
4.10. Molecular Docking
4.11. 3D-QSAR
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schulz, A.; Ort, O.; Beyer, P.; Kleinig, H. SC-0051, a 2-benzoyl-cyclohexane-1,3-dione bleaching herbicide, is a potent inhibitor of the enzyme p-hydroxyphenylpyruvate dioxygenase. FEBS Lett. 1993, 318, 162–166. [Google Scholar] [CrossRef]
- Lee, D.L.; Prisbylla, M.P.; Cromartie, T.H.; Dagarin, D.P.; Howard, S.W.; Provan, W.M.; Ellis, M.K.; Fraser, T.; Mutter, L.C. The discovery and structural requirements of inhibitors of p-hydroxyphenylpyruvate dioxygenase. Weed Sci. 1997, 45, 601–609. [Google Scholar] [CrossRef]
- Pallett, K.E.; Little, J.P.; Sheekey, M.; Veerasekaran, P. The mode of action of isoxaflutole I. Physiological effects, metabolism, and selectivity. Pestic. Biochem. Physiol. 1998, 62, 113–124. [Google Scholar] [CrossRef]
- Viviani, E.; Little, J.P.; Pallett, K.E. The mode of action of isoxaflutole II. Characterization of the inhibition of carrot 4-hydroxyphenylpyruvate dioxygenase by the diketonitrile derivative of isoxaflutole. Pestic. Biochem. Physiol. 1998, 62, 125–134. [Google Scholar] [CrossRef]
- Schultz, T.W.; Bearden, A.P. Structure-toxicity relationships for selected naphthoquinones to Tetrahymena pyriformis. Bull. Environment. Contam. Toxicol. 1998, 61, 405–410. [Google Scholar]
- Dayan, F.E.; Barker, A.; Takano, H.; Bough, R.; Ortiz, M.; Duke, S.O. Herbicide mechanisms of action and resistance. In Comprehensive Biotechnology, 3rd ed.; Grodzinski, B., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 4, pp. 36–48. [Google Scholar]
- Williams, M.M.; Pataky, J.K. Genetic basis of sensitivity in sweet corn to tembotrione. Weed Sci. 2008, 56, 364–370. [Google Scholar] [CrossRef]
- Norris, S.R.; Barrette, T.R.; DellaPenna, D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 1995, 7, 2139–2149. [Google Scholar]
- Gray, R.A.; Tseng, C.K.; Rusay, R.J. 1-Hydroxy-2-(alkylketo)-4,4,6,6-tetramethyl Cyclohexen-3,5-dione Herbicides. U.S. Patent 4,202,840, 14 October 1980. [Google Scholar]
- Dayan, F.E.; Duke, S.O.; Sauldubois, A.; Singh, N.; McCurdy, C.R.; Cantrell, C.L. p-Hydroxyphenylpyruvate dioxygenase is a herbicidal target site for β-triketones from Leptospermum scoparium. Phytochemistry 2007, 68, 2004–2014. [Google Scholar] [CrossRef]
- Dayan, F.E.; Howell, J.L.; Marais, J.M.; Ferreira, D.; Koivunen, M.E. Manuka oil, a natural herbicide with preemergence activity. Weed Sci. 2011, 59, 464–469. [Google Scholar] [CrossRef]
- Owens, D.K.; Nanayakkara, N.P.D.; Dayan, F.E. In planta mechanism of action of leptospermone: Impact of its physico-chemical properties on uptake, translocation, and metabolism. J. Chem. Ecol. 2013, 39, 262–270. [Google Scholar] [CrossRef]
- Wanke, S.; Samain, M.S.; Vanderschaeve, L.; Mathieu, G.; Goetghebeur, P.; Neinhuis, C. Phylogeny of the genus Peperomia (Piperaceae) inferred from the trnK/matK region (cpDNA). Plant Biol. 2006, 8, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.A.; Manos, P.S.; Zimmer, E.A. Phylogenetic relationships of the perianthless Piperales: Reconstructing the evolution of floral development. Internat. J. Plant Sci. 2004, 165, 403–416. [Google Scholar] [CrossRef]
- Malquichagua Salazar, K.J.; Delgado Paredes, G.E.; Lluncor, L.R.; Young, M.C.M.; Kato, M.J. Chromenes of polyketide origin from Peperomia villipetiola. Phytochemistry 2005, 66, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Jacobs, H.; McLean, S.; Reynolds, W.F. A prenylated benzopyran derivative from Peperomia clusiifolia. Phytochemistry 1998, 49, 1389–1391. [Google Scholar] [CrossRef]
- Tanaka, T.; Asai, F.; Iinuma, M. Phenolic compounds from Peperomia obtusifolia. Phytochemistry 1998, 49, 229–232. [Google Scholar] [CrossRef]
- Mbah, J.A.; Tchuendem, M.H.K.; Tane, P.; Sterner, O. Two chromones from Peperomia vulcanica. Phytochemistry 2002, 60, 799–801. [Google Scholar] [CrossRef]
- Bayma, J.D.; Arruda, M.S.P.; Müller, A.H.; Arruda, A.C.; Canto, W.C. A dimeric ArC2 compound from Peperomia pellucida. Phytochemistry 2000, 55, 779–782. [Google Scholar] [CrossRef]
- Govindachari, T.R.; Krishna Kumari, G.N.; Partho, P.D. Two secolignans from Peperomia dindigulensis. Phytochemistry 1998, 49, 2129–2131. [Google Scholar] [CrossRef]
- Monache, F.D.; Compagnone, R.S. A secolignan from Peperomia glabella. Phytochemistry 1996, 43, 1097–1098. [Google Scholar] [CrossRef]
- Xu, S.; Li, N.; Ning, M.-M.; Zhou, C.-H.; Yang, Q.-R.; Wang, M.-W. Bioactive compounds from Peperomia pellucida. J. Nat. Prod. 2006, 69, 247–250. [Google Scholar] [CrossRef]
- Wu, J.-l.; Li, N.; Hasegawa, T.; Sakai, J.-i.; Kakuta, S.; Tang, W.; Oka, S.; Kiuchi, M.; Ogura, H.; Kataoka, T.; et al. Bioactive tetrahydrofuran lignans from Peperomia dindygulensis. J. Nat. Prod. 2005, 68, 1656–1660. [Google Scholar] [CrossRef] [PubMed]
- Mahiou, V.; Roblot, F.; Hocquemiller, R.; Cavé, A.; Rojas De Arias, A.; Inchausti, A.; Yaluff, G.; Fournet, A. New prenylated quinones from Peperomia galioides. J. Nat. Prod. 1996, 59, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Soares, M.G.; Felippe, A.P.V.d.; Guimarães, E.F.; Kato, M.J.; Ellena, J.; Doriguetto, A.C. 2-Hydroxy-4,6-dimethoxyacetophenone from leaves of Peperomia glabella. J. Brazil. Chem. Soc. 2006, 17, 1205–1210. [Google Scholar] [CrossRef]
- Li, N.; Wu, J.-l.; Hasegawa, T.; Sakai, J.-i.; Bai, L.-m.; Wang, L.-y.; Kakuta, S.; Furuya, Y.; Ogura, H.; Kataoka, T.; et al. Bioactive polyketides from Peperomia duclouxii. J. Nat. Prod. 2007, 70, 998–1001. [Google Scholar] [CrossRef]
- Lago, J.H.G.; Oliveira, A.d.; Guimarães, E.F.; Kato, M.J. 3-Ishwarone and 3-ishwarol, rare sesquiterpenes in essential oil from leaves of Peperomia oreophila Hensch. J. Brazil. Chem. Soc. 2007, 18, 638–642. [Google Scholar] [CrossRef]
- Gutierrez, Y.V.; Yamaguchi, L.F.; de Moraes, M.M.; Jeffrey, C.S.; Kato, M.J. Natural products from Peperomia: Occurrence, biogenesis and bioactivity. Phytochem. Rev. 2016, 15, 1009–1033. [Google Scholar] [CrossRef]
- Ferreira, E.A.; Reigada, J.B.; Correia, M.V.; Young, M.C.M.; Guimarães, E.F.; Franchi, G.C.; Nowill, A.E.; Lago, J.H.G.; Yamaguchi, L.F.; Kato, M.J. Antifungal and cytotoxic 2-acylcyclohexane-1,3-diones from Peperomia alata and P. trineura. J. Nat. Prod. 2014, 77, 1377–1382. [Google Scholar] [CrossRef]
- Nemoto, T.; Shibuya, M.; Kuwahara, Y.; Suzuki, T. New 2-acylcyclohexane-1,3-diones: Kairomone components against a parasitic wasp, Venturia canescens, from feces of the almond moth, Cadra cautella, and the Indian meal moth, Plodia interpunctella. Agric. Biol. Chem. 1987, 51, 1805–1810. [Google Scholar] [CrossRef]
- Mudd, A. Further novel 2-acylcyclohexane-l,3-diones from lepidopteran larvae. J. Chem. Soc. Perkin Trans. 1983, 1, 2161–2164. [Google Scholar] [CrossRef]
- Kuwahara, Y.; Nemoto, T.; Shibuya, M.; Matsuura, H.; Shiraiwa, Y. 2-Palmitoyl- and 2-oleoyl-cyclohexane-1,3-dione from feces of the Indian meal moth, Plodia interpunctella: Kairomone components against a parasitic wasp, Venturia canescen. Agric. Biol. Chem. 1983, 47, 1929–1931. [Google Scholar]
- Denny, C.; Zacharias, M.E.; Ruiz, A.L.T.G.; Amaral, M.d.C.E.d.; Bittrich, V.; Kohn, L.K.; Sousa, I.M.d.O.; Rodrigues, R.A.F.; Carvalho, J.E.d.; Foglio, M.A. Antiproliferative properties of polyketides isolated from Virola sebifera leaves. Phytotherapy Res. 2008, 22, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-W.; Yu, D.-H.; Lin, M.-G.; Zhao, M.; Zhu, W.-J.; Lu, Q.; Li, G.-X.; Wang, C.; Yang, Y.-F.; Qin, X.-M.; et al. Antiangiogenic polyketides from Peperomia dindygulensis Miq. Molecules 2012, 17, 4474–4483. [Google Scholar] [CrossRef]
- Beaudegnies, R.; Edmunds, A.J.F.; Fraser, T.E.M.; Hall, R.G.; Hawkes, T.R.; Mitchell, G.; Schaetzer, J.; Wendeborn, S.; Wibley, J. Herbicidal 4-hydroxyphenylpyruvate dioxygenase inhibitors—A review of the triketone chemistry story from a Syngenta perspective. Bioorg. Med. Chem. 2009, 17, 4134–4152. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, C.G.; Lee, D.L.; Michaely, W.J.; Chin, H.-L.; Nguyen, N.H.; Rusay, R.J.; Cromartie, T.H.; Gray, R.; Lake, B.H.; Fraser, T.E.M.; et al. Discovery of the triketone class of HPPD inhibiting herbicides and their relationship to naturally occurring β-triketones. In Allelopathy in Ecological Agriculture and Forestry; Narwal, S.S., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; pp. 101–111. [Google Scholar]
- Mayo, S.L.; Olafson, B.D.; Goddard, W.A. DREIDING: A generic force field for molecular simulations. J. Physic. Chem. 1990, 94, 8897–8909. [Google Scholar] [CrossRef]
- Ellis, M.K.; Whitfield, A.C.; Gowans, L.A.; Auton, T.R.; Provan, W.M.; Lock, E.A.; Lee, D.L.; Smith, L.L. Characterization of the interaction of 2-[2-nitro-4-(trifluoromethyl)benzoyl]-4,4,6,6-tetramethylcyclohexane-1,3,5-trione with rat hepatic 4-hydroxyphenylpyruvate dioxygenase. Chem. Res. Toxicol. 1996, 9, 24–27. [Google Scholar] [CrossRef]
- Purpero, V.M.; Moran, G.R. Catalytic, noncatalytic, and inhibitory phenomena: Kinetic analysis of (4-hydroxyphenyl)pyruvate dioxygenase from Arabidopsis thaliana. Biochemistry 2006, 2006, 6044–6055. [Google Scholar] [CrossRef]
- Hawkes, T.R. Hydroxyphenylpyruvate dioxygenase (HPPD)—The herbicide target. In Modern Crop Protection Compounds; Krämer, W., Schirmer, U., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2007; Volume 1, pp. 211–220. [Google Scholar]
- Meazza, G.; Scheffler, B.E.; Tellez, M.R.; Rimando, A.M.; Nanayakkara, N.P.D.; Khan, I.A.; Abourashed, E.A.; Romagni, J.G.; Duke, S.O.; Dayan, F.E. The inhibitory activity of natural products on plant p-hydroxyphenylpyruvate dioxygenase. Phytochemistry 2002, 59, 281–288. [Google Scholar] [CrossRef]
- Lee, D.L.; Knudsen, C.G.; Michaely, W.J.; Chin, H.-L.; Nguyen, N.H.; Carter, C.G.; Cromartie, T.H.; Lake, B.H.; Shribbs, J.M.; Fraser, T. The structure-activity relationships of the triketone class of HPPD herbicides. Pestic. Sci. 1998, 54, 377–384. [Google Scholar] [CrossRef]
- Dayan, F.E.; Singh, N.; McCurdy, C.; Godfrey, C.A.; Larsen, L.; Weavers, R.T.; Van Klink, J.W.; Perry, N.B. β-triketone inhibitors of plant p-hydroxyphenylpyruvate dioxygenase: Modeling and comparative molecular field analysis of their interactions. J. Agric. Food Chem. 2009, 57, 5194–5200. [Google Scholar] [CrossRef]
- Wang, D. Design, Synthesis and herbicidal activity of novel quinazoline-2, 4-diones as 4-hydroxyphenylpyruvate sioxygenase inhibitors. Pest Manag. Sci. 2015, 71, 1122–1132. [Google Scholar] [CrossRef]
- Hagar, M.; Ahmed, H.A.; Aouad, M.R. Mesomorphic and DFT diversity of schiff base derivatives bearing protruded methoxy groups. Liq. Cryst. 2020, 47, 2222–2233. [Google Scholar] [CrossRef]
- Laane, J. Vibrational potential energy surfaces in electronic excited states. In Frontiers of Molecular Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2009; pp. 63–132. [Google Scholar]
- Goncalves, S.; Nicolas, M.; Wagner, A.; Baati, R. Exploring the one-pot C-acylation of cyclic 1,3-diones with unactivated carboxylic acid. Tetrahedron Lett. 2010, 51, 2348–2350. [Google Scholar] [CrossRef]
- Rubinov, D.B.; Rubinova, I.L.; Lakhvich, F.A. Synthesis of exo- and endocyclic enamino derivatives of 2-(3-arylprop-2-enoyl)cyclohexane-1,3-diones. Russian J. Org. Chem. 2011, 47, 319–330. [Google Scholar] [CrossRef]
- Hodgson, D.M.; Galano, J.-M.; Christlieb, M. Synthesis of (−)-xialenon A by enantioselective α-deprotonation-rearrangement of a meso-epoxide. Tetrahedron 2003, 59, 9719–9728. [Google Scholar] [CrossRef]
- Oliver, J.E.; Lusby, W.R. Synthesis of 2-acyl-3,6-dihydroxy-2-cyclohexen-1-ones. Tetrahedron 1988, 44, 1591–1596. [Google Scholar] [CrossRef]
- Dayan, F.E.; Owens, D.K.; Corniani, N.; Silva, F.M.L.; Watson, S.B.; Howell, J.L.; Shaner, D.L. Biochemical markers and enzyme assays for herbicide mode of action and resistance studies. Weed Sci. 2015, 63, 23–63. [Google Scholar] [CrossRef]
- Garcia, I.; Rodgers, M.; Pepin, R.; Hsieh, T.-Z.; Matringe, M. Characterization and subcellular compartmentation of recombinant 4-hydroxyphenylpyruvate dioxygenase from Arabidopsis in transgenic tobacco. Plant Physiol. 1999, 119, 1507–1516. [Google Scholar] [CrossRef]
- Ritz, C.; Streibig, J.C. Bioassay analysis using R. J. Stat. Softw. 2005, 12, 1–22. [Google Scholar] [CrossRef]
- R-Development-Core-Team. R: A Language and Environment for Statistical Computing, 3.3.3; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Ellis, M.K.; Whitfield, A.C.; Gowans, L.A.; Auton, T.R.; Provan, W.M.; Lock, E.A.; Smith, L.L. Inhibition of 4-hydroxyphenylpyruvate dioxygenase by 2-(2-nitro-4-trifluoromethylbenzoyl)-cyclohexane-1,3-dione and 2-(2-chloro-4-methanesulfonylbenzoyl)-cyclohexane-1,3-dione. Toxicol. Appl. Pharmacol. 1995, 133, 12–19. [Google Scholar] [CrossRef]
- Spartan’18 Wavefunction, Inc. Wavefunction, Inc.: Irvine, CA, USA. 2018. Available online: https://downloads.wavefun.com/FAQ/Spartan18Manual.pdf (accessed on 14 March 2022).
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Dewar, M.J.S.; Zoebisch, E.G.; Healy, E.F.; Stewart, J.J.P. Development and use of quantum mechanical molecular models. 76. AM1: A new general purpose quantum mechanical molecular model. J. Am. Chem. Soc. 1985, 107, 3902–3909. [Google Scholar] [CrossRef]
- Metropolis, N.; Ulam, S. The monte carlo method. J. Am. Statist. Assoc. 1949, 44, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.; Baroni, M.; Goracci, L.; Cruciani, G. GRID-based three-dimensional pharmacophores I: FLAPpharm, a novel approach for pharmacophore elucidation. J. Chem. Inform. Model. 2012, 52, 2587–2598. [Google Scholar] [CrossRef] [PubMed]
- Strassner, T.; Busold, M.; Herrmann, W.A. Investigation of the performance and optimal composition of a genetic algorithm for the parametrization of the MM3 forcefield. In Abstracts of Papers of the American Chemical Society; American Chemical Society: Washington, DC, USA, 2020; p. U396. [Google Scholar]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
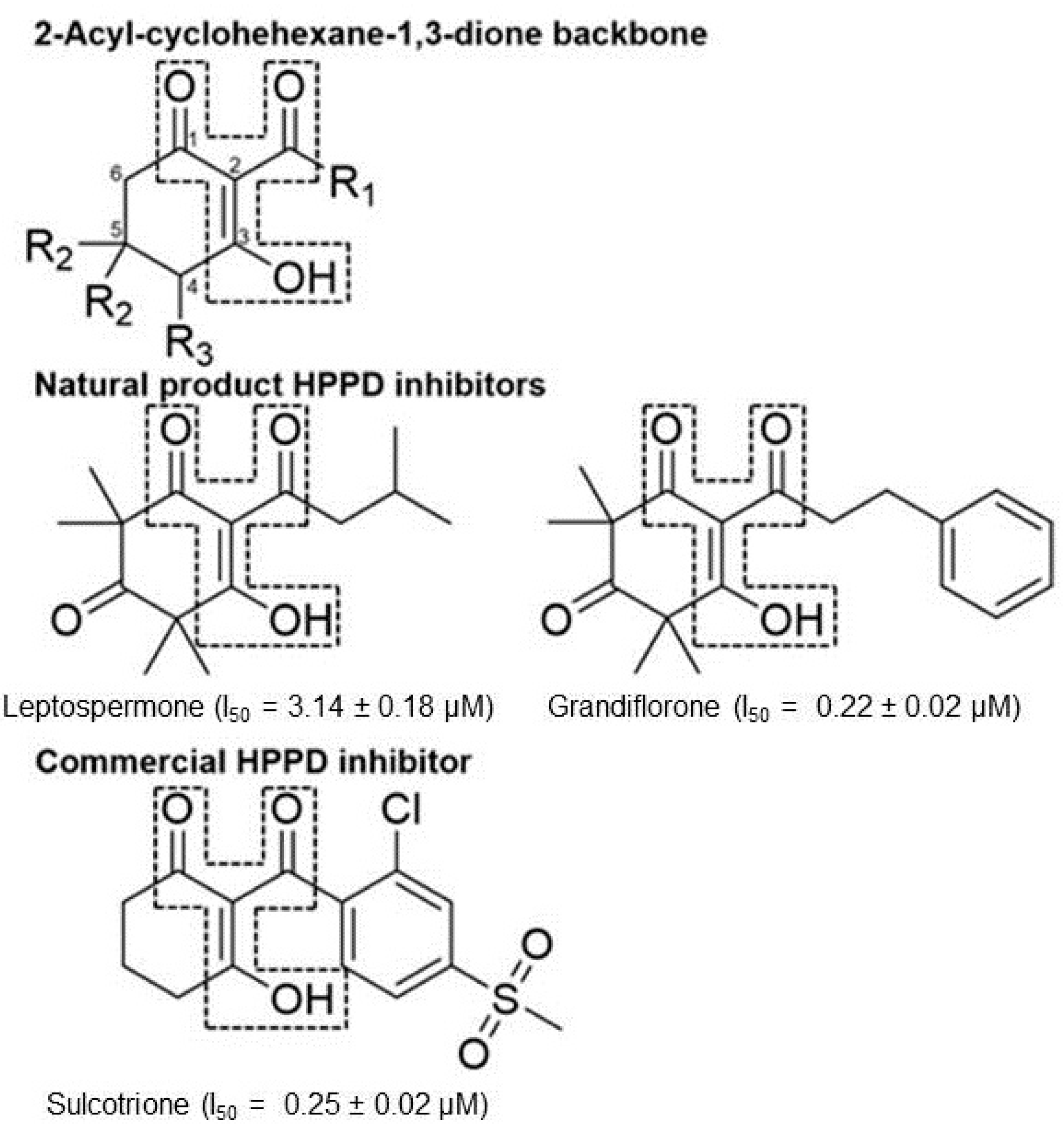
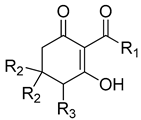
| Cpd # | R1 | R2 | R3 | mw | I50 (μM) ab |
|---|---|---|---|---|---|
| 1bc | methyl | H | H | 154 | 23.09 ± 4.34 |
| 1d | methyl | H | OH | 170 | >1000 |
| 1f | methyl | methyl | H | 182 | 53.12 ± 17.09 |
| 1h | methyl | methyl | OH | 198 | 166.03 ± 19.66 |
| 2a | propyl | H | H | 182 | 35.12 ± 6.24 |
| 2b | propyl | methyl | H | 210 | 16.59 ± 4.36 |
| 3b | pentyl | H | H | 210 | 2.93 ± 0.47 |
| 3d | pentyl | H | OH | 226 | 15.84 ± 4.9 |
| 3e | pentyl | methyl | H | 238 | 4.17 ± 0.45 |
| 4b | nonyl | H | H | 266 | 0.46 ± 0.06 |
| 4d | nonyl | H | OH | 282 | 1.25 ± 0.40 |
| 4e | nonyl | methyl | H | 294 | 2.66 ± 0.95 |
| 5b | undecyl | H | H | 294 | 0.30 ± 0.03 |
| 5d | undecyl | H | OH | 310 | 0.18 ± 0.02 |
| 5f | undecyl | methyl | H | 322 | 0.77 ± 0.11 |
| 6a | hexadecyl | H | H | 364 | 1.58 ± 0.20 |
| 6b | hexadecyl | methyl | H | 392 | 1.70 ± 0.31 |
| 7a | 4-oxopentyl | H | H | 224 | 9.6 ± 1.4 |
| 7b | 4-oxopentyl | methyl | H | 252 | 17.2 ± 2.6 |
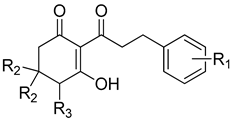
| Cpd # | R1 | R2 | R3 | mw | I50 (μM) ab |
|---|---|---|---|---|---|
| 8bc | H | H | H | 244 | 1.27 ± 0.09 |
| 8e | H | methyl | H | 272 | 2.17 ± 0.37 |
| 9b | 2-methyl | H | H | 258 | 0.97 ± 0.09 |
| 9d | 2-methyl | methyl | H | 286 | nd d |
| 10b | 3-methoxy | H | H | 274 | 0.31 ± 0.04 |
| 10d | 3-methoxy | methyl | H | 302 | 1.50 ± 0.17 |
| 11b | 4-methoxy | H | H | 274 | 2.36 ± 0.30 |
| 11c | 4-methoxy | H | OH | 290 | 59.5 ± 11.8 |
| 11e | 4-methoxy | methyl | H | 302 | 12.9 ± 2.18 |
| 12b | 3,4-dimethoxy | H | H | 304 | 1.35 ± 0.18 |
| 12d | 3,4-dimethoxy | methyl | H | 332 | 8.69 ± 1.32 |
| 14b | 3,4,5-trimethoxy | H | H | 334 | nd |
| 14d | 3,4,5-trimethoxy | methyl | H | 362 | 2.29 ± 0.25 |
| 15b | 4-dimethylamino | H | H | 287 | 4.47 ± 0.51 |
| 15d | 4-dimethylamino | methyl | H | 315 | 3.46 ± 0.64 |
| 16b | 4-chloro | H | H | 278 | 0.24 ± 0.02 |
| 16d | 4-chloro | methyl | H | 307 | 2.05 ± 0.19 |
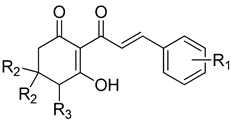
| Cpd # | R1 | R2 | R3 | mw | I50 (μM) ab |
|---|---|---|---|---|---|
| 8ac | H | H | H | 242 | 5.43 ± 0.58 |
| 8c | H | H | OH | 258 | 59.0 ± 3.16 |
| 8d | H | methyl | H | 270 | 58.6 ± 8.67 |
| 9a | 2-methyl | H | H | 256 | 1.20 ± 0.14 |
| 9c | 2-methyl | methyl | H | 284 | 28.7 ± 8.7 |
| 10a | 3-methoxy | H | H | 272 | 0.89 ± 0.10 |
| 10c | 3-methoxy | methyl | H | 300 | 6.4 ± 1.2 |
| 11a | 4-methoxy | H | H | 272 | 12.3 ± 1.6 |
| 11d | 4-methoxy | methyl | H | 300 | 141.4 ± 27 |
| 12a | 3,4-dimethoxy | H | H | 302 | 9.16 ± 1.21 |
| 12c | 3,4-dimethoxy | methyl | H | 330 | 14.1 ± 2.21 |
| 14a | 3,4,5-trimethoxy | H | H | 332 | 2.06 ± 0.13 |
| 14c | 3,4,5-trimethoxy | methyl | H | 360 | 2.22 ± 0.50 |
| 15a | 4-dimethylamino | H | H | 285 | nd d |
| 15c | 4-dimethylamino | methyl | H | 313 | nd |
| 16a | 4-chloro | H | H | 276 | nd |
| 16c | 4-chloro | methyl | H | 305 | nd |
| 17a | 4-bromo | H | H | 320 | 7.55 ± 0.93 |
| 17b | 4-bromo | H | OH | 336 | 28.7 ± 4.82 |
| 17c | 4-bromo | methyl | H | 349 | 8.34 ± 1.57 |
| 18a | 3-bromo | H | H | 321 | 0.82 ± 0.08 |
| 18b | 3-bromo | methyl | H | 349 | 2.42 ± 0.25 |
| 19a | 4-nitro | H | H | 287 | 7.61 ± 0.89 |
| 19b | 4-nitro | methyl | H | 315 | 27.5 ± 2.39 |
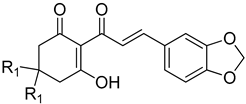 | |||
| Cpd # | R1 | mw | I50(μM) a,b |
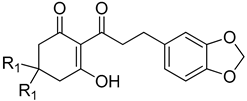
| 13ac | H | 286 | 9.51 ± 1.84 |
| 13c | methyl | 314 | 28.7 ± 3.72 |
 | |||
| Cpd # | R1 | mw | I50(μM) a,b |
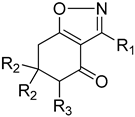
| 13b | H | 288 | 0.97 ± 0.16 |
| 13d | methyl | 316 | 3.25 ± 0.60 |
| Cpd # | R1 | R2 | R3 | mw | I50 (μM) ab |
|---|---|---|---|---|---|
| 1a | methyl | H | H | 151 | >100 |
| 1c | methyl | H | OH | 167 | >100 |
| 1e | methyl | methyl | H | 179 | >100 |
| 1g | methyl | methyl | OH | 195 | >100 |
| 3a | pentyl | H | H | 207 | >100 |
| 3c | pentyl | H | OH | 223 | >100 |
| 4a | nonyl | H | H | 263 | >100 |
| 4c | nonyl | H | OH | 279 | >100 |
| 5a | undecyl | H | H | 291 | >100 |
| 5c | undecyl | H | OH | 307 | >100 |
| 5e | undecyl | methyl | H | 319 | >100 |
| 5g | undecyl | methyl | OH | 335 | >100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ooka, J.K.; Correia, M.V.; Scotti, M.T.; Fokoue, H.H.; Yamaguchi, L.F.; Kato, M.J.; Dayan, F.E.; Owens, D.K. Synthesis and Activity of 2-Acyl-cyclohexane-1,3-dione Congeners Derived from Peperomia Natural Products against the Plant p-Hydroxyphenylpyruvate Dioxygenase Herbicidal Molecular Target Site. Plants 2022, 11, 2269. https://doi.org/10.3390/plants11172269
Ooka JK, Correia MV, Scotti MT, Fokoue HH, Yamaguchi LF, Kato MJ, Dayan FE, Owens DK. Synthesis and Activity of 2-Acyl-cyclohexane-1,3-dione Congeners Derived from Peperomia Natural Products against the Plant p-Hydroxyphenylpyruvate Dioxygenase Herbicidal Molecular Target Site. Plants. 2022; 11(17):2269. https://doi.org/10.3390/plants11172269
Chicago/Turabian StyleOoka, Joey K., Mauro V. Correia, Marcus T. Scotti, Harold H. Fokoue, Lydia F. Yamaguchi, Massuo J. Kato, Franck E. Dayan, and Daniel K. Owens. 2022. "Synthesis and Activity of 2-Acyl-cyclohexane-1,3-dione Congeners Derived from Peperomia Natural Products against the Plant p-Hydroxyphenylpyruvate Dioxygenase Herbicidal Molecular Target Site" Plants 11, no. 17: 2269. https://doi.org/10.3390/plants11172269
APA StyleOoka, J. K., Correia, M. V., Scotti, M. T., Fokoue, H. H., Yamaguchi, L. F., Kato, M. J., Dayan, F. E., & Owens, D. K. (2022). Synthesis and Activity of 2-Acyl-cyclohexane-1,3-dione Congeners Derived from Peperomia Natural Products against the Plant p-Hydroxyphenylpyruvate Dioxygenase Herbicidal Molecular Target Site. Plants, 11(17), 2269. https://doi.org/10.3390/plants11172269









