Phylogeny and Taxonomic Synopsis of the Genus Bougainvillea (Nyctaginaceae)
Abstract
1. Introduction
2. Results
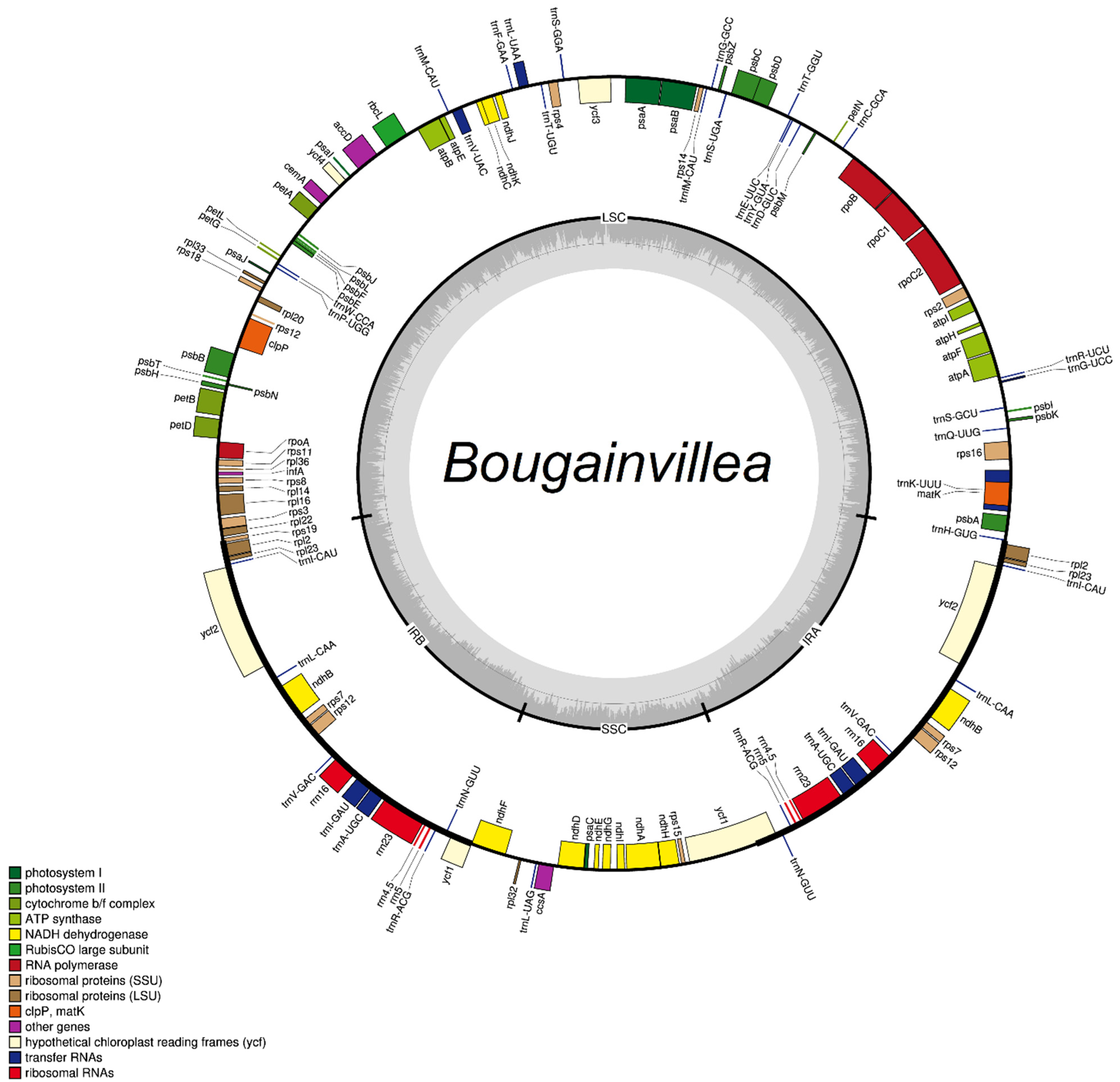
2.1. Sequence Characteristics
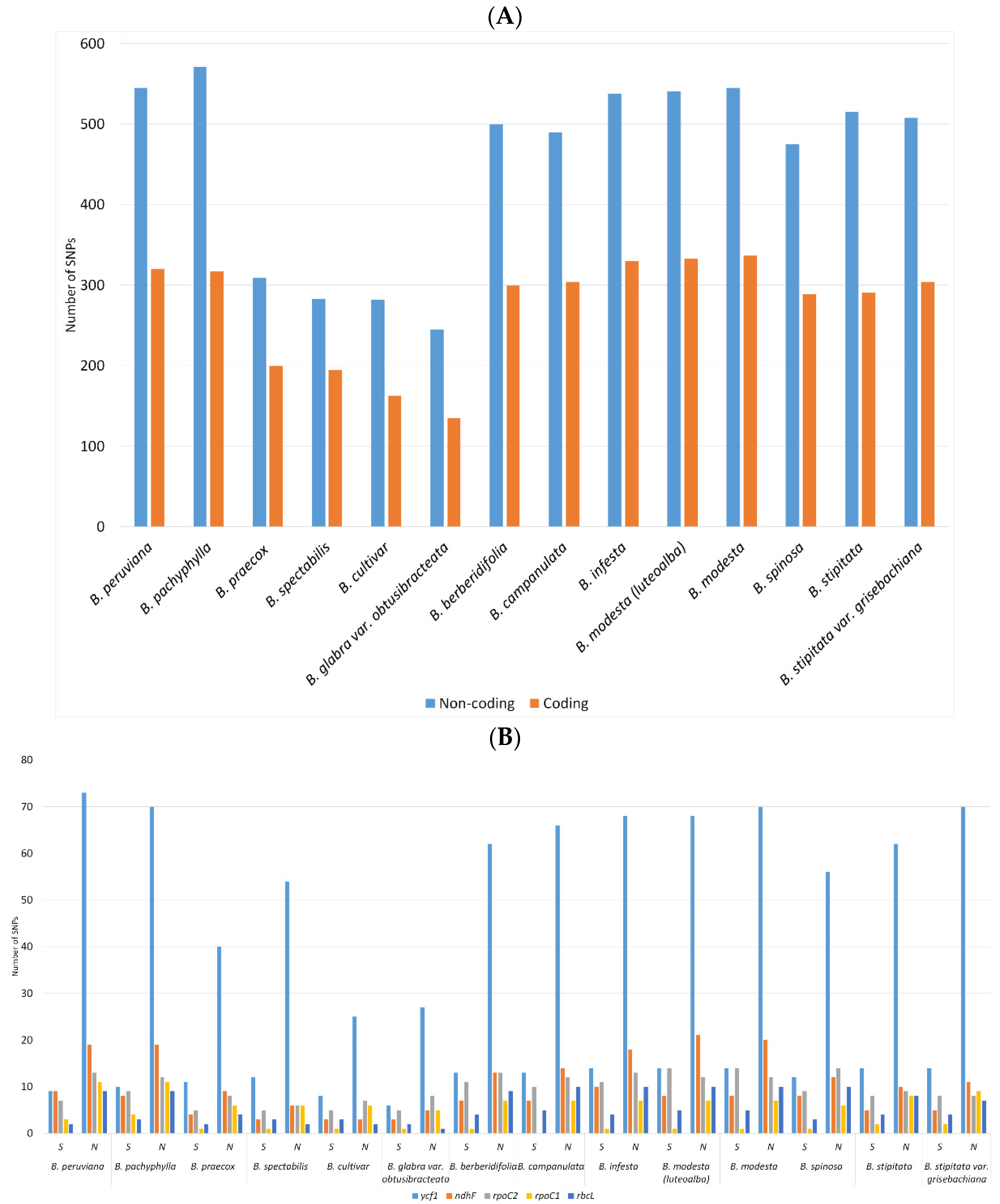
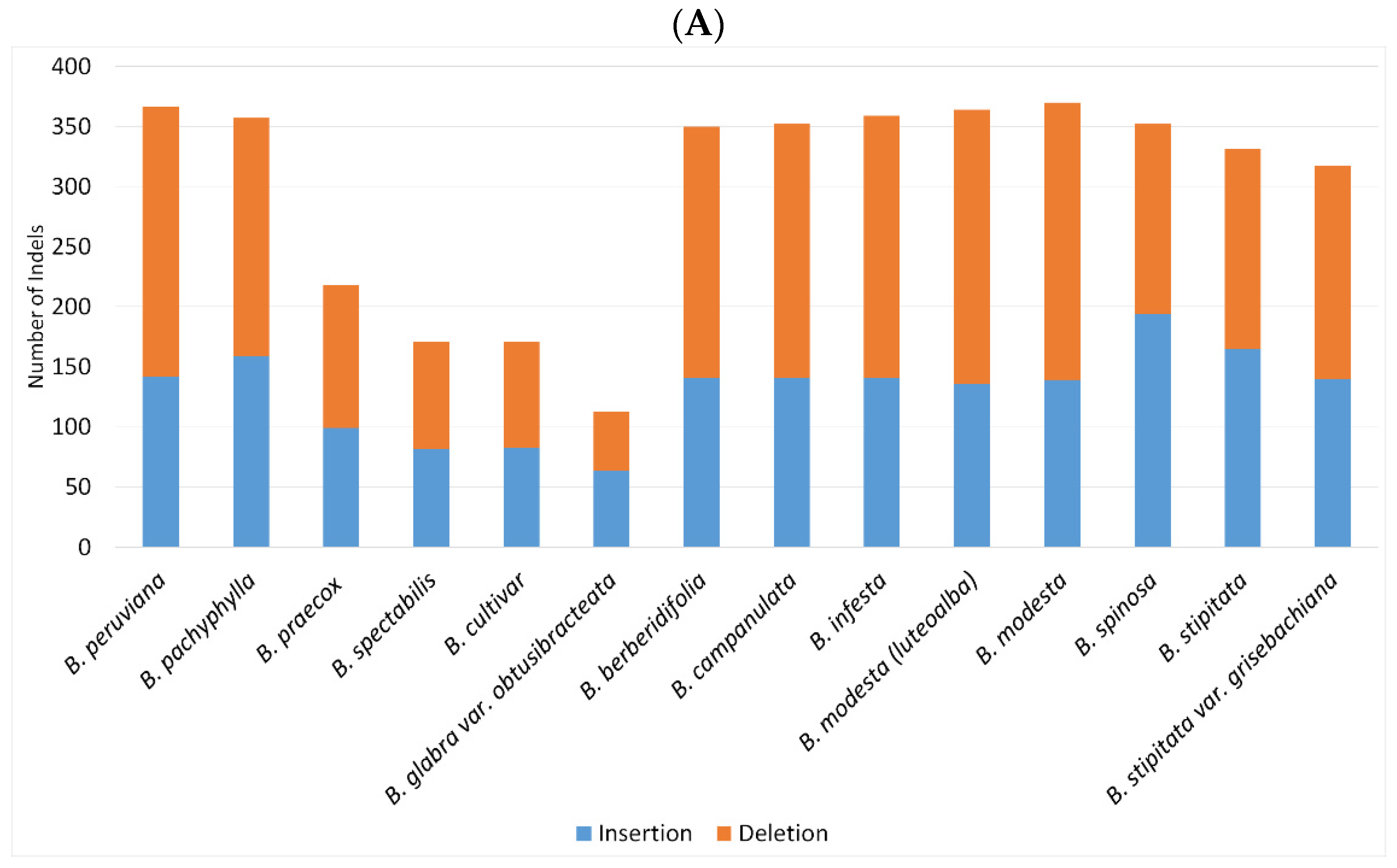
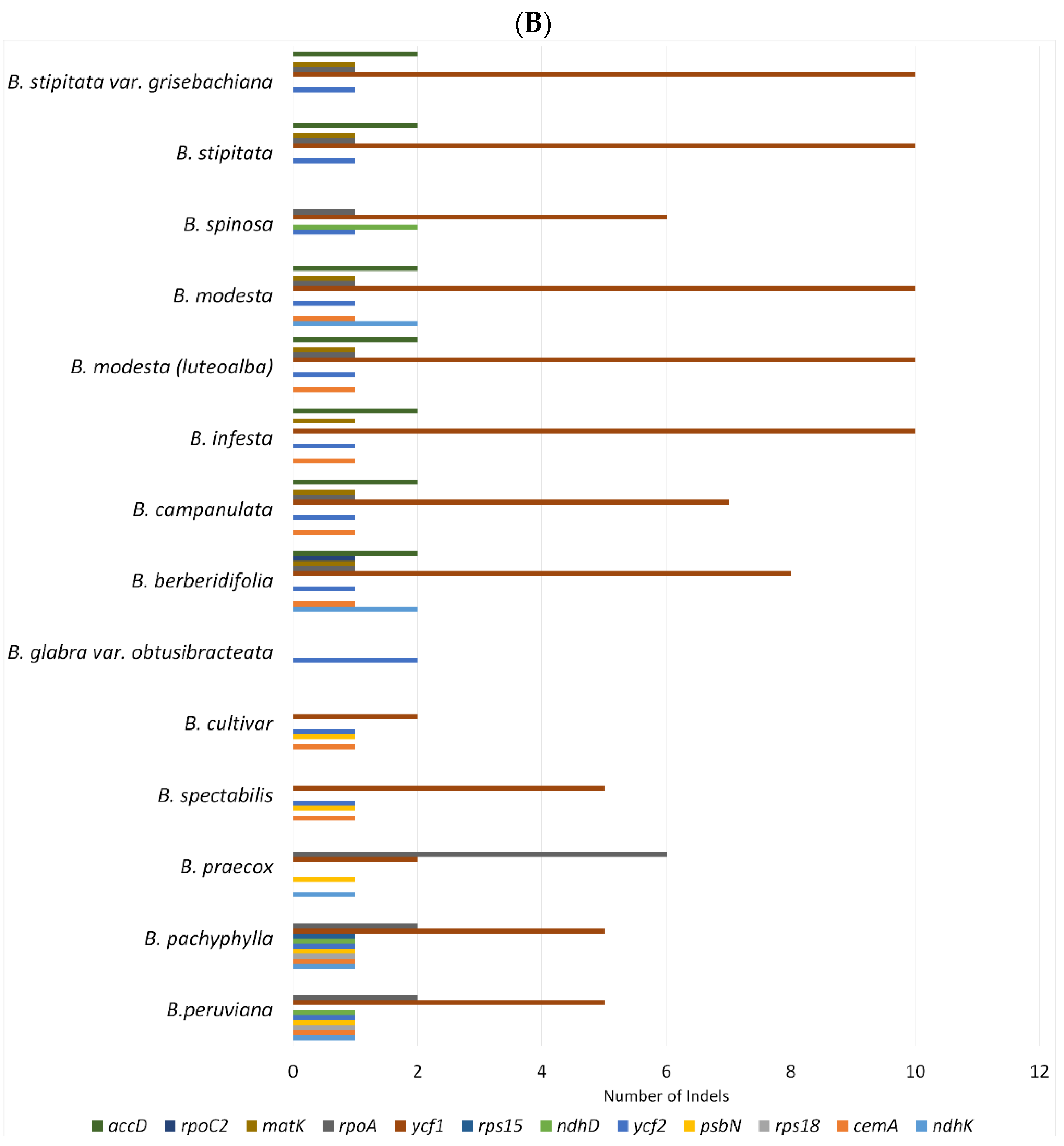
2.2. SNPs and Indels Analysis
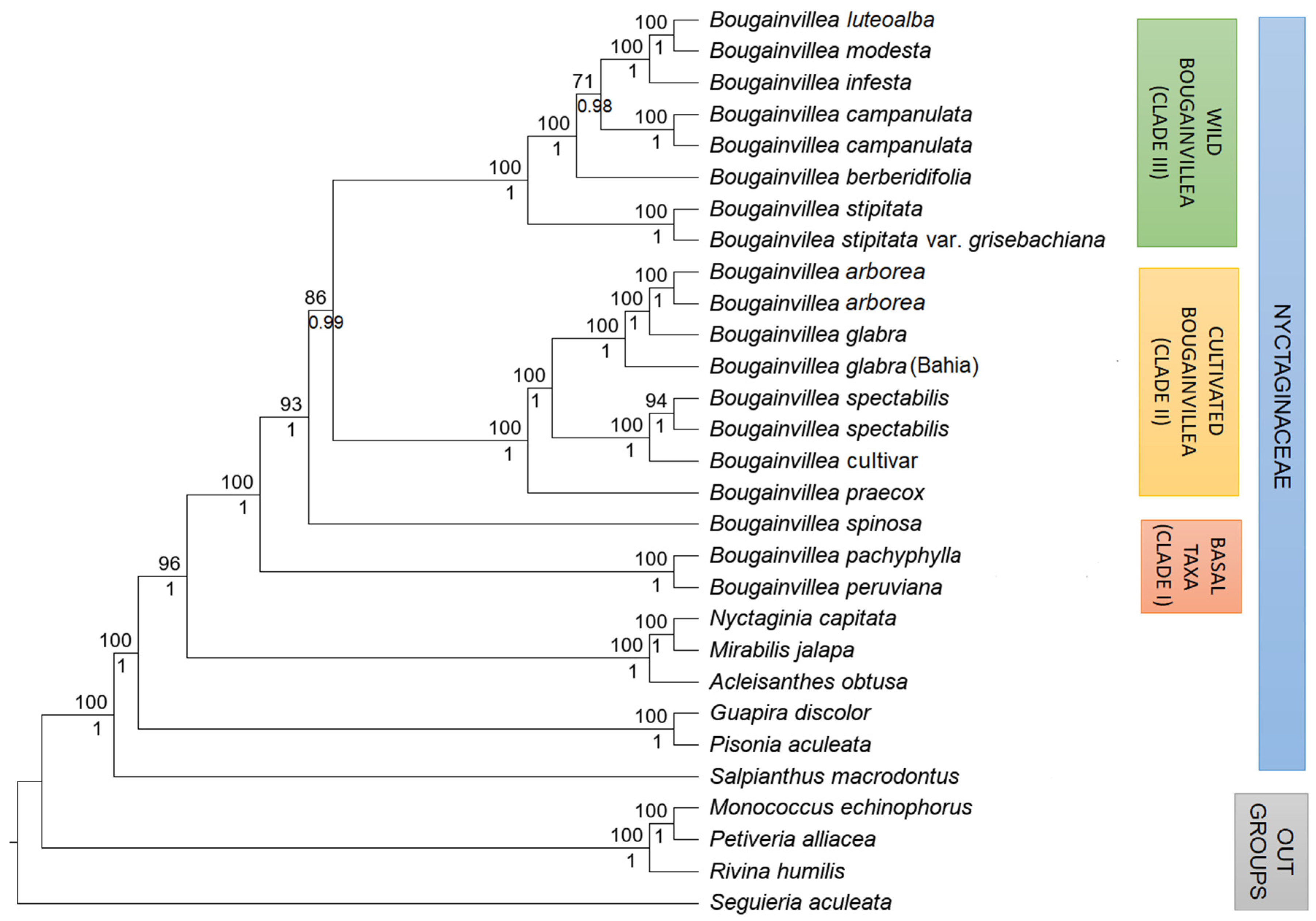
2.3. Phylogenetic Analysis
3. Discussion
3.1. Taxonomic Treatment
| 1. Leaves less than 3 mm wide; inflorescences 1-flowered, surrounded by 3 bracts; thorns furcate or forked at apex..............B. spinosa |
| 1. Leaves more than 3 mm wide; inflorescences 3-flowered, surrounded by 3 bracts; thorns simple. |
| 2. Bracts 2.5–4.5 cm long, usually strikingly colored; perianth lobes inconspicuous, white or cream. |
| 3. Leaves thick, leathery .........................................................................................................................................................B. pachyphylla |
| 3. Leaves thin and papery, especially when dry. |
| 4. Leaves and perianth tube glabrous; apex of bracts usually obtuse or rounded...........................................................B. peruviana |
| 4. Leaves and perianth tube variously pubescent; apex of bracts acute or acuminate. |
| 5. Leaves densely villous; perianth tube short villous...................................................................................................B. spectabilis |
| 5. Leaves merely puberulent; perianth tube sparsely to densely puberulent |
| 6. Anthocarp oblong...............................................................................................................................................................B. glabra |
| 6. Anthocarp obconical-obturbinoid to fusiform............................................................................B. glabra var. obtusibracteata |
| 2. Bracts 1–2.5 cm long, normally unostentatious; perianth lobes somewhat conspicuous, bright yellow, green, or red. |
| 7. Perianth tube 6–7 mm long, campanulate, gradually widening from base to apex...................................................B. campanulata |
| 7. Perianth tube 9–25 mm long, hypocrateriform or infundibuliform, slightly or highly constricted distally or in middle |
| 8. Perianth tube ca. 1.5 mm wide, glabrous, reddish....................................................................................................B. berberidifolia |
| 8. Perianth tube broader >1.5 mm wide, variously pubescent, greenish-brown or greenish-yellow. |
| 9. Perianth tube 9–11 mm long, densely tomentulose |
| 10. Perianth tube base ellipsoid, constricted in the middle...........................................................................................B. praecox |
| 10. Perianth tube almost straight or slightly constricted..............................................................................................B. modesta |
| 9. Perianth tube 12–25 mm long, densely pubescent or puberulent |
| 11. Perianth tube densely pubescent; ovary navicular....................................................................................................B. infesta |
| 11. Perianth tube puberulent; ovary spindle-shaped...................................................................................................B. stipitata |
3.2. Cultivated Hybrid
4. Materials and Methods
4.1. Sample Collection and DNA Extraction
4.2. Sequencing, Assembly, and Annotation of Bougainvillea Plastid Genomes
4.3. SNPs and Indels Analysis
4.4. Phylogenetic Analysis
4.5. Morphological Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lack, H.W. The discovery, naming and typification of Bougainvillea spectabilis (Nyctaginaceae). Willdenowia 2012, 42, 117–126. [Google Scholar] [CrossRef]
- Jussieu, A.L. Genera Plantarum: Secundum Ordines Naturales Disposita, Juxta Methodum in Horto Regio Parisiensi Exaratam, Anno M.DCC.LXXIV; Apud viduam Herissant et Theophilum Barrois: Parisiis, France, 1789; pp. 1–498. [Google Scholar]
- Spach, E. Histoire Naturelle des Végétaux; Librairie encyclopédique de Roret: Paris, France, 1841; Volume 10, p. 516. [Google Scholar]
- Sprague, T.A. Proposal to conserve 90 additional generic names. In International Botanical Congress: Nomenclature Proposals by British Botanists; Wyman & Sons, Ltd.: London, England, 1930; pp. 49–54. [Google Scholar]
- Rickett, H.W.; Stafleu, F.A. Nomina generica conservanda et rejicienda spermatophytorum. Taxon 1959, 8, 213–243. [Google Scholar] [CrossRef][Green Version]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.H.; Li, D.Z.; Marhold, K.; et al. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code), Regnum Vegetabile 159; Koeltz Botanical Books: Glashütten, Germany, 2018. [Google Scholar]
- Heimerl, A. Nyctaginaceae. In Engler & Prantl, Naturl. Pflanzenfam, 2nd ed.; W. Engelmann: Leipzig, Germany, 1934; Volume 4, pp. 86–134. [Google Scholar]
- Standley, P.C. The Nyctaginaceae and Chenopodiaceae of Northwestern South America; Field Museum of Natural History-Botanical Series: Chicago, IL, USA, 1931; Volume 11, pp. 73–114. [Google Scholar]
- Standley, P.C. Studies of American Plants; Field Museum of Natural History-Botanical Series: Chicago, IL, USA, 1931; Volume 8, pp. 44–48. [Google Scholar]
- Standley, P.C. Nyctaginaceae. In Flora of Peru; Macbride, J.F., Ed.; Field Museum of Natural History-Botanical Series: Chicago, IL, USA, 1937; Volume 13, pp. 518–546. [Google Scholar]
- Toursarkissian, M. Las Nictaginaceas argentinas. Revista Museo Argentino de Ciencias Naturales Bernardino Rivadavia. Botanica 1975, 5, 1–83. [Google Scholar]
- Kobayashi, K.D.; McConnell, J.; Griffis, J. Bougainvillea. In Ornamentals and Flowers; College of Tropical Agriculture and Human Resources, University of Hawaii: Honolulu, HI, USA, 2007; Volume OF-38, pp. 1–12. [Google Scholar]
- Cuénoud, P.; Savolainen, V.; Chatrou, L.W.; Powell, M.; Grayer, R.J.; Chase, M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef]
- Brockington, S.F.; Alexandre, R.; Ramdial, J.; Moore, M.J.; Crawley, S.; Dhingra, A.; Hilu, K.; Soltis, D.E.; Soltis, P.S. Phylogeny of the Caryophyllales sensu lato: Revisiting hypotheses on pollination biology and perianth differentiation in the core Caryophyllales. Int. J. Plant Sci. 2009, 170, 627–643. [Google Scholar] [CrossRef]
- Yao, G.; Jin, J.J.; Lia, H.T.; Yanga, J.B.; Mandalad, V.S.; Croleyd, M.; Mostowd, R.; Douglas, N.A.; Chase, M.W.; Christenhuszg, M.J.M.; et al. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol. Phylogenet. Evol. 2019, 134, 74–86. [Google Scholar] [CrossRef]
- Douglas, N.A.; Manos, P.S. Molecular phylogeny of Nyctaginaceae: Taxonomy, biogeography and characters associated with a radiation of xerophytic genera in North America. Am. J. Bot. 2007, 94, 856–872. [Google Scholar] [CrossRef]
- Douglas, N.; Spellenberg, R. A new tribal classification of Nyctaginaceae. Taxon 2010, 59, 905–910. [Google Scholar] [CrossRef]
- Bautista, M.A.C.; Zheng, Y.; Hu, Z.; Deng, Y.F.; Chen, T. Comparative analysis of complete chloroplast genome sequences of wild and cultivated Bougainvillea (Nyctaginaceae). Plants 2020, 9, 1671. [Google Scholar] [CrossRef]
- Marcais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Singh, S.; Roy, R.K. Pollen morphology of Bougainvillea (Nyctaginaceae): A popular ornamental plant of tropical and sub-tropical gardens of the world. Rev. Palaeobot. Palynol. 2017, 239, 31–46. [Google Scholar] [CrossRef]
- Flora of Argentina. Available online: http://conosur.floraargentina.edu.ar/ (accessed on 20 November 2020).
- Flora of the World. Available online: https://www.floraoftheworld.org/ (accessed on 20 November 2020).
- de Oliveira Chagas, E.C.; da Costa-Lima, J.L. (2826) Proposal to conserve the name Bougainvillea spectabilis against B. brasiliensis (Nyctaginaceae). Taxon 2021, 70, 900–901. [Google Scholar] [CrossRef]
- Kellogg, E.A. Nyctaginaceae. In Flora of the Lesser Antilles, Leeward and Windward Islands; Howard, R.A., Ed.; Arnold Arboretum, Harvard University: Jamaica Plain, MA, USA, 1988; Volume 4, pp. 173–186. [Google Scholar]
- Holttum, R.E. The cultivated Bougainvilleas. Bougainvillea × buttiana, its variety and hybrids. Malay. Agri-hort. Ass. Mag. 1955, 12, 2–11. [Google Scholar]
- Gillis, W.T. Bougainvilleas of cultivation (Nyctaginaceae). Baileya 1976, 20, 34–41. [Google Scholar]
- Chen, T. Bougainvillea, 1st ed.; China Agriculture Press: Beijing, China, 2008; pp. 1–118. [Google Scholar]
- Singh, B.; Panwar, R.S.; Voleti, S.R.; Sharma, V.K.; Thakur, S. The New International Bougainvillea Check List; Division of Floriculture and Landscaping, Indian Agriculture Research Institute: New Delhi, India, 1999; pp. 1–76. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Ruby, J.G.; Bellare, P.; Derisi, J. PRICE: Software for the targeted assembly of components of (meta) genomic sequence data. G3 (Bethseda, Md.) 2013, 5, 865–880. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Huang, D.I.; Cronk, Q. Plann: A command-line application for annotating plastome sequences. Appl. Plant Sci. 2015, 3, 1500026. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.F.; Rodland, E.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Stern, A.; Doron-Faigenboim, A.; Erez, E.; Martz, E.; Bacharach, E.; Pupko, T. Selecton 2007: Advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007, 35, 506–511. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Beentje, H. The Kew Plant Glossary, 2nd ed.; Kew Publishing: Royal Botanic Gardens, Kew, UK, 2016; pp. 1–184. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bautista, M.A.C.; Zheng, Y.; Boufford, D.E.; Hu, Z.; Deng, Y.; Chen, T. Phylogeny and Taxonomic Synopsis of the Genus Bougainvillea (Nyctaginaceae). Plants 2022, 11, 1700. https://doi.org/10.3390/plants11131700
Bautista MAC, Zheng Y, Boufford DE, Hu Z, Deng Y, Chen T. Phylogeny and Taxonomic Synopsis of the Genus Bougainvillea (Nyctaginaceae). Plants. 2022; 11(13):1700. https://doi.org/10.3390/plants11131700
Chicago/Turabian StyleBautista, Mary Ann C., Yan Zheng, David E. Boufford, Zhangli Hu, Yunfei Deng, and Tao Chen. 2022. "Phylogeny and Taxonomic Synopsis of the Genus Bougainvillea (Nyctaginaceae)" Plants 11, no. 13: 1700. https://doi.org/10.3390/plants11131700
APA StyleBautista, M. A. C., Zheng, Y., Boufford, D. E., Hu, Z., Deng, Y., & Chen, T. (2022). Phylogeny and Taxonomic Synopsis of the Genus Bougainvillea (Nyctaginaceae). Plants, 11(13), 1700. https://doi.org/10.3390/plants11131700






