Proteomic Analysis Reveals Salicylic Acid as a Pivotal Signal Molecule in Rice Response to Blast Disease Infection
Abstract
1. Introduction
2. Results
2.1. Identification and Relative Quantification of Proteins
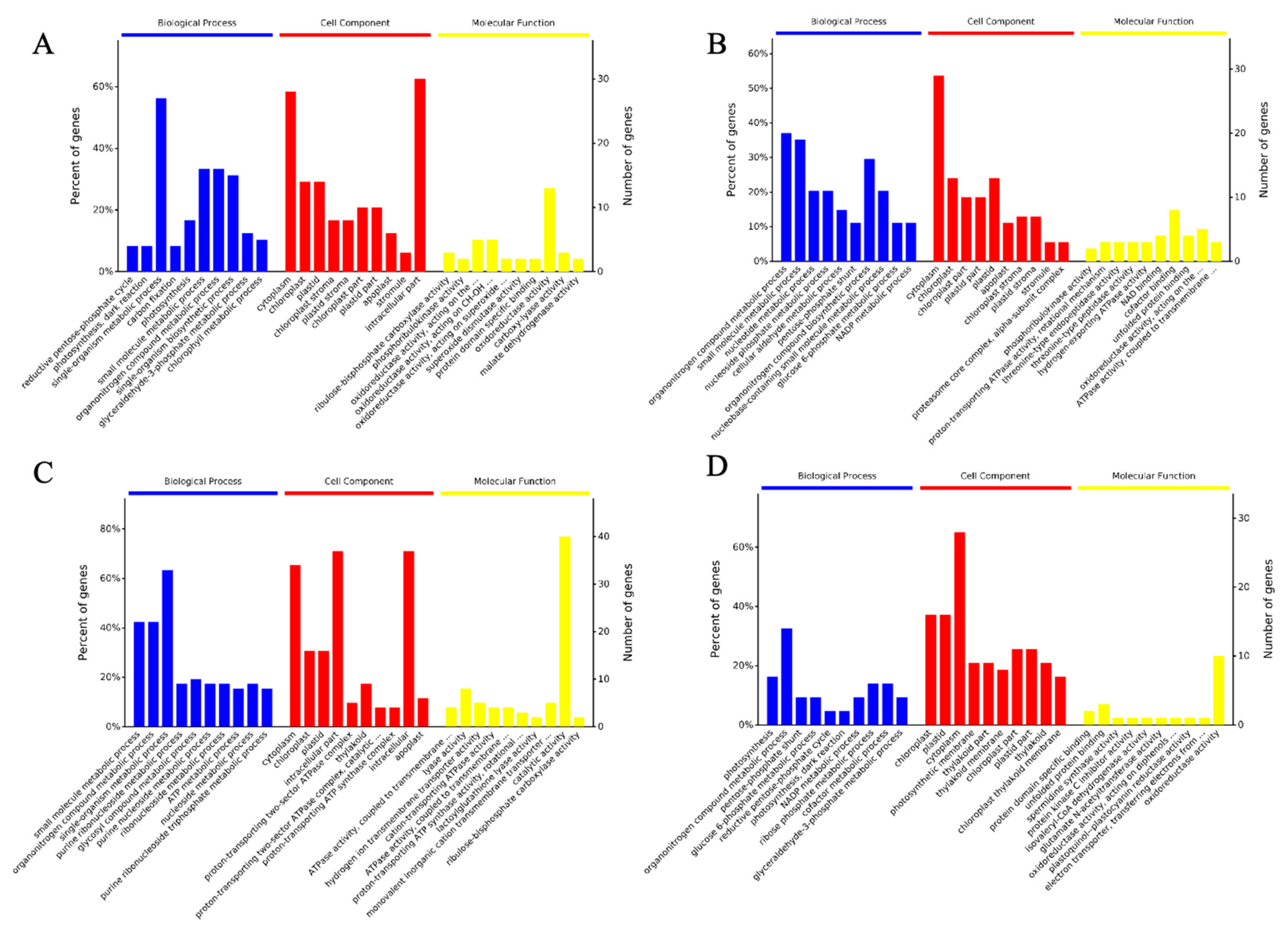
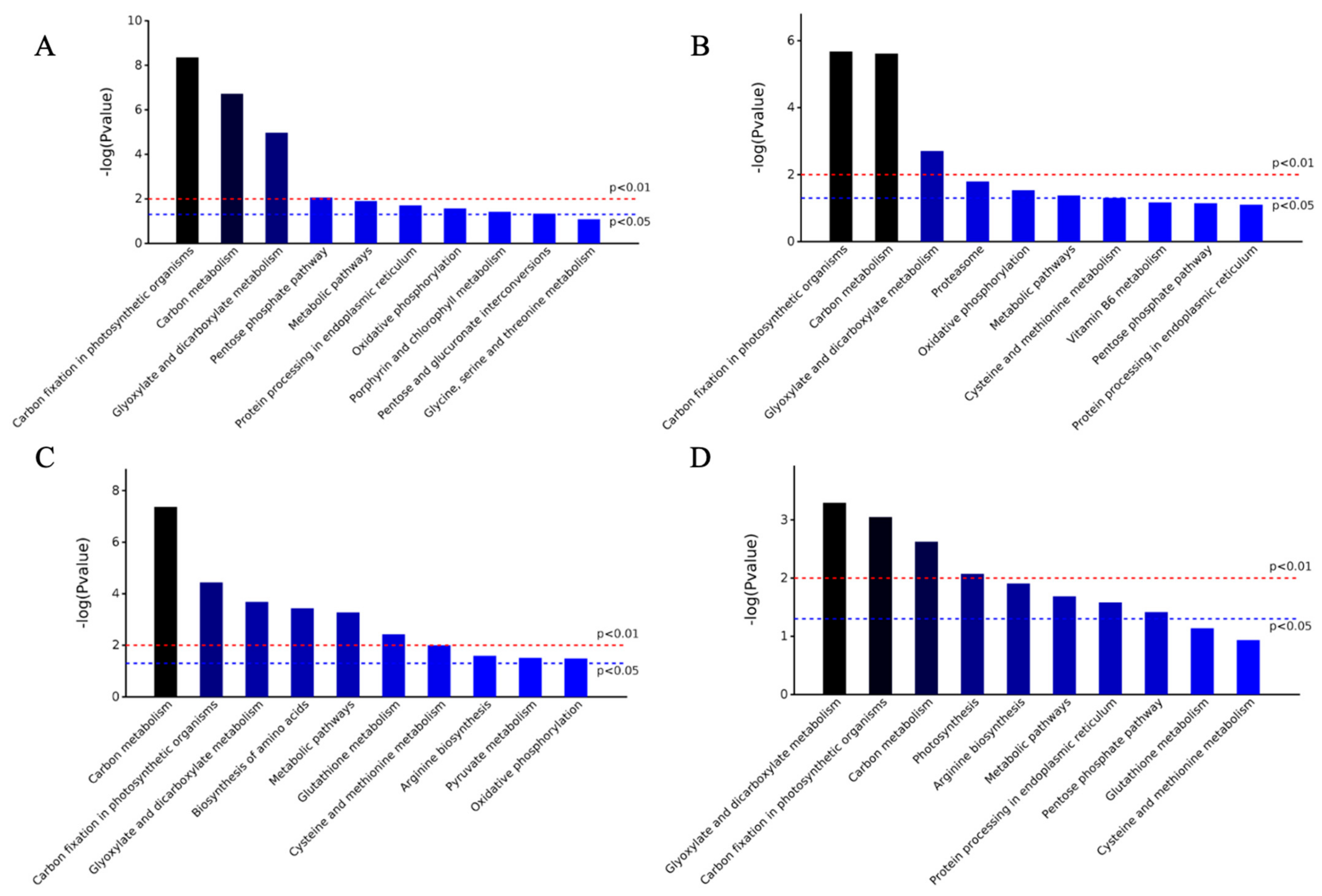
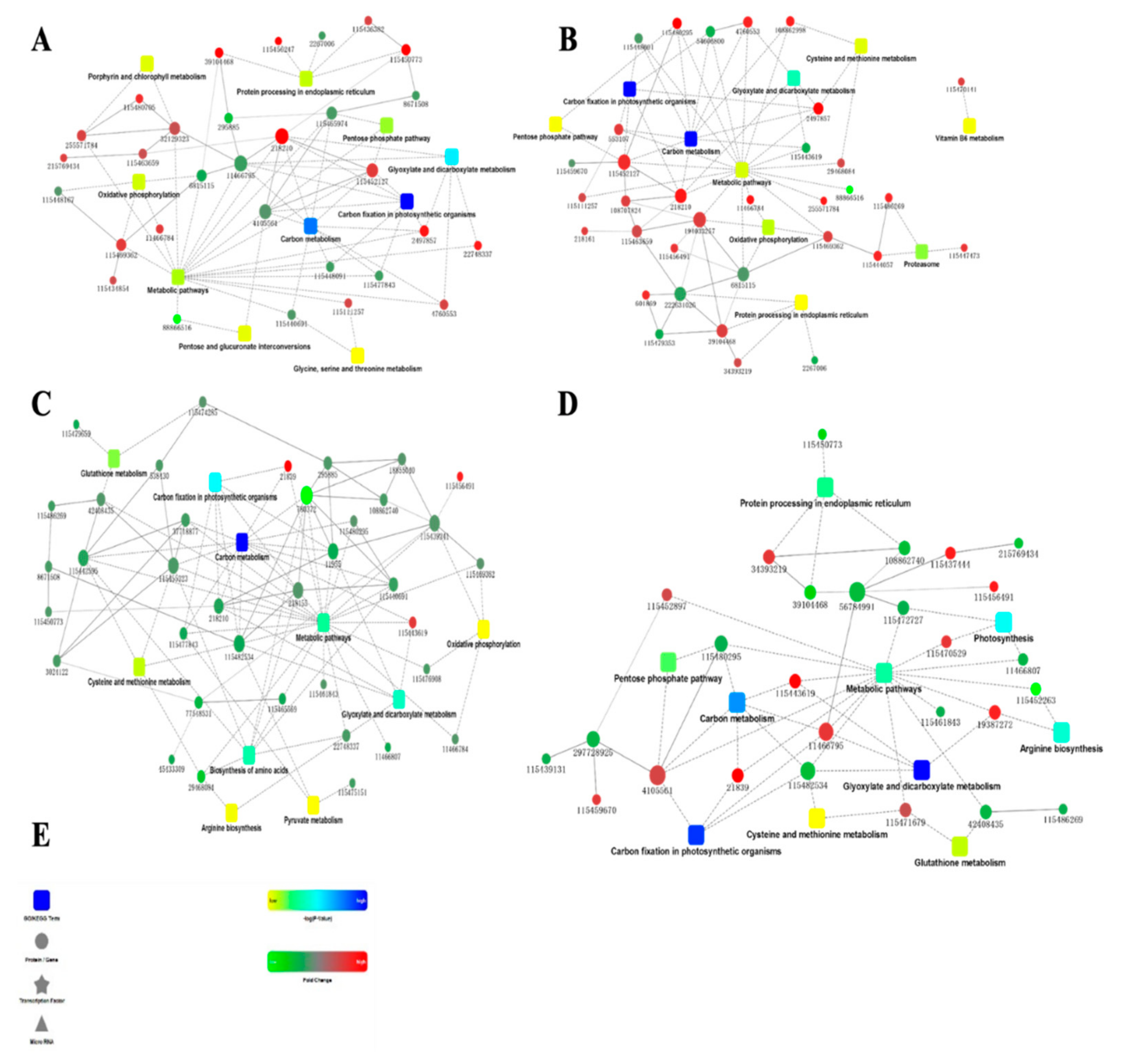
2.2. Bioinformatics Analysis of Differentially Expressed Proteins
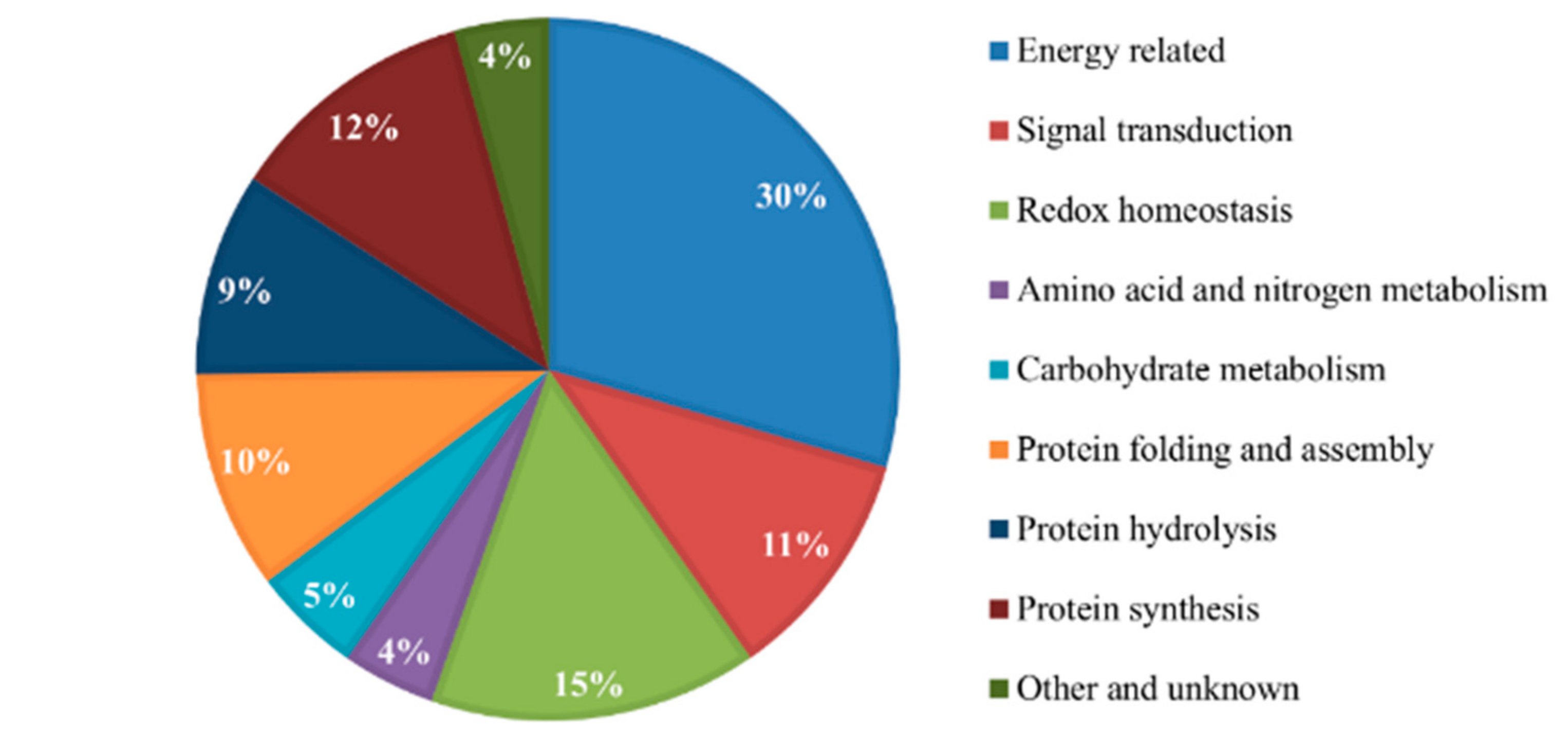
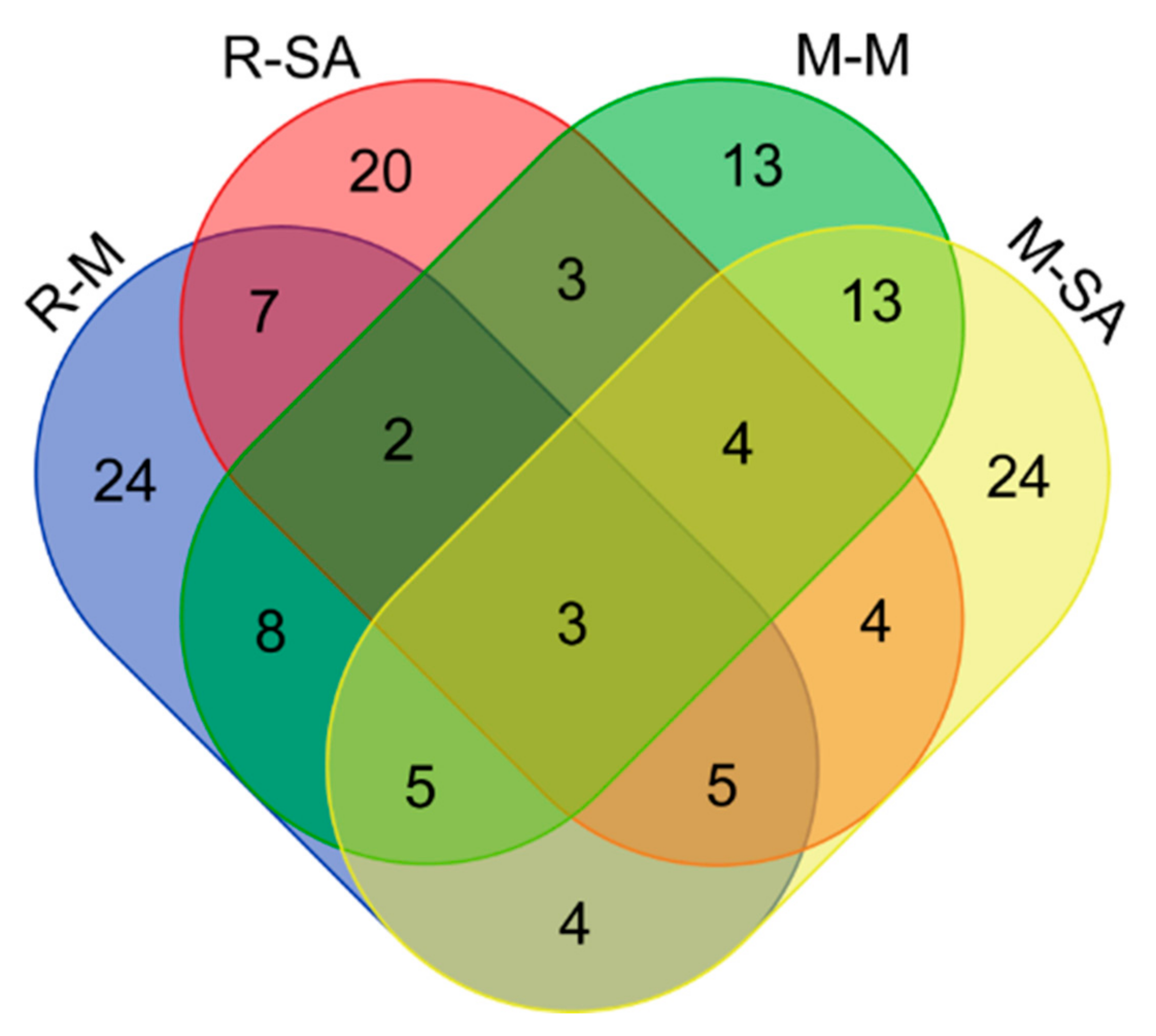
2.3. Comprehensive Inventory of the Differentially Expressed Proteins
2.4. Specific Metabolic Processes in Minghui and Nipponbare Are Involved in the Response against M. grisea
2.5. Protein-Protein Interactions
3. Discussion
3.1. Differentially Expressed Proteins Related to Energy
3.2. Proteins Associated with Signal Transduction
3.3. Differentially Expressed Proteins with Redox Homeostasis
3.4. Proteins Associated with Amino Acid and Nitrogen Metabolism
3.5. Proteins Associated with Carbohydrate Metabolism
3.6. Differentially Expressed Proteins Related to Protein Metabolism
4. Materials and Methods
4.1. Plant Materials
4.2. Protein Extraction
4.3. Trypsin Digestion, iTRAQ™ Labels, Off-Line Strong Cation Exchange Chromatography, and Online nano-LC-MALDI-TOF-TOF MS Analysis
4.4. Database Search and iTRAQ Quantification
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pingali, P. Agricultural policy and nutrition outcomes—Getting beyond the preoccupation with staple grains. Food Secur. 2015, 7, 583–591. [Google Scholar] [CrossRef]
- Oerke, E.C.; Dehne, H.W. Safeguarding production—Losses in major crops and the role of crop protection. Crop Prot. 2004, 23, 275–285. [Google Scholar] [CrossRef]
- Chen, D.; Chen, X.W.; Lei, C.L.; Bing-Tian, M.A. Rice Blast Resistance of Transgenic Rice Plants with Pi-d2 Gene. Rice Sci. 2010, 17, 179–184. [Google Scholar] [CrossRef]
- Veneault-Fourrey, C.; Talbot, N.J. Moving toward a systems biology approach to the study of fungal pathogenesis in the rice blast fungus Magnaporthe grisea. Adv. Appl. Microbiol. 2005, 57, 177–215. [Google Scholar]
- Liu, X.; Li, W.; Hu, B.; Wang, M.; Wang, J. Identification of isobavachalcone as a potential drug for rice blast disease caused by the fungus Magnaporthe grisea. J. Biomol. Struct. Dyn. 2019, 37, 3399–3409. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, X.; Zhang, Z. The Magnaporthe grisea species complex and plant pathogenesis. Mol. Plant. Pathol. 2016, 17, 796–804. [Google Scholar] [CrossRef]
- Zhang, K.L.; Liu, Q.S.; Kang, H.X.; Liu, X.M. Herbivore-induced rice resistance against rice blast mediated by salicylic acid. Insect Sci. 2020, 27, 49–57. [Google Scholar] [CrossRef]
- Takatsuji, H. Development of disease-resistant rice using regulatory components of induced disease resistance. Front. Plant Sci. 2014, 5, 630. [Google Scholar] [CrossRef]
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef]
- Sun, R.; Qin, S.; Zhang, T.; Wang, Z. Comparative phosphoproteomic analysis of blast resistant and susceptible rice cultivars in response to salicylic acid. BMC Plant Biol. 2019, 19, 454. [Google Scholar] [CrossRef]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C. Rice WRKY45 Plays a Crucial Role in Benzothiadiazole-Inducible Blast Resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar] [CrossRef] [PubMed]
- Sugano, S.; Jiang, C.J.; Miyazawa, S.I.; Masumoto, C. Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol. Biol. 2010, 74, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Inoue, H.; Tang, X.; Tan, Y.; Xu, X. Rice OsAAA-ATPase1 is Induced during Blast Infection in a Salicylic Acid-Dependent Manner, and Promotes Blast Fungus Resistance. Int. J. Mol. Sci. 2020, 21, 1443. [Google Scholar] [CrossRef]
- Xue, W.; Mao, X.; Wei, Y.; Ling, L. Proteomic analysis of blast-resistant near-isogenic lines derived from japonica rice, var. Yunyin, infected with Magnaporthe oryzae. Chin. Sci. Bull. 2014, 59, 4312–4322. [Google Scholar] [CrossRef]
- Meleady, P. Two-Dimensional Gel Electrophoresis and 2D-DIGE. Methods Mol. Biol. 2018, 1664, 3–14. [Google Scholar] [PubMed]
- Kachroo, P.; Lee, K.H.; Schwerdel, C.; Bailey, J.E.; Chattoo, B.B. Analysis of host-induced response in the rice blast fungus Magnaporthe grisea using two-dimensional polyacrylamide gel electrophoresis. Electrophoresis 1997, 18, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Kim, S.G.; Hwang, D.H.; Kang, S.Y. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 2004, 4, 3569–3578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, H.; Xie, X.; Ji, J.; Dong, Y.; Du, Y. Comparative proteomic analyses reveal that the regulators of G-protein signaling proteins regulate amino acid metabolism of the rice blast fungus Magnaporthe oryzae. Proteomics 2014, 14, 2508–2522. [Google Scholar] [CrossRef]
- Evans, C.; Noirel, J.; Ow, S.Y.; Salim, M.; Pereira-Medrano, A.G. An insight into iTRAQ: Where do we stand now? Anal. Bioanal. Chem. 2012, 404, 1011–1027. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, L.; Zhao, M.; Gu, S.; Wang, C. iTRAQ proteomics reveals the regulatory response to Magnaporthe oryzae in durable resistant vs. susceptible rice genotypes. PLoS ONE 2020, 15, e0227470. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Lei, G.; Zhou, H.W.; He, C.; Liao, J.L. Quantitative iTRAQ-based proteomic analysis of rice grains to assess high night temperature stress. Proteomics 2017, 17, 1600365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, W.; Huang, S.; Xu, P.; Cao, Z. The potential role of plasma membrane proteins in response to Zn stress in rice roots based on iTRAQ and PRM under low Cd condition. J. Hazard Mater. 2022, 429, 128324. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, H.; Qu, Z.; Wang, J.; Wang, X. Transcriptome Sequencing and iTRAQ of Different Rice Cultivars Provide Insight into Molecular Mechanisms of Cold-Tolerance Response in Japonica Rice. Rice 2020, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Zhu, C.; Bai, Z.; Huang, J.; Zhu, L. iTRAQ-Based Protein Profiling and Biochemical Analysis of Two Contrasting Rice Genotypes Revealed Their Differential Responses to Salt Stress. Int. J. Mol. Sci. 2019, 20, 547. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Coleman, H.D.; Yan, J.; Mansfield, S.D. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. USA 2009, 106, 13118–13123. [Google Scholar] [CrossRef]
- An, X.; Zhang, J.; Dai, L.; Deng, G. Isobaric Tags for Relative and Absolute Quantitation (iTRAQ)-Based Comparative Proteome Analysis of the Response of Ramie under Drought Stress. Int. J. Mol. Sci. 2016, 17, 1607. [Google Scholar] [CrossRef]
- Junge, W. Protons, proteins and ATP. Photosynth Res. 2004, 80, 197–221. [Google Scholar] [CrossRef]
- Connelly, J.P.; Connelly, J.P.; Müller, M.G.; Bassi, R.; Croce, R.; Holzwarth, A.R. Femtosecond transient absorption study of carotenoid to chlorophyll energy transfer in the light-harvesting complex II of photosystem II. Biochemistry 1997, 36, 281–287. [Google Scholar] [CrossRef]
- Abe, R.; Sayama, K.; Sugihara, H. Development of new photocatalytic water splitting into H2 and O2 using two different semiconductor photocatalysts and a shuttle redox mediator IO3-/I. J. Phys. Chem. B 2005, 109, 16052–16061. [Google Scholar] [CrossRef]
- Jiang, H.X.; Yang, L.T.; Qi, Y.P.; Lu, Y.B. Root iTRAQ protein profile analysis of two Citrus species differing in aluminum-tolerance in response to long-term aluminum-toxicity. BMC Genom. 2015, 16, 949. [Google Scholar] [CrossRef] [PubMed]
- Przybyło, M.; Pocheć, E.; Link-Lenczowski, P.; Lityńska, A. Beta1-6 branching of cell surface glycoproteins may contribute to uveal melanoma progression by up-regulating cell motility. Mol. Vis. 2008, 14, 625–636. [Google Scholar] [PubMed]
- Krähling, H.; Mally, S.; Eble, J.A.; Noël, J.; Schwab, A.; Stock, C. The glycocalyx maintains a cell surface pH nanoenvironment crucial for integrin-mediated migration of human melanoma cells. Pflugers Arch. 2009, 458, 1069–1083. [Google Scholar] [CrossRef]
- Wu, H.; Min, J.; Ikeguchi, Y.; Zeng, H. Structure and mechanism of spermidine synthases. Biochemistry 2007, 46, 8331–8339. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.A.; Havrilla, C.M.; Brady, T.C.; Abramo, K.H.; Levin, E.D. Oxidative stress in toxicology: Established mammalian and emerging piscine model systems. Env. Health Perspect. 1998, 106, 375–384. [Google Scholar] [CrossRef]
- Lamb, G.D.; Westerblad, H. Acute effects of reactive oxygen and nitrogen species on the contractile function of skeletal muscle. J. Physiol. 2011, 589, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zang, T.Z.; Yang, W.; Sun, Y.; Mu, Y.; Liu, J.Q. Single chain antibody displays glutathione S-transferase activity. J. Biol. Chem. 2006, 281, 12516–12520. [Google Scholar] [CrossRef] [PubMed]
- Board, P.G.; Anders, M.W. Glutathione transferase omega 1 catalyzes the reduction of S-(phenacyl)glutathiones to acetophenones. Chem. Res. Toxicol. 2007, 20, 149–154. [Google Scholar] [CrossRef] [PubMed]
- De Leo, M.E.; Landriscina, M.; Palazzotti, B.; Borrello, S.; Galeotti, T. Iron modulation of LPS-induced manganese superoxide dismutase gene expression in rat tissues. FEBS Lett. 1997, 403, 131–135. [Google Scholar] [CrossRef]
- Marchitti, S.A.; Chen, Y.; Thompson, D.C.; Vasiliou, V. Ultraviolet radiation: Cellular antioxidant response and the role of ocular aldehyde dehydrogenase enzymes. Eye Contact Lens 2011, 37, 206–213. [Google Scholar] [CrossRef]
- Wang, S.; Park, S.; Kodali, V.K.; Han, J.; Yip, T. Identification of protein disulfide isomerase 1 as a key isomerase for disulfide bond formation in apolipoprotein B100. Mol. Biol. Cell. 2015, 26, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Ask, M.; Bettiga, M.; Duraiswamy, V.R.; Olsson, L. Pulsed addition of HMF and furfural to batch-grown xylose-utilizing Saccharomyces cerevisiae results in different physiological responses in glucose and xylose consumption phase. Biotechnol. Biofuels 2013, 6, 181. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.E.; Kwon, K.W.; Lee, J.C.; Woo, S.Y. Expression of the rice cytoplasmic cysteine synthase gene in tobacco reduces ozone-induced damage. Plant Biotech. Rep. 2007, 1, 93–100. [Google Scholar] [CrossRef]
- Zagorchev, L.; Seal, C.E.; Kranner, I.; Odjakova, M. A central role for thiols in plant tolerance to abiotic stress. Int. J. Mol. Sci. 2013, 14, 7405–7432. [Google Scholar] [CrossRef] [PubMed]
- Verdel-Aranda, K.; López-Cortina, S.T.; Hodgson, D.A.; Barona-Gómez, F. Molecular annotation of ketol-acid reductoisomerases from Streptomyces reveals a novel amino acid biosynthesis interlock mediated by enzyme promiscuity. Microb. Biotechnol. 2015, 8, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Gunina, A.; Kuzyakov, Y. Sugars in soil and sweets for microorganisms: Review of origin, content, composition and fate. Soil Biol. Biochem. 2015, 90, 87–100. [Google Scholar] [CrossRef]
- Hung, C.-C.; Guo, L.; Santschi, P.H.; Alvarado-Quiroz, N.; Haye, J.M. Distributions of carbohydrate species in the Gulf of Mexico. Mar. Chem. 2003, 81, 119–135. [Google Scholar] [CrossRef]
- Lu, M.C.; Ji, J.-A.; Jiang, Z.-Y.; You, Q.-D. The Keap1-Nrf2-ARE Pathway As a Potential Preventive and Therapeutic Target: An Update. Med. Res. Rev. 2016, 36, 924–963. [Google Scholar] [CrossRef]
- Takusagawa, F.; Kamitori, S.; Markham, G.D. Structure and function of S-adenosylmethionine synthetase: Crystal structures of S-adenosylmethionine synthetase with ADP, BrADP, and PPi at 28 angstroms resolution. Biochemistry 1996, 35, 2586–2596. [Google Scholar] [CrossRef]
- Silva, D.; Rayol, C.D.; Moura, P.M.M.F.; Andrade, P.P. Partial purification, immunogenicity and putative new localization of a native Leishmania heat shock protein 70. Parasitol. A Latinoam. 2008, 63, 4–11. [Google Scholar] [CrossRef][Green Version]
- Ivanina, A.V.; Taylor, C.; Sokolova, I.M. Effects of elevated temperature and cadmium exposure on stress protein response in eastern oysters Crassostrea virginica (Gmelin). Aquat. Toxicol. 2009, 91, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R.; McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Jin, G.Z.; Cong, Y.L.; Yu, W.M.H.; Dong, H.; Shu, H. iTRAQ-2DLC-ESI-MS/MS based identification of a new set of immunohistochemical biomarkers for classification of dysplastic nodules and small hepatocellular carcinoma. J. Proteome. Res. 2011, 10, 3418–3428. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.S.; Wang, Y.S.; Cao, R.; Jin, H.; Huang, Y.; Zhang, X.Y. Altered protein expression in serum from endometrial hyperplasia and carcinoma patients. J. Hematol. Oncol. 2011, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, Y.; Li, X.; Wei, L.L.; Pan, Z. Serum protein S100A9, SOD3, and MMP9 as new diagnostic biomarkers for pulmonary tuberculosis by iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics 2015, 15, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, Y.; Jin, X.; Zhang, L.; Zhou, Y. Urinary proteomics analysis for renal injury in hypertensive disorders of pregnancy with iTRAQ labeling and LC-MS/MS. Proteom. Clin. Appl. 2011, 5, 300–310. [Google Scholar] [CrossRef]
- Jing, M.; Ma, H.; Li, H.; Guo, B.; Zhang, X. Differential regulation of defense-related proteins in soybean during compatible and incompatible interactions between Phytophthora sojae and soybean by comparative proteomic analysis. Plant Cell. Rep. 2015, 34, 1263–1280. [Google Scholar] [CrossRef]
- Ma, H.; Song, L.; Shu, Y.; Wang, S.; Niu, J. Comparative proteomic analysis of seedling leaves of different salt tolerant soybean genotypes. J. Proteom. 2012, 75, 1529–1546. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, L.; Hua, Y.; Jia, X.; Li, J.; Hu, S. Comparative proteomic analysis of plasma membrane proteins between human osteosarcoma and normal osteoblastic cell lines. BMC Cancer 2010, 10, 206. [Google Scholar] [CrossRef]

| a Spot No. | b Accession | c Protein name | d M | e SC | f MP | g TpI | h Tmw | Nipponbare | Minghui | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i R-CK | i R-M | i R-SA | i M-CK | i M-M | i M-SA | ||||||||
| Energy related | |||||||||||||
| 1 | gi|115470529 | 23 kDa polypeptide of photosystem II, rice | 237 | 19% | 4 | 8.66 | 27.1 | 1.0 | 0.96 | 2.82 * | 1.0 | 1.42 | 1.84 |
| 2 | gi|19387272 | Chloroplastic glutamine synthetase, rice | 405 | 14% | 6 | 6.18 | 49.8 | 1.0 | 1.02 | 3.68 * | 1.0 | 1.19 | 1.28 |
| 3 | gi|54606800 | NADP dependent malic enzyme, rice | 274 | 9% | 5 | 5.79 | 65.8 | 1.0 | 0.52 | 1.26 | 1.0 | 0.97 | 0.19 * |
| 4 | gi|115475413 | Oxalate oxidase-like protein, rice | 227 | 13% | 2 | 5.48 | 24.7 | 1.0 | 0.88 | 0.48 * | 1.0 | 0.83 | 2.13 * |
| 5 | gi|115452897 | Uroporphyrinogen decarboxylase, rice | 96 | 5% | 3 | 6.15 | 43.0 | 1.0 | 0.73 | 2.19 * | 1.0 | 1.36 | 0.96 |
| 6 | gi|115440691 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, rice | 645 | 17% | 8 | 5.42 | 61.0 | 1.0 | 0.34 * | 1.34 | 1.0 | 1.66 | 1.35 |
| 7 | 88 | 8% | 3 | 5.42 | 61.0 | 1.0 | 0.85 | 1.14 | 1.0 | 0.44 * | 1.48 | ||
| 8 | gi|115480295 | 6-phosphogluconolactonase, rice | 262 | 20% | 5 | 5.46 | 29.1 | 1.0 | 0.5 * | 0.48 * | 1.0 | 1.29 | 5.44 * |
| 9 | gi|46391135 | ATP synthase beta chain, rice | 277 | 17% | 5 | 5.66 | 43.1 | 1.0 | 0.33 * | 0.56 | 1.0 | 0.79 | 0.69 |
| 10 | gi|56784991 | ATP synthase beta subunit, rice | 241 | 19% | 6 | 5.33 | 45.9 | 1.0 | 0.87 | 0.38 * | 1.0 | 0.98 | 0.75 |
| 11 | gi|6815115 | ATP synthase beta subunit, rice | 700 | 21% | 8 | 5.38 | 54.0 | 1.0 | 0.90 | 1.12 | 1.0 | 0.25 * | 0.43 * |
| 12 | gi|11466784 | ATP synthase CF1 alpha subunit, rice | 85 | 4% | 2 | 5.95 | 55.7 | 1.0 | 0.46 * | 0.70 | 1.0 | 0.63 | 0.45 * |
| 13 | gi|11466784 | ATP synthase CF1 alpha subunit, rice | 655 | 23% | 10 | 5.95 | 55.7 | 1.0 | 1.25 | 1.43 | 1.0 | 2.08 * | 1.05 |
| 14 | gi|11466784 | ATP synthase CF1 alpha subunit, rice | 512 | 21% | 8 | 5.95 | 55.7 | 1.0 | 0.16 * | 0.91 | 1.0 | 2.74 * | 3.82 * |
| 15 | gi|115476908 | ATP synthase D chain, mitochondrial, rice | 77 | 21% | 3 | 5.19 | 19.7 | 1.0 | 0.40 * | 0.61 | 1.0 | 0.80 | 1.06 |
| 16 | gi|194033257 | ATP synthase F0 subunit 1, rice | 330 | 16% | 6 | 5.85 | 55.7 | 1.0 | 0.88 | 1.84 | 1.0 | 1.93 | 2.38 * |
| 17 | gi|110289207 | Chaperonin CPN60-1, rice | 158 | 5% | 4 | 6.95 | 67.6 | 1.0 | 0.85 | 1.19 | 1.0 | 1.30 | 3.72 * |
| 18 | gi|115465974 | 6-phosphogluconate dehydrogenase, rice | 376 | 13% | 6 | 5.85 | 52.9 | 1.0 | 0.86 | 1.74 | 1.0 | 0.41 * | 1.25 |
| 19 | gi|2267006 | Endosperm lumenal binding protein, rice | 102 | 3% | 2 | 5.30 | 73.7 | 1.0 | 1.01 | 1.23 | 1.0 | 0.37 * | 0.22 * |
| 20 | gi|780372 | Enolase, rice | 321 | 12% | 7 | 5.42 | 48.3 | 1.0 | 0.00 * | 0.65 | 1.0 | 0.72 | 0.76 |
| 21 | gi|115468758 | Enoyl-CoA hydratase/isomerase, rice | 152 | 8% | 5 | 6.48 | 47.3 | 1.0 | 1.07 | 4.21 * | 1.0 | 0.95 | 1.08 |
| 22 | gi|115452127 | Fructose-1,6-bisphosphatase, rice | 216 | 8% | 4 | 5.00 | 44.1 | 1.0 | 0.55 | 0.98 | 1.0 | 2.77 * | 2.96 * |
| 23 | gi|115448167 | Inorganic pyrophosphatase-like protein, rice | 202 | 23% | 4 | 5.56 | 24.4 | 1.0 | 0.98 | 1.12 | 1.0 | 0.48 * | 0.67 |
| 24 | gi|34393836 | Ribose-5-phosphate isomerase, rice | 249 | 22% | 4 | 4.91 | 27.1 | 1.0 | 0.99 | 1.14 | 1.0 | 1.37 | 2.17 * |
| 25 | gi|27261025 | Thiamine biosynthesis protein, rice | 284 | 20% | 7 | 5.44 | 37.2 | 1.0 | 1.13 | 0.01 * | 1.0 | 1.32 | 1.68 |
| 26 | gi|553107 | Triosephosphate isomerase, rice | 231 | 15% | 4 | 6.60 | 27.8 | 1.0 | 1.07 | 1.44 | 1.0 | 1.40 | 2.48 * |
| 27 | gi|115469362 | Vacuolar H+-ATPase subunit A, rice | 410 | 9% | 6 | 5.20 | 68.7 | 1.0 | 0.50 | 1.39 | 1.0 | 2.81 * | 2.30 * |
| 28 | gi|62733870 | Chlorophyll a/b-binding protein CP26, rice | 243 | 28% | 5 | 5.95 | 24.3 | 1.0 | 0.42 * | 0.85 | 1.0 | 1.12 | 1.03 |
| 29 | gi|255571784 | Glutamate-1-semialdehyde 2,1-aminomutase, rice | 77 | 8% | 3 | 5.99 | 50.8 | 1.0 | 0.62 | 0.93 | 1.0 | 2.51 * | 3.69 * |
| 30 | gi|32129323 | Magnesium chelatase subunit chlD, rice | 170 | 8% | 6 | 5.38 | 81.0 | 1.0 | 0.68 | 0.99 | 1.0 | 2.05 * | 1.23 |
| 31 | gi|115448091 | Phosphoribulokinase, chloroplast, rice | 214 | 8% | 4 | 5.68 | 45.2 | 1.0 | 0.56 | 0.71 | 1.0 | 0.37 * | 0.40 * |
| 32 | gi|21839 | Phosphoribulokinase, rice | 45 | 2% | 2 | 5.84 | 45.4 | 1.0 | 3.73 * | 6.29 * | 1.0 | 2.15 * | 6.65 * |
| 33 | gi|34394725 | Photosystem I reaction center subunit IV, rice | 57 | 22% | 2 | 9.64 | 15.5 | 1.0 | 0.29 * | 0.30 * | 1.0 | 0.99 | 2.07 * |
| 34 | gi|11466807 | Photosystem II protein V, rice | 93 | 26% | 2 | 4.64 | 9.4 | 1.0 | 0.24 * | 0.44 * | 1.0 | 1.10 | 1.45 |
| 35 | gi|11955 | Ribulose-1,5-bisphosphate carboxylase, rice | 103 | 6% | 3 | 6.13 | 53.4 | 1.0 | 0.23 * | 0.77 | 1.0 | 0.43 * | 1.43 |
| 36 | gi|11466795 | Ribulose-1,5-bisphosphate carboxylase, rice | 122 | 10% | 4 | 6.22 | 53.4 | 1.0 | 0.65 | 2.82 * | 1.0 | 0.35 | 1.15 |
| 37 | gi|115439241 | Similar to ATP synthase beta chain, rice | 376 | 17% | 7 | 6.10 | 59.6 | 1.0 | 0.45 * | 1.05 | 1.0 | 0.79 | 1.19 |
| 38 | gi|218210 | Ribulose-1,5-bisphosphate carboxylase, rice | 156 | 20% | 3 | 9.11 | 19.8 | 1.0 | 0.29 * | 1.59 | 1.0 | 6.43 * | 3.78 * |
| 39 | gi|115456265 | NAD-dependent epimerase/dehydratase, rice | 124 | 11% | 2 | 6.34 | 27.9 | 1.0 | 1.33 | 2.18 * | 1.0 | 1.39 | 1.11 |
| 40 | gi|4760553 | Nad-dependent formate dehydrogenase, rice | 207 | 14% | 4 | 6.87 | 41.4 | 1.0 | 0.97 | 1.01 | 1.0 | 2.43 * | 3.23 * |
| 41 | gi|115443619 | NADH-hydroxypyruvate reductase, rice | 237 | 14% | 6 | 6.56 | 42.3 | 1.0 | 2.03 * | 5.27 * | 1.0 | 1.14 | 0.20 * |
| Signal transduction | |||||||||||||
| 42 | gi|115458806 | 14-3-3-like protein GF14-6, rice | 376 | 27% | 7 | 4.76 | 29.9 | 1.0 | 0.69 | 0.42 * | 1.0 | 0.83 | 1.64 |
| 43 | gi|108712139 | Ankyrin repeat domain protein 2, rice | 291 | 20% | 5 | 4.64 | 37.3 | 1.0 | 0.86 | 1.68 | 1.0 | 1.22 | 2.21 * |
| 44 | gi|303859 | Brain specific protein, rice | 77 | 11% | 2 | 4.77 | 29.2 | 1.0 | 0.69 | 1.05 | 1.0 | 0.81 | 7.12 * |
| 45 | gi|303859 | Brain specific protein, rice | 438 | 32% | 6 | 4.77 | 29.2 | 1.0 | 0.48 * | 0.35 * | 1.0 | 2.01 * | 2.63 * |
| 46 | gi|303859 | Brain specific protein, rice | 204 | 17% | 5 | 4.77 | 29.2 | 1.0 | 0.35 * | 0.40 * | 1.0 | 1.33 | 3.09 * |
| 47 | gi|34393219 | Calreticulin precursor, rice | 221 | 7% | 4 | 4.49 | 49.0 | 1.0 | 0.82 | 2.76 * | 1.0 | 1.48 | 2.04 * |
| 48 | gi|115445587 | GTP-binding protein typA, rice | 186 | 7% | 4 | 7.08 | 74.0 | 1.0 | 0.92 | 2.65 * | 1.0 | 0.68 | 0.76 |
| 49 | gi|12957710 | GTP-binding protein, rice | 460 | 18% | 8 | 7.03 | 46.9 | 1.0 | 1.14 | 2.66 * | 1.0 | 0.90 | 1.17 |
| 50 | gi|6682927 | Importin alpha 1b, rice | 194 | 10% | 4 | 5.18 | 59.1 | 1.0 | 0.67 | 1.17 | 1.0 | 1.02 | 4.29 * |
| 51 | gi|115471679 | Spermidine synthase, rice | 90 | 8% | 2 | 5.23 | 35.6 | 1.0 | 0.75 | 2.01 * | 1.0 | 1.00 | 1.39 |
| 52 | gi|115476928 | TaWIN2, rice | 290 | 21% | 4 | 4.85 | 29.1 | 1.0 | 0.89 | 1.92 | 1.0 | 3.00 * | 1.29 |
| 53 | gi|115456491 | TolB, C-terminal domain protein, rice | 191 | 5% | 3 | 5.45 | 73.9 | 1.0 | 2.60 * | 1.93 | 1.0 | 1.32 | 2.60 * |
| 54 | gi|115452585 | Glutamyl endopeptidase, rice | 162 | 5% | 4 | 5.66 | 104.4 | 1.0 | 0.99 | 0.28 * | 1.0 | 2.32 * | 3.55 * |
| 55 | gi|115456491 | TolB, C-terminal domain protein, rice | 121 | 7% | 4 | 5.45 | 73.9 | 1.0 | 1.27 | 4.11 * | 1.0 | 1.10 | 1.47 |
| 56 | gi|115458498 | UVB-resistance protein UVR8, rice | 79 | 5% | 2 | 5.37 | 48.7 | 1.0 | 0.97 | 1.82 | 1.0 | 0.87 | 0.50 * |
| Redox homeostasis | |||||||||||||
| 57 | gi|115479659 | Glutathione S-transferase GST 23, rice | 111 | 15% | 4 | 5.50 | 25.3 | 1.0 | 0.24 * | 0.80 | 1.0 | 0.23 * | 1.30 |
| 58 | gi|11177845 | Glutathione S-transferase OsGSTF3, rice | 228 | 15% | 4 | 5.81 | 25.1 | 1.0 | 0.86 | 0.65 | 1.0 | 1.37 | 2.21 * |
| 59 | gi|115477793 | Glutathione S-transferase, rice | 101 | 9% | 3 | 6.84 | 37.5 | 1.0 | 0.32 * | 0.45 * | 1.0 | 0.77 | 0.76 |
| 60 | gi|601869 | Manganese superoxide dismutase, rice | 469 | 35% | 7 | 6.50 | 24.9 | 1.0 | 0.95 | 0.82 | 1.0 | 3.28 * | 2.96 * |
| 61 | gi|50252391 | Putative glyoxalase I, rice | 103 | 8% | 3 | 5.82 | 32.4 | 1.0 | 0.37 * | 1.29 | 1.0 | 1.02 | 1.39 |
| 62 | gi|538430 | superoxide dismutase, rice | 58 | 19% | 2 | 5.71 | 15.3 | 1.0 | 0.42 * | 0.73 | 1.0 | 2.41 * | 1.94 |
| 63 | gi|115475151 | Lactoylglutathione lyase, rice | 266 | 20% | 5 | 5.51 | 32.9 | 1.0 | 0.38 * | 0.63 | 1.0 | 0.59 | 1.05 |
| 64 | gi|115439131 | Peroxiredoxin, rice | 87 | 16% | 2 | 5.58 | 17.3 | 1.0 | 0.77 | 0.48 * | 1.0 | 1.44 | 1.75 |
| 65 | gi|115476190 | Quinone oxidoreductase-like protein, rice | 163 | 8% | 3 | 7.63 | 39.7 | 1.0 | 0.43 * | 1.04 | 1.0 | 0.36 * | 0.38 * |
| 66 | gi|115111257 | Betaine aldehyde dehydrogenase, rice | 147 | 7% | 3 | 5.29 | 55.4 | 1.0 | 0.75 | 0.95 | 1.0 | 2.37 * | 2.06 * |
| 67 | gi|53370754 | C1-like domain containing protein, rice | 241 | 7% | 5 | 5.37 | 85.6 | 1.0 | 0.71 | 0.81 | 1.0 | 0.94 | 5.31 * |
| 68 | gi|115456828 | 2-hydroxyacid dehydrogenase, rice | 84 | 7% | 2 | 6.00 | 34.7 | 1.0 | 0.96 | 4.20 * | 1.0 | 0.30 * | 0.47 * |
| 69 | gi|41052915 | Ferredoxin-NADP(H) oxidoreductase, rice | 222 | 19% | 6 | 7.98 | 41.1 | 1.0 | 0.69 | 0.88 | 1.0 | 1.19 | 0.10 * |
| 70 | gi|41052915 | Ferredoxin-NADP(H) oxidoreductase, rice | 206 | 21% | 6 | 7.98 | 41.1 | 1.0 | 0.34 * | 0.92 | 1.0 | 1.98 | 1.13 |
| 71 | gi|115461843 | Isovaleryl-CoA dehydrogenase, rice | 152 | 10% | 5 | 6.52 | 45.1 | 1.0 | 0.46 * | 0.48 * | 1.0 | 1.82 | 1.89 |
| 72 | gi|115474285 | L-ascorbate peroxidase, rice | 500 | 35% | 6 | 5.21 | 27.2 | 1.0 | 0.50 * | 0.61 | 1.0 | 0.84 | 0.62 |
| 73 | gi|37718877 | Methylenetetrahydrofolate reductase, rice | 201 | 14% | 4 | 6.10 | 42.2 | 1.0 | 0.37 * | 0.74 | 1.0 | 0.95 | 1.25 |
| 74 | gi|115453457 | PDI-like protein, rice | 270 | 12% | 5 | 4.95 | 64.0 | 1.0 | 0.13 * | 0.20 * | 1.0 | 5.09 * | 13.92 * |
| 75 | gi|115472727 | Rieske iron-sulfur protein, rice | 180 | 11% | 3 | 8.54 | 24.2 | 1.0 | 0.53 | 0.47 * | 1.0 | 1.50 | 1.26 |
| 76 | gi|115436382 | Thioredoxin domain 2 containing protein, rice | 90 | 7% | 2 | 6.43 | 41.1 | 1.0 | 1.23 | 0.84 | 1.0 | 2.45 * | 1.52 |
| 77 | gi|297728925 | Thioredoxin, rice | 294 | 23% | 4 | 8.16 | 18.9 | 1.0 | 0.95 | 0.43 * | 1.0 | 1.35 | 0.96 |
| Amino acid and nitrogen metabolism | |||||||||||||
| 78 | gi|115455323 | Cysteine synthase, rice | 155 | 12% | 3 | 5.35 | 34.4 | 1.0 | 0.42 * | 0.92 | 1.0 | 1.24 | 1.36 |
| 79 | gi|108862998 | Cysteine synthase, rice | 59 | 4% | 1 | 8.76 | 43.8 | 1.0 | 1.08 | 0.97 | 1.0 | 0.92 | 3.38 * |
| 80 | gi|115442595 | Cysteine synthase, rice | 531 | 22% | 7 | 6.28 | 42.1 | 1.0 | 0.30 * | 0.51 | 1.0 | 1.23 | 1.09 |
| 81 | gi|22748337 | Glutamate ammonia ligase, rice | 139 | 6% | 2 | 5.73 | 38.8 | 1.0 | 0.41 * | 0.95 | 1.0 | 4.37 * | 1.36 |
| 82 | gi|115465569 | Ketol-acid reductoisomerase, chloroplast, rice | 225 | 9% | 4 | 6.01 | 62.7 | 1.0 | 0.26 * | 1.48 | 1.0 | 1.89 | 1.47 |
| 83 | gi|115452263 | Ornithine acetyltransferase, rice | 118 | 6% | 2 | 6.45 | 48.3 | 1.0 | 0.71 | 0.12 * | 1.0 | 1.38 | 1.10 |
| Carbohydrate metabolism | |||||||||||||
| 84 | gi|218155 | Chloroplastic aldolase, rice | 267 | 11% | 5 | 7.60 | 42.4 | 1.0 | 0.46 * | 0.64 | 1.0 | 1.06 | 1.40 |
| 85 | gi|218155 | Chloroplastic aldolase, rice | 456 | 14% | 6 | 7.60 | 42.4 | 1.0 | 0.09 * | 0.62 | 1.0 | 1.57 | 1.39 |
| 86 | gi|115482534 | Cytoplasmic malate dehydrogenase, rice | 171 | 18% | 4 | 5.75 | 35.9 | 1.0 | 0.26 * | 0.37 * | 1.0 | 1.66 | 1.97 |
| 87 | gi|2497857 | Malate dehydrogenase, mitochondrial, rice | 320 | 12% | 4 | 8.81 | 35.9 | 1.0 | 1.34 | 0.98 | 1.0 | 4.66 * | 4.70 * |
| 88 | gi|115477843 | Malate dehydrogenase, NADP-dependent, rice | 188 | 6% | 2 | 6.96 | 47.5 | 1.0 | 0.27 * | 1.00 | 1.0 | 0.36 * | 0.76 |
| 89 | gi|4105561 | Ribulose-5-phosphate-3-epimerase, rice | 173 | 11% | 2 | 8.93 | 29.2 | 1.0 | 0.75 | 2.30 * | 1.0 | 0.48 * | 0.77 |
| 90 | gi|3024122 | S-adenosylmethionine synthase 2, rice | 378 | 18% | 5 | 5.68 | 43.3 | 1.0 | 0.41 * | 1.02 | 1.0 | 1.59 | 0.77 |
| Protein folding and assembly | |||||||||||||
| 91 | gi|115479353 | 20 kDa chaperonin, chloroplast, rice | 173 | 36% | 5 | 5.97 | 25.5 | 1.0 | 0.81 | 0.96 | 1.0 | 1.24 | 0.23 * |
| 92 | gi|222631026 | Chloroplast, Hsp70, rice | 336 | 7% | 4 | 5.12 | 73.7 | 1.0 | 1.00 | 1.59 | 1.0 | 1.95 | 0.30 * |
| 93 | gi|218161 | Elongation factor 1 beta, rice | 329 | 23% | 5 | 4.86 | 23.8 | 1.0 | 1.28 | 1.00 | 1.0 | 1.99 | 2.36 * |
| 94 | gi|115489714 | Glycine-rich RNA-binding protein 1, rice | 506 | 45% | 6 | 6.32 | 16.1 | 1.0 | 0.64 | 0.49 * | 1.0 | 0.75 | 0.61 |
| 95 | gi|27476086 | Hsp 70, mitochondrial precursor, rice | 288 | 8% | 6 | 5.46 | 70.7 | 1.0 | 0.36 * | 0.58 | 1.0 | 1.14 | 1.35 |
| 96 | gi|115456247 | Heat shock 70 kDa protein, rice | 523 | 14% | 9 | 5.10 | 71.7 | 1.0 | 0.98 | 1.16 | 1.0 | 5.64 * | 1.18 |
| 97 | gi|115448989 | Heat shock 70 kDa protein, rice | 406 | 13% | 8 | 5.49 | 73.1 | 1.0 | 0.97 | 1.44 | 1.0 | 3.07 * | 4.74 * |
| 98 | gi|18855040 | Heat shock protein 90, rice | 439 | 15% | 9 | 4.89 | 93.0 | 1.0 | 0.48 * | 1.21 | 1.0 | 1.33 | 1.27 |
| 99 | gi|39104468 | Heat shock protein 90, rice | 579 | 19% | 10 | 4.98 | 80.4 | 1.0 | 0.75 | 0.94 | 1.0 | 1.83 | 0.45 * |
| 100 | gi|39104468 | Heat shock protein 90, rice | 434 | 12% | 8 | 4.98 | 80.4 | 1.0 | 0.70 | 0.26 * | 1.0 | 6.29 * | 2.32 * |
| 101 | gi|108862740 | Hsp 90 protein, rice | 122 | 5% | 3 | 5.02 | 79.7 | 1.0 | 0.41 * | 0.37 * | 1.0 | 1.48 | 1.40 |
| 102 | gi|115459670 | Lambda integrase-like, rice | 333 | 26% | 6 | 5.57 | 25.8 | 1.0 | 0.96 | 2.64 * | 1.0 | 0.76 | 0.49 * |
| 103 | gi|77557101 | Methyl-CpG binding domain protein, rice | 156 | 17% | 4 | 4.74 | 31.5 | 1.0 | 0.91 | 1.55 | 1.0 | 2.07 * | 1.26 |
| 104 | gi|115470141 | PDX1-like protein 4, rice | 185 | 12% | 5 | 6.41 | 33.9 | 1.0 | 0.82 | 1.02 | 1.0 | 0.72 | 2.31 * |
| Protein hydrolysis | |||||||||||||
| 105 | gi|42408435 | Aminopeptidase N, rice | 296 | 11% | 7 | 5.42 | 98.6 | 1.0 | 0.75 | 0.42 * | 1.0 | 1.02 | 1.23 |
| 106 | gi|42408435 | Aminopeptidase N, rice | 152 | 4% | 3 | 5.42 | 98.6 | 1.0 | 0.47 * | 1.98 | 1.0 | 1.16 | 1.86 |
| 107 | gi|42408435 | Aminopeptidase N, rice | 65 | 4% | 2 | 5.42 | 98.6 | 1.0 | 0.35 * | 0.37 * | 1.0 | 1.51 | 1.30 |
| 108 | gi|215694277 | Aminopeptidase, rice | 96 | 4% | 3 | 5.38 | 100.1 | 1.0 | 2.46 * | 1.14 | 1.0 | 1.30 | 1.35 |
| 109 | gi|29468084 | Aspartate aminotransferase, rice | 391 | 18% | 6 | 5.90 | 46.0 | 1.0 | 0.08 * | 0.86 | 1.0 | 1.59 | 2.07 * |
| 110 | gi|115434854 | Similar to Ubiquitin-specific protease 14, rice | 94 | 1% | 1 | 5.08 | 89.2 | 1.0 | 0.62 | 1.05 | 1.0 | 2.20 * | 1.77 |
| 111 | gi|115449043 | Subtilisin-like protease, rice | 192 | 7% | 4 | 5.95 | 81.4 | 1.0 | 0.19 * | 0.47 * | 1.0 | 2.45 * | 1.21 |
| 112 | gi|115482934 | Glycine cleavage system H protein, rice | 88 | 8% | 1 | 4.92 | 17.5 | 1.0 | 1.30 | 2.39 * | 1.0 | 0.69 | 0.79 |
| 113 | gi|8671508 | Beta 4 subunit of 20S proteasome, rice | 160 | 16% | 3 | 5.42 | 23.6 | 1.0 | 0.41 * | 1.07 | 1.0 | 0.37 * | 1.22 |
| 114 | gi|115444057 | Proteasome subunit alpha type 1, rice | 215 | 16% | 3 | 5.37 | 29.9 | 1.0 | 0.71 | 0.99 | 1.0 | 1.08 | 3.16 * |
| 115 | gi|115447473 | Proteasome subunit alpha type 2, rice | 196 | 15% | 4 | 5.39 | 25.8 | 1.0 | 0.76 | 0.87 | 1.0 | 1.05 | 2.60 * |
| 116 | gi|115486269 | Proteasome subunit alpha type 5, rice | 250 | 31% | 5 | 4.70 | 26.1 | 1.0 | 0.37 * | 0.48 * | 1.0 | 1.58 | 3.14 * |
| 117 | gi|14091862 | Putative hydrolase, rice | 233 | 9% | 3 | 9.17 | 41.4 | 1.0 | 0.93 | 0.64 | 1.0 | 1.86 | 2.37 * |
| Protein synthesis | |||||||||||||
| 118 | gi|115469770 | 60S acidic ribosomal protein P3, rice | 86 | 22% | 2 | 4.40 | 11.9 | 1.0 | 0.69 | 0.37 * | 1.0 | 0.78 | 2.14 * |
| 119 | gi|6525065 | Translational elongation factor Tu, rice | 84 | 4% | 2 | 6.05 | 50.5 | 1.0 | 0.41 * | 0.65 | 1.0 | 4.07 * | 7.31 * |
| 120 | gi|88866516 | UDP-glucose pyrophosphorylase, rice | 103 | 4% | 2 | 5.43 | 51.8 | 1.0 | 1.15 | 1.78 | 1.0 | 2.21 | 1.56 |
| 121 | gi|115449577 | 29 kDa ribonucleoprotein, chloroplast, rice | 235 | 12% | 4 | 5.17 | 34.9 | 1.0 | 0.78 | 0.72 | 1.0 | 0.40 * | 0.63 |
| 122 | gi|108707824 | 30S ribosomal protein S1, chloroplast, rice | 380 | 18% | 8 | 4.70 | 43.6 | 1.0 | 1.08 | 1.15 | 1.0 | 1.39 | 2.33 * |
| 123 | gi|115456525 | 30S ribosomal protein S6, chloroplast, rice | 82 | 12% | 2 | 7.79 | 23.4 | 1.0 | 0.48 * | 0.80 | 1.0 | 2.10 * | 1.06 |
| 124 | gi|115456525 | 30S ribosomal protein S6, chloroplast, rice | 121 | 16% | 3 | 7.79 | 23.4 | 1.0 | 1.87 | 2.76 * | 1.0 | 2.11 * | 2.11 * |
| 125 | gi|115463659 | 50S ribosomal protein L1, rice | 268 | 14% | 3 | 6.86 | 38.9 | 1.0 | 0.71 | 0.80 | 1.0 | 2.08 * | 2.06 * |
| 126 | gi|77548531 | 60S acidic ribosomal protein P0, rice | 127 | 9% | 2 | 5.38 | 34.5 | 1.0 | 0.15 * | 0.82 | 1.0 | 1.60 | 1.79 |
| 127 | gi|115442127 | NAC-alpha-like protein 3, rice | 88 | 16% | 2 | 4.39 | 22.1 | 1.0 | 0.51 | 0.92 | 1.0 | 0.48 * | 1.06 |
| 128 | gi|115480705 | Nucleic acid-binding protein precursor, rice | 254 | 21% | 6 | 4.41 | 35.4 | 1.0 | 0.77 | 1.09 | 1.0 | 3.36 * | 1.97 |
| 129 | gi|45433309 | Nucleosome assembly protein 1, rice | 125 | 10% | 3 | 4.34 | 42.7 | 1.0 | 0.25 * | 0.45 * | 1.0 | 0.75 | 1.13 |
| 130 | gi|115477393 | Peptidylprolyl isomerase, rice | 223 | 9% | 4 | 5.15 | 64.4 | 1.0 | 1.04 | 1.01 | 1.0 | 1.39 | 0.34 * |
| 131 | gi|115437444 | RNA helicase, rice | 78 | 7% | 3 | 7.03 | 56.5 | 1.0 | 0.81 | 4.01 * | 1.0 | 1.97 | 0.83 |
| 132 | gi|215769434 | RNA-binding protein 8A, rice | 196 | 30% | 5 | 5.18 | 22.1 | 1.0 | 1.02 | 0.34 * | 1.0 | 2.54 * | 1.47 |
| 133 | gi|125541223 | Zn-dependent peptidases, rice | 140 | 4% | 4 | 5.41 | 121.9 | 1.0 | 0.74 | 1.40 | 1.0 | 0.19 * | 0.14 * |
| Others and unknown | |||||||||||||
| 134 | gi|115477851 | CaLB domain containing protein, rice | 150 | 29% | 9 | 4.77 | 30.4 | 1.0 | 0.76 | 1.02 | 1.0 | 0.91 | 2.70 * |
| 135 | gi|115450773 | Cell division control protein 48 homolog A, rice | 314 | 7% | 6 | 5.12 | 90.4 | 1.0 | 0.43 * | 0.24 * | 1.0 | 6.91 * | 1.55 |
| 136 | gi|295885 | Actin, rice | 140 | 13% | 4 | 5.29 | 42.2 | 1.0 | 0.44 * | 0.86 | 1.0 | 0.10 * | 1.12 |
| 137 | gi|115434036 | Isoflavone reductase, rice | 205 | 15% | 4 | 5.69 | 33.5 | 1.0 | 0.45 * | 0.86 | 1.0 | 0.67 | 0.78 |
| 138 | gi|115457122 | O-methyltransferase, rice | 376 | 17% | 6 | 5.33 | 40.9 | 1.0 | 0.23 * | 0.77 | 1.0 | 0.66 | 0.32 * |
| 139 | gi|4158221 | Reversibly glycosylated polypeptide, rice | 123 | 6% | 2 | 5.82 | 41.9 | 1.0 | 0.16 * | 0.67 | 1.0 | 1.67 | 0.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Hwarari, D.; Zhang, Y.; Mo, X.; Luo, Y.; Ma, H. Proteomic Analysis Reveals Salicylic Acid as a Pivotal Signal Molecule in Rice Response to Blast Disease Infection. Plants 2022, 11, 1702. https://doi.org/10.3390/plants11131702
Zhou H, Hwarari D, Zhang Y, Mo X, Luo Y, Ma H. Proteomic Analysis Reveals Salicylic Acid as a Pivotal Signal Molecule in Rice Response to Blast Disease Infection. Plants. 2022; 11(13):1702. https://doi.org/10.3390/plants11131702
Chicago/Turabian StyleZhou, Haiying, Delight Hwarari, Yunhui Zhang, Xiaosong Mo, Yuming Luo, and Hongyu Ma. 2022. "Proteomic Analysis Reveals Salicylic Acid as a Pivotal Signal Molecule in Rice Response to Blast Disease Infection" Plants 11, no. 13: 1702. https://doi.org/10.3390/plants11131702
APA StyleZhou, H., Hwarari, D., Zhang, Y., Mo, X., Luo, Y., & Ma, H. (2022). Proteomic Analysis Reveals Salicylic Acid as a Pivotal Signal Molecule in Rice Response to Blast Disease Infection. Plants, 11(13), 1702. https://doi.org/10.3390/plants11131702






