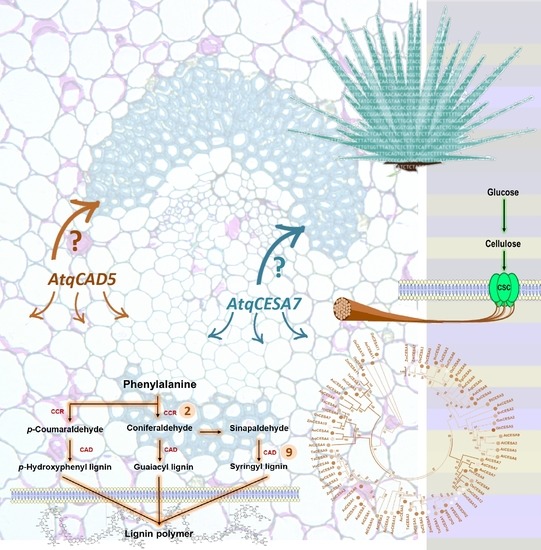
Transcriptome Mining Provides Insights into Cell Wall Metabolism and Fiber Lignification in Agave tequilana Weber
Abstract
1. Introduction
2. Results
2.1. Identification of Orthologous Biosynthetic Genes for Cellulose and Lignin in A. tequilana
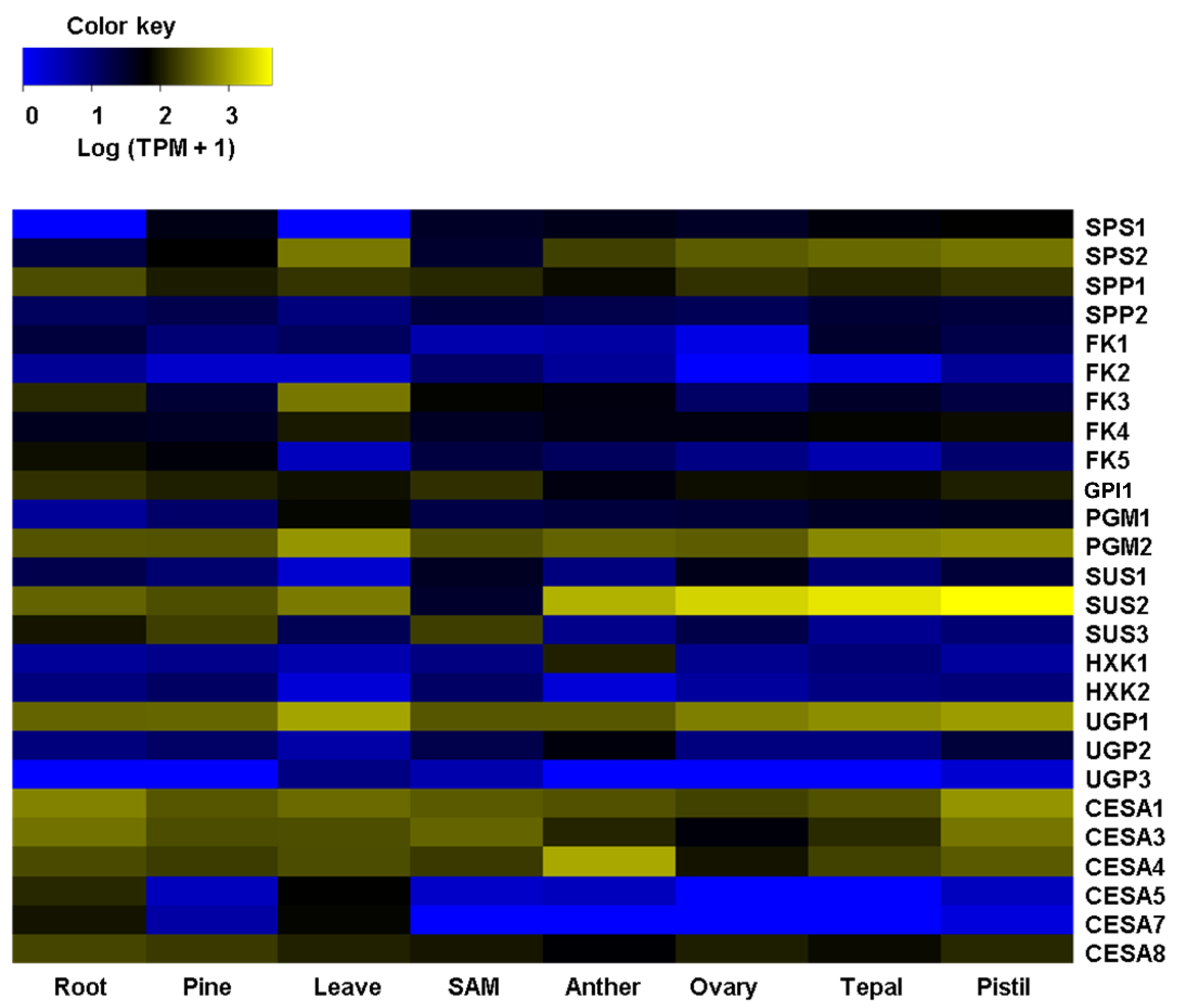
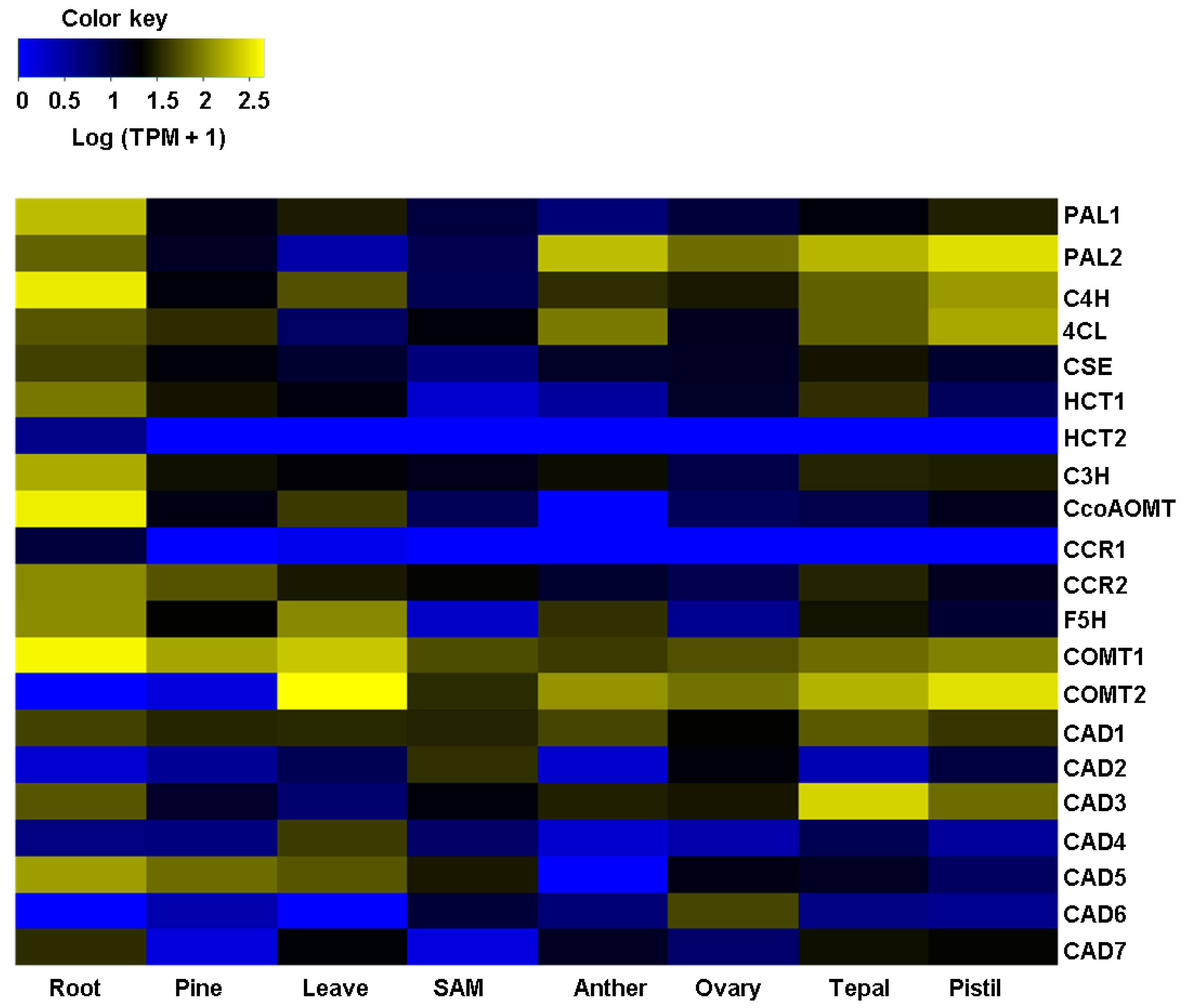
2.2. Agave Cellulose and Lignin-Related Predicted Genes Are Differentially Expressed in Different Agave Tissues
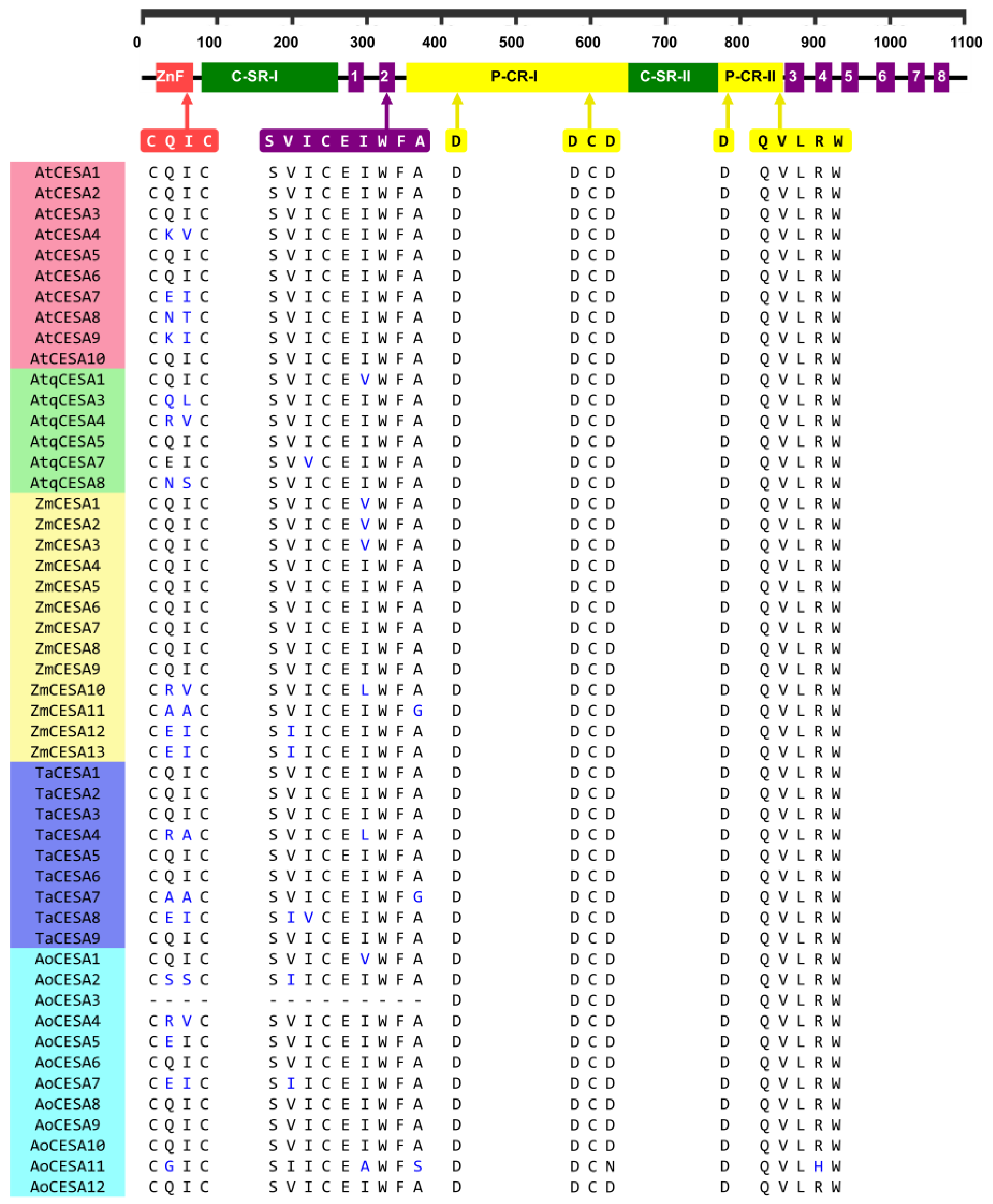
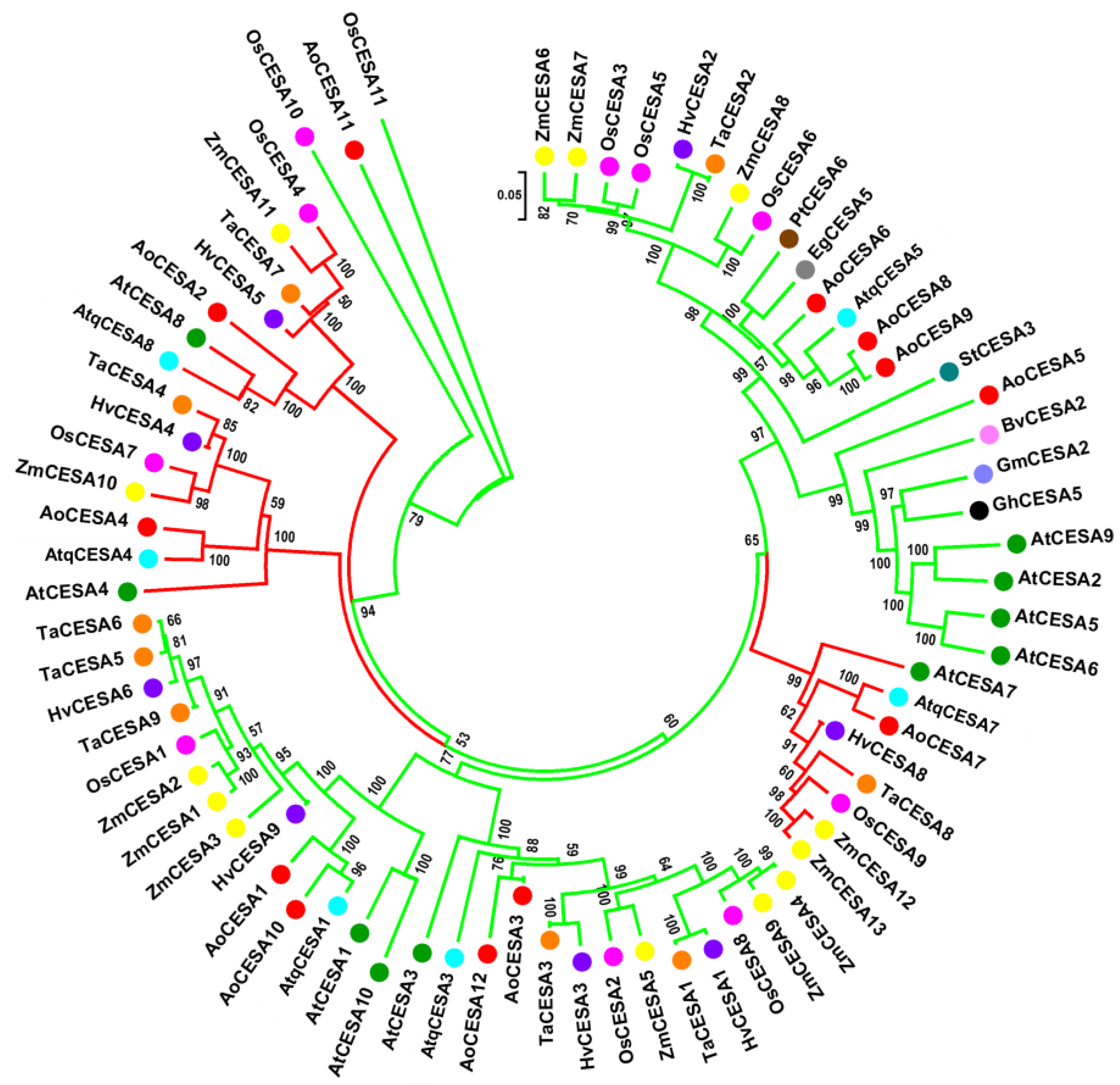
2.3. Structural and Phylogenetic Characteristics of AtqCESA and AtqCAD Proteins
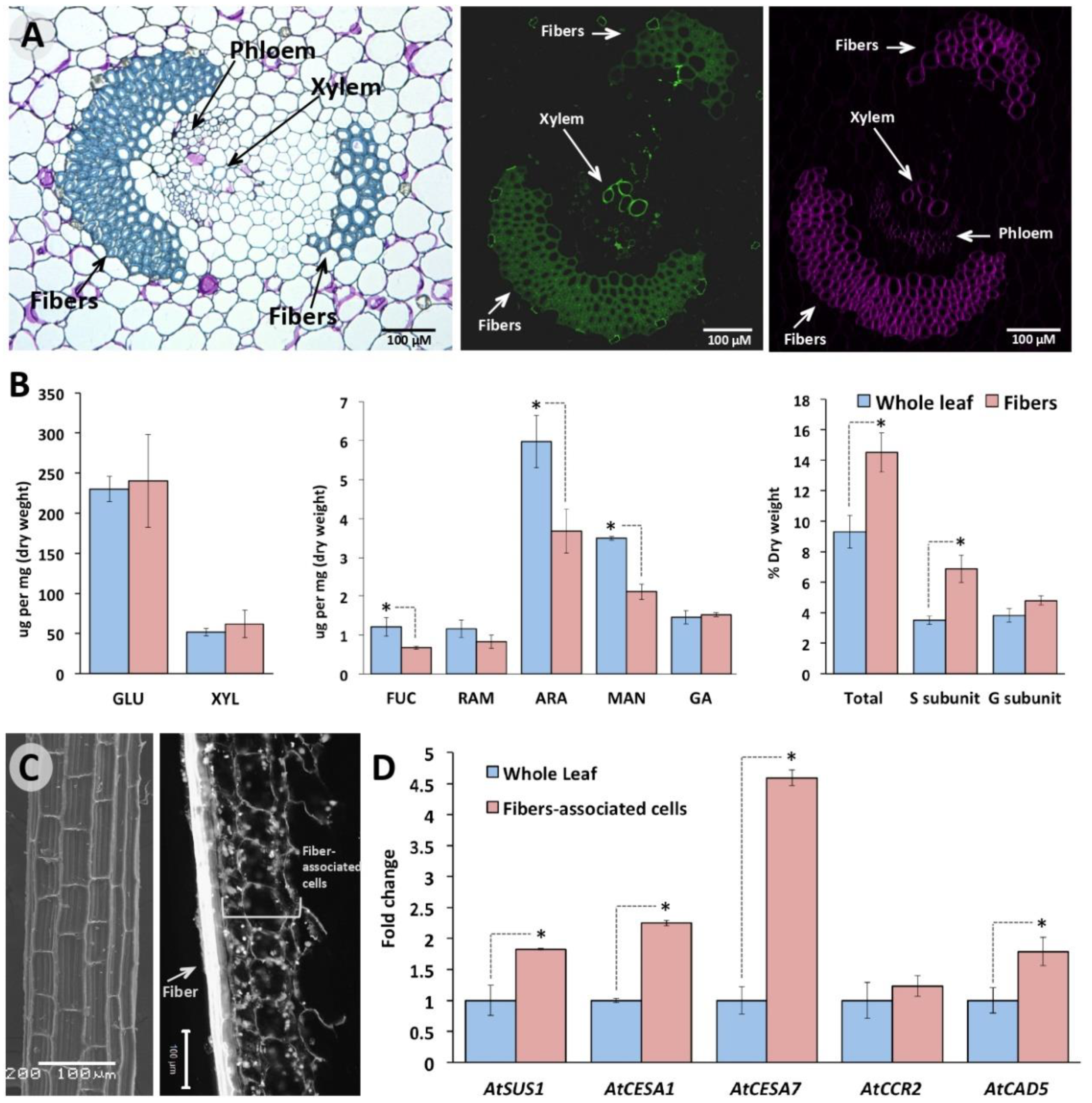
2.4. Retrieved Agave Orthologous Genes for Cellulose and Lignin Biosynthesis Are Preferentially Expressed in Cells Surrounding Leaf Sclerenchyma Fibers
3. Discussion
4. Materials and Methods
4.1. Data Mining for Cell Wall-Related Genes
4.2. Orthologous Groups Identification and Protein Subcellular Localization Prediction
4.3. Sequence Alignment and Phylogenetics of AtqCESA and AtqCAD Families
4.4. In Silico Gene Expression Analysis of Cellulose and Lignin Biosynthesis
4.5. Samples Collection and Processing
4.6. Monosaccharide and Lignin Composition
4.7. Histochemistry and Microscopy
4.8. RT-qPCR Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lampugnani, E.R.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131, jcs207373. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Keegstra, K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008, 54, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.O.G.; Vaz, R.P.; Filho, E.X.F. Bringing plant cell wall-degrading enzymes into the lignocellulosic biorefinery concept. Biofuel Bioprod. Biorefin. 2018, 12, 277–289. [Google Scholar] [CrossRef]
- Prasad, R.K.; Chatterjee, S.; Mazumder, P.B.; Gupta, S.K.; Sharma, S.; Vairale, M.G.; Datta, S.; Dwivedi, S.K.; Gupta, D.K. Bioethanol production from waste lignocelluloses: A review on microbial degradation potential. Chemosphere 2019, 231, 588–606. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Zarra, I.; Revilla, G.; Sampedro, J.; Valdivia, E.R. Biosynthesis and regulation of secondary cell wall. In Progress in Botany; Cánovas, F.M., Lüttge, U., Leuschner, C., Risueño, M.-C., Eds.; Springer International Publishing: Cham, Switzerland, 2020; Volume 81, pp. 189–226. [Google Scholar] [CrossRef]
- Davis, S.C.; Dohleman, F.G.; Long, S.P. The global potential for agave as a biofuel feedstock. GCB Bioenergy 2011, 3, 68–78. [Google Scholar] [CrossRef]
- Escamilla-treviño, L.L. potential of plants from the genus agave as bioenergy crops. Bioenergy Res. 2012, 5, 1–9. [Google Scholar] [CrossRef]
- Gross, S.M.; Martin, J.A.; Simpson, J.; Abraham-Juarez, M.J.; Wang, Z.; Visel, A. De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genom. 2013, 14, 563. [Google Scholar] [CrossRef]
- Corbin, K.R.; Byrt, C.S.; Bauer, S.; DeBolt, S.; Chambers, D.; Holtum, J.A.M.; Karem, G.; Henderson, M.; Lahnstein, J.; Beahan, C.T.; et al. Prospecting for energy-rich renewable raw materials: Agave leaf case study. PLoS ONE 2015, 10, e0135382. [Google Scholar] [CrossRef]
- Lu, M.L.; Wyman, C.E. Elucidation of native California Agave americana and Agave deserti biofuel potential: Compositional analysis. PLoS ONE 2021, 16, e0252201. [Google Scholar] [CrossRef]
- Avila de Dios, E.; Gomez Vargas, A.D.; Damián Santos, M.L.; Simpson, J. New insights into plant glycoside hydrolase family 32 in agave species. Front. Plant Sci. 2015, 6, 594. [Google Scholar] [CrossRef][Green Version]
- Abraham, P.E.; Yin, H.; Borland, A.M.; Weighill, D.; Lim, S.D.; De Paoli, H.C.; Engle, N.; Jones, P.C.; Agh, R.; Weston, D.J.; et al. Transcript, protein and metabolite temporal dynamics in the CAM plant agave. Nat. Plants 2016, 2, 16178. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Pérez, S.A.; Espinal-Centeno, A.; Oropeza-Aburto, A.; Caballero-Pérez, J.; Falcon, F.; Aragón-Raygoza, A.; Sánchez-Segura, L.; Herrera-Estrella, L.; Cruz-Hernández, A.; Cruz-Ramírez, A. Transcriptional profiling of the CAM plant Agave salmiana reveals conservation of a genetic program for regeneration. Dev. Biol. 2018, 442, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, B.; Xi, J.; Zhang, Y.; He, C.; Zheng, J.; Gao, J.; Chen, H.; Zhang, S.; Wu, W.; et al. Transcriptome comparison reveals distinct selection patterns in domesticated and wild agave species, the important cam plants. Int. J. Genom. 2018, 2018, 5716518. [Google Scholar] [CrossRef] [PubMed]
- de Dios, E.A.; Delaye, L.; Simpson, J. Transcriptome analysis of bolting in A. tequilana reveals roles for florigen, MADS, fructans and gibberellins. BMC Genom. 2019, 20, 473. [Google Scholar] [CrossRef]
- Huang, X.; Xiao, M.; Xi, J.; He, C.; Zheng, J.; Chen, H.; Gao, J.; Zhang, S.; Wu, W.; Liang, Y.; et al. De novo transcriptome assembly of Agave H11648 by Illumina sequencing and identification of cellulose synthase genes in agave species. Genes 2019, 10, 103. [Google Scholar] [CrossRef]
- Sarwar, M.B.; Ahmad, Z.; Rashid, B.; Hassan, S.; Gregersen, P.L.; De la O Leyva, M.L.; Nagy, I.; Asp, T.; Husnain, T. De novo assembly of Agave sisalana transcriptome in response to drought stress provides insight into the tolerance mechanisms. Sci. Rep. 2019, 9, 396. [Google Scholar] [CrossRef]
- Trigo Raya, F.; Marone Pupke, M.; Carvalho, L.M.; Rabelo, S.C.; de Paula, M.S.; Zaneli Campanari, M.F.; Freschi, L.; Sampaio Mayer, J.L.; Ribeiro Ferreira Silva, O.R.; Mieczkowski, P.; et al. Extreme physiology: Biomass and transcriptional profiling of three abandoned Agave cultivars. Ind. Crops Prod. 2021, 172, 114043. [Google Scholar] [CrossRef]
- Penning, B.W.; Hunter, C.T., III; Tayengwa, R.; Eveland, A.L.; Dugard, C.K.; Olek, A.T.; Vermerris, W.; Koch, K.E.; McCarty, D.R.; Davis, M.F.; et al. Genetic resources for maize cell wall biology. Plant Physiol. 2009, 151, 1703–1728. [Google Scholar] [CrossRef]
- Wang, L.; Guo, K.; Li, Y.; Tu, Y.; Hu, H.; Wang, B.; Cui, X.; Peng, L. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010, 10, 282. [Google Scholar] [CrossRef]
- Courtial, A.; Soler, M.; Chateigner Boutin, A.L.; Reymond, M.; Mechin, V.; Wang, H.; Grima-Pettenati, J.; Barriere, Y. Breeding grasses for capacity to biofuel production or silage feeding value: An updated list of genes involved in maize secondary cell wall biosynthesis and assembly. Maydica 2013, 58, 67–102. [Google Scholar]
- Penning, B.W.; McCann, M.C.; Carpita, N.C. Evolution of the cell wall gene families of grasses. Front. Plant Sci. 2019, 10, 1205. [Google Scholar] [CrossRef] [PubMed]
- Penning, B.W.; Shiga, T.M.; Klimek, J.F.; SanMiguel, P.J.; Shreve, J.; Thimmapuram, J.; Sykes, R.W.; Davis, M.F.; McCann, M.C.; Carpita, N.C. Expression profiles of cell-wall related genes vary broadly between two common maize inbreds during stem development. BMC Genom. 2019, 20, 785. [Google Scholar] [CrossRef]
- Zhao, K.; Lin, F.; Romero-Gamboa, S.P.; Saha, P.; Goh, H.-J.; An, G.; Jung, K.-H.; Hazen, S.P.; Bartley, L.E. Rice genome-scale network integration reveals transcriptional regulators of grass cell wall synthesis. Front. Plant Sci. 2019, 10, 1275. [Google Scholar] [CrossRef]
- Abraham-Juarez, M.J.; Martinez-Hernandez, A.; Leyva-Gonzalez, M.A.; Herrera-Estrella, L.; Simpson, J. Class I KNOX genes are associated with organogenesis during bulbil formation in Agave tequilana. J. Exp. Bot. 2010, 61, 4055–4067. [Google Scholar] [CrossRef] [PubMed]
- Abraham Juárez, M.J.; Hernández Cárdenas, R.; Santoyo Villa, J.N.; O’Connor, D.; Sluis, A.; Hake, S.; Ordaz-Ortiz, J.; Terry, L.; Simpson, J. Functionally different PIN proteins control auxin flux during bulbil development in Agave tequilana. J. Exp. Bot. 2015, 66, 3893–3905. [Google Scholar] [CrossRef]
- Liepman, A.H.; Wightman, R.; Geshi, N.; Turner, S.R.; Scheller, H.V. Arabidopsis a powerful model system for plant cell wall research. Plant J. 2010, 61, 1107–1121. [Google Scholar] [CrossRef]
- Li, S.; Bashline, L.; Lei, L.; Gu, Y. Cellulose synthesis and its regulation. Arab. Book 2014, 12, e0169. [Google Scholar] [CrossRef]
- Maloney, V.J.; Park, J.Y.; Unda, F.; Mansfield, S.D. Sucrose phosphate synthase and sucrose phosphate phosphatase interact in planta and promote plant growth and biomass accumulation. J. Exp. Bot. 2015, 66, 4383–4394. [Google Scholar] [CrossRef]
- Stein, O.; Granot, D. Plant fructokinases: Evolutionary, developmental, and metabolic aspects in sink tissues. Front. Plant Sci. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Bilska-Kos, A.; Mytych, J.; Suski, S.; Magoń, J.; Ochodzki, P.; Zebrowski, J. Sucrose phosphate synthase (SPS), sucrose synthase (SUS) and their products in the leaves of Miscanthus × giganteus and Zea mays at low temperature. Planta 2020, 252, 23. [Google Scholar] [CrossRef] [PubMed]
- Muhlbach, H.; Schnarrenberger, C. Properties and intracellular distribution of two phosphoglucomutases from spinach leaves. Planta 1978, 141, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Periappuram, C.; Steinhauer, L.; Barton, D.L.; Taylor, D.C.; Chatson, B.; Zou, J. The plastidic phosphoglucomutase from Arabidopsis. A reversible enzyme reaction with an important role in metabolic control. Plant Physiol. 2000, 122, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Maceda-López, L.F.; Villalpando-Aguilar, J.L.; García-Hernández, E.; Ávila de Dios, E.; Andrade-Canto, S.B.; Morán-Velázquez, D.C.; Rodríguez-López, L.; Hernández-Díaz, D.; Chablé-Vega, M.A.; Trejo, L.; et al. Improved method for isolation of high quality total RNA from Agave tequilana Weber roots. 3 Biotech 2021, 11, 75. [Google Scholar] [CrossRef]
- Lacombe, E.; Hawkins, S.; Van Doorsselaere, J.; Piquemal, J.; Goffner, D.; Poeydomenge, O.; Boudet, A.-M.; Grima-Pettenati, J. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: Cloning, expression and phylogenetic relationships. Plant J. 1997, 11, 429–441. [Google Scholar] [CrossRef]
- Kaur, S.; Dhugga, K.S.; Gill, K.; Singh, J. Novel structural and functional motifs in cellulose synthase (CesA) genes of bread wheat (Triticum aestivum, L.). PLoS ONE 2016, 11, e0147046. [Google Scholar] [CrossRef][Green Version]
- Scavuzzo-Duggan, T.R.; Chaves, A.M.; Singh, A.; Sethaphong, L.; Slabaugh, E.; Yingling, Y.G.; Haigler, C.H.; Roberts, A.W. Cellulose synthase ‘class specific regions’ are intrinsically disordered and functionally undifferentiated. J. Integr. Plant Biol. 2018, 60, 481–497. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Lin, X.; Chan, T.-F.; Imtiaz, M.; Rehman, H.M.; Ali, M.A.; Baloch, F.S.; Atif, R.M.; Yang, S.H.; Chung, G. Characterization of cellulose synthase A (CESA) gene family in Eudicots. Biochem. Genet. 2019, 57, 248–272. [Google Scholar] [CrossRef]
- Vergara, C.E.; Carpita, N.C. β-D-Glycan synthases and the CesA gene family: Lessons to be learned from the mixed-linkage (1→3),(1→4)β-D-glucan synthase. In Plant Cell Walls; Carpita, N.C., Campbell, M., Tierney, M., Eds.; Springer: Dordrecht, The Netherlands, 2001. [Google Scholar] [CrossRef]
- Beeckman, T.; Przemeck, G.K.H.; Stamatiou, G.; Lau, R.; Terryn, N.; De Rycke, R.; Inzé, D.; Berleth, T. Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol. 2002, 130, 1883–1893. [Google Scholar] [CrossRef]
- Xu, L. Overexpression of a new cellulose synthase gene (‘PuCesA6’) from Ussuri poplar (‘Populus ussuriensis’) exhibited a dwarf phenotype in transgenic tobacco. Plant Omics 2014, 7, 54. [Google Scholar]
- Cao, S.; Cheng, H.; Zhang, J.; Aslam, M.; Yan, M.; Hu, A.; Lin, L.; Ojolo, S.P.; Zhao, H.; Priyadarshani, S.V.G.N.; et al. Genome-wide identification, expression pattern analysis and evolution of the Ces/Csl gene superfamily in pineapple (Ananas comosus). Plants 2019, 8, 275. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Zhang, X.Q.; Li, Y.H.; Xie, X.M. Cloning and in silico analysis of a cinnamyl alcohol dehydrogenase gene in Pennisetum purpureum. J. Genet. 2014, 93, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Youn, B.; Camacho, R.; Moinuddin, S.G.A.; Lee, C.; Davin, L.B.; Lewis, N.G.; Kang, C.H. Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org. Biomol. Chem. 2006, 4, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.-M.; Ran, J.-H.; Wang, X.-Q. Evolution of the cinnamyl/sinapyl clcohol dehydrogenase (CAD/SAD) gene family: The emergence of real lignin is associated with the origin of bona fide CAD. J. Mol. Evol. 2010, 71, 202–218. [Google Scholar] [CrossRef] [PubMed]
- Bukh, C.; Nord-Larsen, P.H.; Rasmussen, S.K. Phylogeny and structure of the cinnamyl alcohol dehydrogenase gene family in Brachypodium distachyon. J. Exp. Bot. 2012, 63, 6223–6236. [Google Scholar] [CrossRef]
- Park, H.L.; Kim, T.L.; Bhoo, S.H.; Lee, T.H.; Lee, S.-W.; Cho, M.-H. Biochemical characterization of the rice cinnamyl alcohol dehydrogenase gene family. Molecules 2018, 23, 2659. [Google Scholar] [CrossRef]
- Bomati, E.K.; Noel, J.P. Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase. Plant Cell. 2005, 17, 1598–1611. [Google Scholar] [CrossRef][Green Version]
- Cheng, X.; Li, M.; Li, D.; Zhang, J.; Jin, Q.; Sheng, L.; Cai, Y.; Lin, Y. Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol. Open. 2017, 6, 1602–1613. [Google Scholar] [CrossRef]
- Evert, R.F. Esau’s Plant Anatomy, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef]
- Lledías, F.; Gutiérrez, J.; Martínez-Hernández, A.; García-Mendoza, A.; Sosa, E.; Hernández-Bermúdez, F.; Dinkova, T.D.; Reyes, S.; Cassab, G.I.; Nieto-Sotelo, J. Mayahuelin, a type I ribosome inactivating protein: Characterization, evolution, and utilization in phylogenetic analyses of Agave. Front. Plant Sci. 2020, 11, 573. [Google Scholar] [CrossRef]
- Appenzeller, L.; Doblin, M.; Barreiro, R.; Wang, H.; Niu, X.; Kollipara, K.; Carrigan, L.; Tomes, D.; Chapman, M.; Dhugga, K.S. Cellulose synthesis in maize: Isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 2004, 11, 287–299. [Google Scholar] [CrossRef]
- Wang, D.; Qin, Y.; Fang, J.; Yuan, S.; Peng, L.; Zhao, J.; Li, X. A missense mutation in the zinc finger domain of OsCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice. PLoS ONE 2016, 11, e0153993. [Google Scholar] [CrossRef] [PubMed]
- Mazarei, M.; Baxter, H.L.; Li, M.; Biswal, A.K.; Kim, K.; Meng, X.; Pu, Y.; Wuddineh, W.A.; Zhang, J.-Y.; Turner, G.B.; et al. Functional analysis of cellulose synthase CesA4 and CesA6 genes in switchgrass (Panicum virgatum) by overexpression and RNAi-mediated gene silencing. Front. Plant Sci. 2018, 9, 1114. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, S.; Lu, X.; Li, C.; Liu, X.; Dong, G.; Xia, T. Tissue-specific transcriptome analysis reveals lignocellulose synthesis regulation in elephant grass (Pennisetum purpureum Schum). BMC Plant Biol. 2020, 20, 528. [Google Scholar] [CrossRef] [PubMed]
- Guillaumie, S.; Goffner, D.; Barbier, O.; Martinant, J.-P.; Pichon, M.; Barrière, Y. Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT) down-regulated, and normal maize plants. BMC Plant Biol. 2008, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Jardim-Messeder, D.; da Franca Silva, T.; Fonseca, J.P.; Junior, J.N.; Barzilai, L.; Felix-Cordeiro, T.; Pereira, J.C.; Rodrigues-Ferreira, C.; Bastos, I.; da Silva, T.C.; et al. Identification of genes from the general phenylpropanoid and monolignol-specific metabolism in two sugarcane lignin-contrasting genotypes. Mol. Genet. Genom. 2020, 295, 717–739. [Google Scholar] [CrossRef]
- Hirano, K.; Aya, K.; Kondo, M.; Okuno, A.; Morinaka, Y.; Matsuoka, M. OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep. 2012, 31, 91–101. [Google Scholar] [CrossRef]
- Ferreira, S.S.; Simoes, M.S.; Carvalho, G.G.; De Lima, L.G.A.; Svartman, R.M.A.; Cesarino, I. The lignin toolbox of the model grass Setaria viridis. Plant Mol. Biol. 2019, 101, 235–255. [Google Scholar] [CrossRef]
- Lv, L.; Sun, G.; Xie, J.; Zang, X.; Hu, Y.; Duan, J. Determination of chromosomal ploidy in Agave ssp. Afr. J. Biotechnol. 2009, 8, 5248–5252. [Google Scholar]
- Simpson, J.; Martínez Hernández, A.; Abraham Juárez, M.J.; Delgado Sandoval, S.; Sánchez Villarreal, A.; Cortés Romero, C. Genomic resources and transcriptome mining in Agave tequilana. GCB Bioenergy 2011, 3, 25–36. [Google Scholar] [CrossRef]
- Tamayo-Ordóñez, M.C.; Rodriguez-Zapata, L.C.; Narváez-Zapata, J.A.; Tamayo-Ordóñez, Y.J.; Ayil-Gutiérrez, B.A.; Barredo-Pool, F.; Sánchez-Teyer, L.F. Morphological features of different polyploids for adaptation and molecular characterization of CC-NBS-LRR and LEA gene families in Agave L. J. Plant Physiol. 2016, 195, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl Shikimate Esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.M.; Escamilla-Trevino, L.; Yarce, J.C.S.; Kim, H.; Ralph, J.; Chen, F.; Dixon, R.A. An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J. 2016, 86, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013, 4, 62. [Google Scholar] [CrossRef]
- Xue, J.-S.; Zhang, B.; Zhan, H.; Lv, Y.-L.; Jia, X.-L.; Wang, T.; Yang, N.-Y.; Lou, Y.-X.; Zhang, Z.-B.; Hu, W.-J.; et al. Phenylpropanoid derivatives are essential components of sporopollenin in vascular plants. Mol. Plant 2020, 13, 1644–1653. [Google Scholar] [CrossRef]
- Subramanian, S.S.; Nair, A.G.R. Chlorogenin and kaempferol glycosides from the flowers of Agave americana. Phytochemistry 1970, 9, 2582. [Google Scholar] [CrossRef]
- Almaraz-Abarca, N.; Delgado-Alvarado, E.A.; Hernández-Vargas, V.; Ortega-Chávez, M.; Orea-Lara, G.; Cifuentes-Díaz de León, A.; Ávila-Reyes, J.A.; Muñiz-Martínez, R. Profiling of phenolic compounds of somatic and reproductive tissues of Agave durangensis Gentry (Agavaceae). Am. J. Appl. Sci. 2009, 6, 1076–1085. [Google Scholar] [CrossRef]
- Chen, Z.; Hong, X.; Zhang, H.; Wang, Y.; Li, X.; Zhu, J.-K.; Gong, Z. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 2005, 43, 273–283. [Google Scholar] [CrossRef]
- Müssig, J.; Slootmaker, T. Types of fibre. In Industrial Application of Natural Fibres: Structure, Properties, and Technical Applications; Müssig, J., Ed.; John Wiley & Sons: Chichester, UK, 2010. [Google Scholar]
- Smith, R.A.; Schuetz, M.; Roach, M.; Mansfield, S.D.; Ellis, B.; Samuels, L. Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell. 2013, 25, 3988–3999. [Google Scholar] [CrossRef]
- Chernova, T.E.; Gorshkova, T.A. Biogenesis of plant fibers. Russ. J. Dev. Biol. 2007, 38, 221–232. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.; Loose, M. AlignWise: A tool for identifying protein-coding sequence and correcting frame-shifts. BMC Bioinforma. 2015, 16, 376. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, S. Performance criteria for graph clustering and Markov cluster experiments. In Technical Report INS-R 0012; National Research Institute for Mathematics and Computer Science: Amsterdam, The Netherlands, 2000; Available online: http://ftp.cwi.nl//CWIreports/INS/INS-R0012.pdf (accessed on 1 April 2022).
- Enright, J.A.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome. Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: Javascript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Morais de Carvalho, D.; Martínez-Abad, A.; Evtuguin, D.V.; Colodette, J.L.; Lindström, M.E.; Vilaplana, F.; Sevastyanova, O. Isolation and characterization of acetylated glucuronoarabinoxylan from sugarcane bagasse and straw. Carbohydr. Polym. 2017, 156, 223–234. [Google Scholar] [CrossRef]
- Morán-Velázquez, D.C.; Monribot-Villanueva, J.L.; Bourdon, M.; Tang, J.Z.; López-Rosas, I.; Maceda-López, L.F.; Villalpando-Aguilar, J.L.; Rodríguez-López, L.; Gauthier, A.; Trejo, L.; et al. Unravelling chemical composition of Agave spines: News from Agave fourcroydes Lem. Plants 2020, 9, 1642. [Google Scholar] [CrossRef] [PubMed]
- Mellado-Mojica, E.; Seeram, N.P.; López, M.G. Comparative analysis of maple syrups and natural sweeteners: Carbohydrates composition and classification (differentiation) by HPAEC-PAD and FTIR spectroscopy-chemometrics. J. Food Compos. Anal. 2016, 52, 1–8. [Google Scholar] [CrossRef]
- Sykes, R.; Yung, M.; Novaes, E.; Kirst, M.; Peter, G.; Davis, M. High-throughput screening of plant cell-wall composition using pyrolysis molecular beam mass spectroscopy. In Biofuels: Methods and Protocols; Mielenz, J.R., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 581, pp. 169–183. [Google Scholar] [CrossRef]
- Sarwar, M.B.; Ahmad, Z.; Anicet, B.A.; Sajid, M.; Rashid, B.; Hassan, S.; Ahmed, M.; Husnain, T. Identification and validation of superior housekeeping gene(s) for qRT-PCR data normalization in Agave sisalana (a CAM-plant) under abiotic stresses. Physiol. Mol. Biol. Plants 2000, 26, 567–584. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]


| Gene | ORF (nt) | Protein (aa) | Calculated Mw (Da) | Theoretical pI | Predicted Subcellular Location and Score * |
|---|---|---|---|---|---|
| AtqSPS1 | 3168 | 1055 | 118,083.33 | 5.77 | nucl: 5, cyto: 4, chlo: 3 |
| AtqSPS2 | 3252 | 1083 | 120,102.55 | 6.11 | nucl: 5, cyto: 5, chlo: 2 |
| AtqSPP1 | 1275 | 424 | 47,733.35 | 5.54 | cyto: 5, nucl: 4, chlo: 2 |
| AtqSPP2 | 1275 | 424 | 48,024.71 | 6.00 | chlo: 7, nucl: 3, mito: 3 |
| AtqFK1 | 651 | 216 | 23,897.62 | 5.62 | chlo: 6, cyto: 5, cysk_nucl: 2 |
| AtqFK2 | 969 | 322 | 34,489.54 | 5.20 | cyto: 8, chlo: 3, pero: 2 |
| AtqFK3 | 996 | 331 | 35,476.84 | 5.03 | pero: 11, chlo: 1, nucl: 1 |
| AtqFK4 | 1224 | 407 | 43,435.34 | 6.13 | chlo: 14 |
| AtqFK5 | 1107 | 368 | 39,509.00 | 5.50 | cysk: 9, cyto: 3, chlo: 1 |
| AtqGPI1 | 1521 | 506 | 55,292.25 | 4.97 | cyto: 9, chlo: 3, mito: 1 |
| AtqPGM1 | 1872 | 623 | 68,121.76 | 6.82 | chlo: 14 |
| AtqPGM2 | 1749 | 582 | 63,450.95 | 6.02 | cyto: 11, chlo: 2, nucl: 1 |
| AtqSUS1 | 2532 | 843 | 96,280.19 | 8.07 | cyto: 12, chlo: 1, plas: 1 |
| AtqSUS2 | 2448 | 815 | 92,525.91 | 6.03 | cyto: 9, chlo: 2, nucl: 1 |
| AtqSUS3 | 2424 | 807 | 92,436.65 | 5.83 | cyto: 5, mito: 4, chlo: 3 |
| AtqHXK1 | 1488 | 495 | 53,179.08 | 6.07 | chlo: 13, mito: 1 |
| AtqHXK2 | 1497 | 498 | 54,454.50 | 5.87 | chlo: 11, nucl: 1, extr: 1 |
| AtqUGP1 | 1422 | 473 | 51,920.96 | 6.09 | cyto: 11, chlo: 2, E.R.: 1 |
| AtqUGP2 | 1575 | 524 | 58,194.63 | 7.74 | chlo: 7, mito: 6, nucl: 1 |
| AtqUGP3 | 2262 | 853 | 93,824.45 | 6.52 | chlo: 10, mito: 2, nucl: 1.5 |
| AtqCESA1 | 3264 | 1087 | 122,300.01 | 6.46 | plas: 13, E.R.: 1 |
| AtqCESA3 | 3180 | 1059 | 119,098.30 | 8.14 | plas: 10, nucl: 2, mito: 1 |
| AtqCESA4 | 3174 | 1057 | 120,445.31 | 6.90 | plas: 11, nucl: 2, E.R.: 1 |
| AtqCESA5 | 3273 | 1090 | 122,477.03 | 6.85 | plas: 13, E.R.: 1 |
| AtqCESA7 | 3153 | 1058 | 118,824.62 | 6.34 | plas: 12, nucl: 1, E.R.: 1 |
| AtqCESA8 | 3063 | 1020 | 114,790.86 | 6.57 | plas: 12, nucl: 1, vacu: 1 |
| Gene | ORF (nt) | Protein Size (aa) | Calculated Mw (Da) | Theoretical pI | Predicted Subcellular Location and Score * |
|---|---|---|---|---|---|
| AtqPAL1 | 2103 | 700 | 75,908.60 | 5.60 | chlo: 4, plas: 3, cyto: 2 |
| AtqPAL2 | 2112 | 703 | 75,997.81 | 5.92 | chlo: 4, plas: 3, cyto: 2 |
| AtqC4H | 1518 | 505 | 57,804.27 | 9.19 | plas: 10, E.R.: 3, vacu: 1 |
| Atq4CL | 1689 | 562 | 59,781.88 | 5.52 | E.R.: 5.5, E.R._plas: 3.5, cyto: 3 |
| AtqCSE | 990 | 329 | 36,767.97 | 6.09 | nucl: 4, cyto: 4, mito: 2, |
| AtqHCT1 | 1326 | 441 | 48,196.19 | 6.22 | cyto: 7, chlo: 4, nucl: 2 |
| AtqHCT2 | 1302 | 433 | 47,684.51 | 6.02 | cyto: 10, chlo: 3, mito: 1 |
| AtqC3H | 1539 | 512 | 58,118.34 | 7.23 | chlo: 13, mito: 1 |
| AtqCCoAOMT | 771 | 256 | 28,666.75 | 5.30 | cyto: 7, chlo: 3, cysk: 2 |
| AtqCCR1 | 981 | 326 | 35,680.92 | 6.18 | chlo: 6, plas: 3, vacu: 2 |
| AtqCCR2 | 1017 | 338 | 36,883.26 | 5.73 | chlo: 6, plas: 4, vacu: 2 |
| AtqF5H | 1545 | 514 | 57,197.12 | 6.81 | chlo: 5, plas: 4, nucl: 2.5 |
| AtqCOMT1 | 1092 | 363 | 39,707.87 | 5.58 | cyto: 9, chlo: 2, cysk: 2 |
| AtqCOMT2 | 1095 | 364 | 39,372.20 | 4.76 | cyto: 10, cysk: 2, chlo: 1 |
| AtqCAD1 | 1062 | 353 | 38,482.99 | 7.05 | cysk: 8, extr: 3, chlo: 1 |
| AtqCAD2 | 1107 | 368 | 39,485.53 | 6.75 | cysk: 11, cyto: 2, pero: 1 |
| AtqCAD3 | 1071 | 356 | 38,497.48 | 6.16 | cyto: 8, cysk: 4, plas: 1 |
| AtqCAD4 | 1065 | 354 | 37,854.72 | 6.86 | cyto: 11, cysk: 2, plas: 1 |
| AtqCAD5 | 1074 | 357 | 38,286.33 | 5.82 | cyto: 10, mito: 1, plas: 1 |
| AtqCAD6 | 1113 | 370 | 39,936.18 | 6.60 | cyto: 4, chlo: 3, nucl: 2 |
| AtqCAD7 | 1065 | 354 | 38,779.55 | 7.55 | cysk: 9, cyto: 2, extr: 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maceda-López, L.F.; Góngora-Castillo, E.B.; Ibarra-Laclette, E.; Morán-Velázquez, D.C.; Girón Ramírez, A.; Bourdon, M.; Villalpando-Aguilar, J.L.; Toomer, G.; Tang, J.Z.; Azadi, P.; et al. Transcriptome Mining Provides Insights into Cell Wall Metabolism and Fiber Lignification in Agave tequilana Weber. Plants 2022, 11, 1496. https://doi.org/10.3390/plants11111496
Maceda-López LF, Góngora-Castillo EB, Ibarra-Laclette E, Morán-Velázquez DC, Girón Ramírez A, Bourdon M, Villalpando-Aguilar JL, Toomer G, Tang JZ, Azadi P, et al. Transcriptome Mining Provides Insights into Cell Wall Metabolism and Fiber Lignification in Agave tequilana Weber. Plants. 2022; 11(11):1496. https://doi.org/10.3390/plants11111496
Chicago/Turabian StyleMaceda-López, Luis F., Elsa B. Góngora-Castillo, Enrique Ibarra-Laclette, Dalia C. Morán-Velázquez, Amaranta Girón Ramírez, Matthieu Bourdon, José L. Villalpando-Aguilar, Gabriela Toomer, John Z. Tang, Parastoo Azadi, and et al. 2022. "Transcriptome Mining Provides Insights into Cell Wall Metabolism and Fiber Lignification in Agave tequilana Weber" Plants 11, no. 11: 1496. https://doi.org/10.3390/plants11111496
APA StyleMaceda-López, L. F., Góngora-Castillo, E. B., Ibarra-Laclette, E., Morán-Velázquez, D. C., Girón Ramírez, A., Bourdon, M., Villalpando-Aguilar, J. L., Toomer, G., Tang, J. Z., Azadi, P., Santamaría, J. M., López-Rosas, I., López, M. G., Simpson, J., & Alatorre-Cobos, F. (2022). Transcriptome Mining Provides Insights into Cell Wall Metabolism and Fiber Lignification in Agave tequilana Weber. Plants, 11(11), 1496. https://doi.org/10.3390/plants11111496