ZmLBD5 Increases Drought Sensitivity by Suppressing ROS Accumulation in Arabidopsis
Abstract
1. Introduction
2. Results
2.1. ZmLBD5 Was Induced by Osmotic Stress in Maize
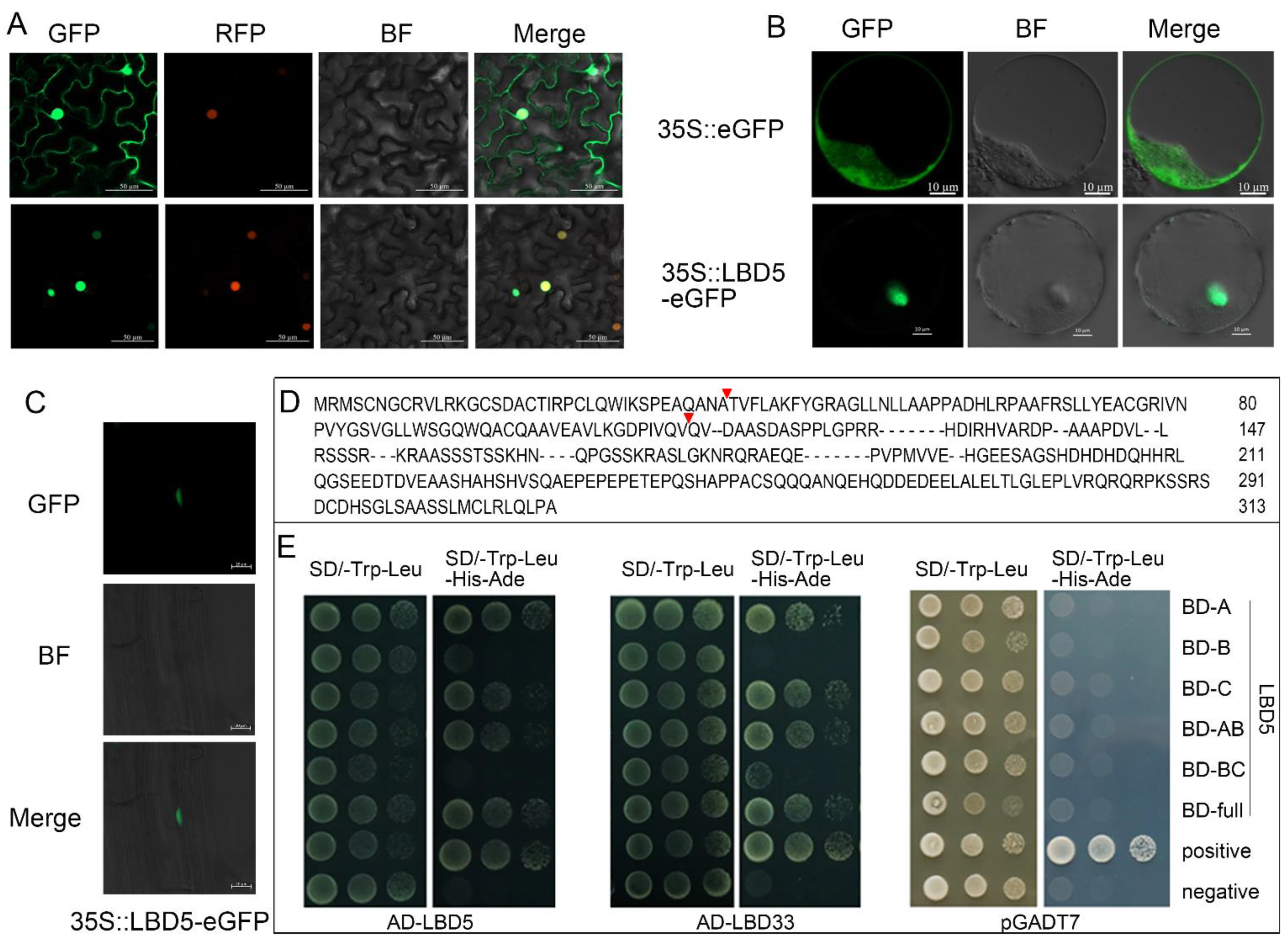
2.2. ZmLBD5 Is Localized in the Nucleus, and Could Form Dimers
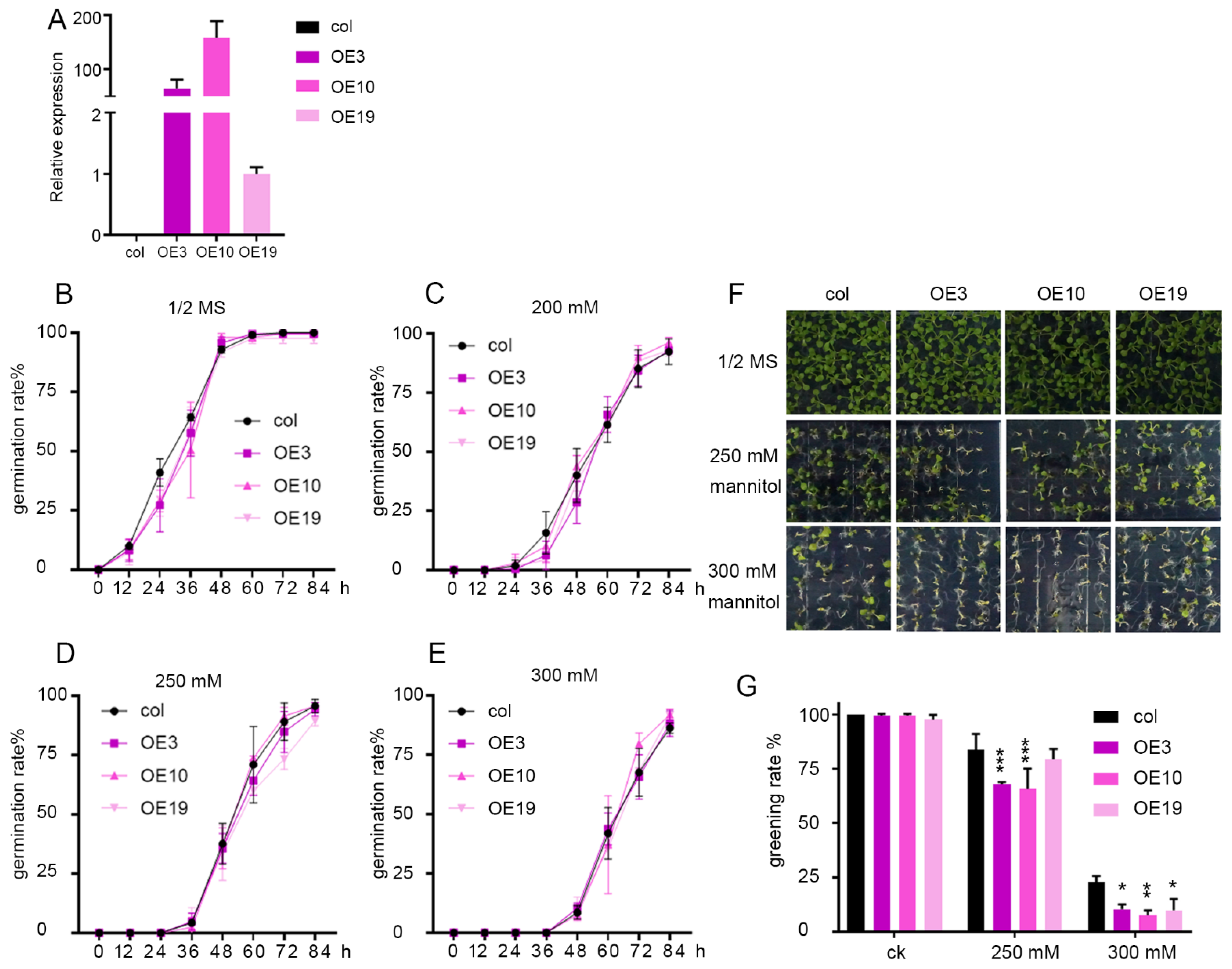
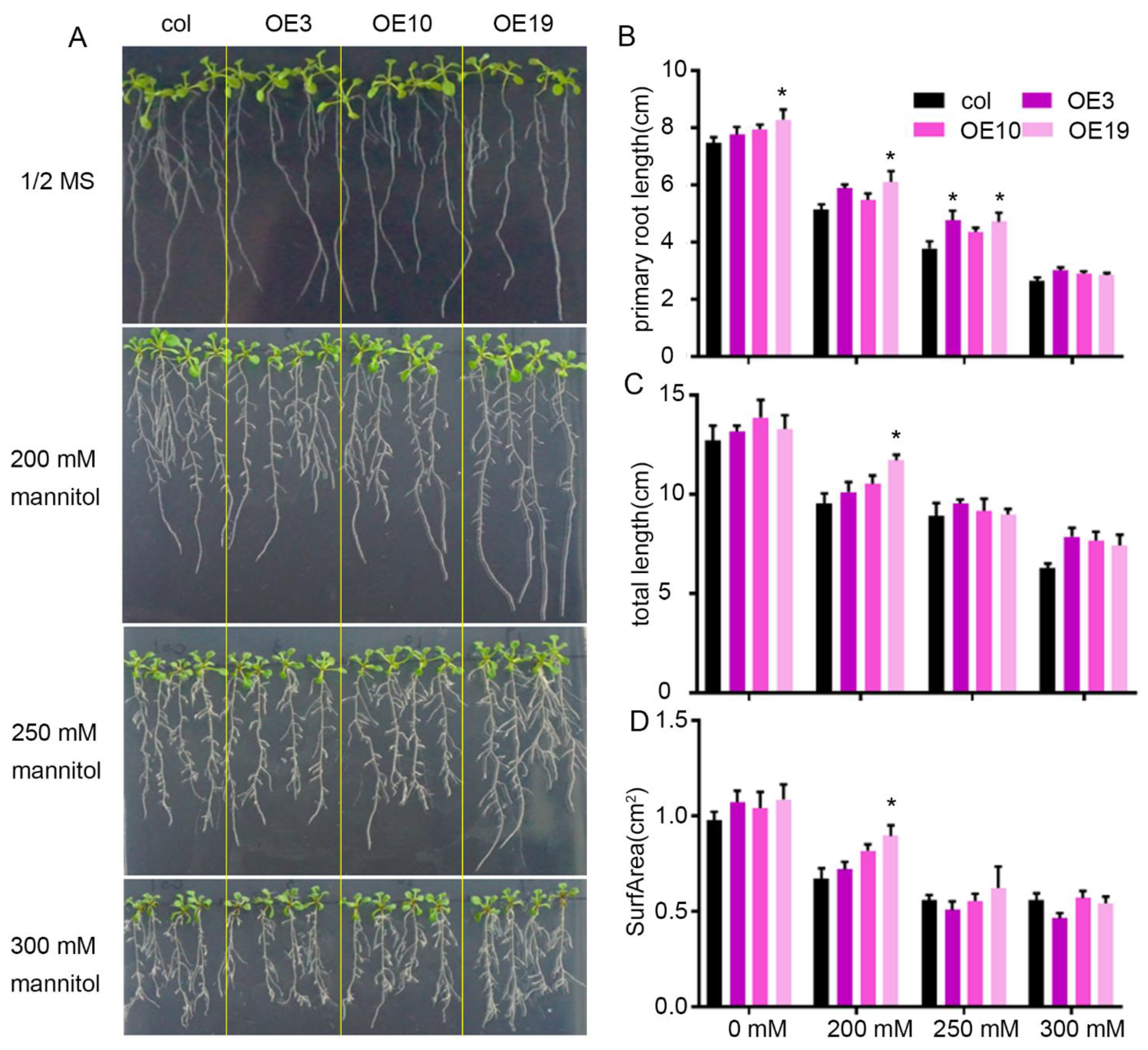
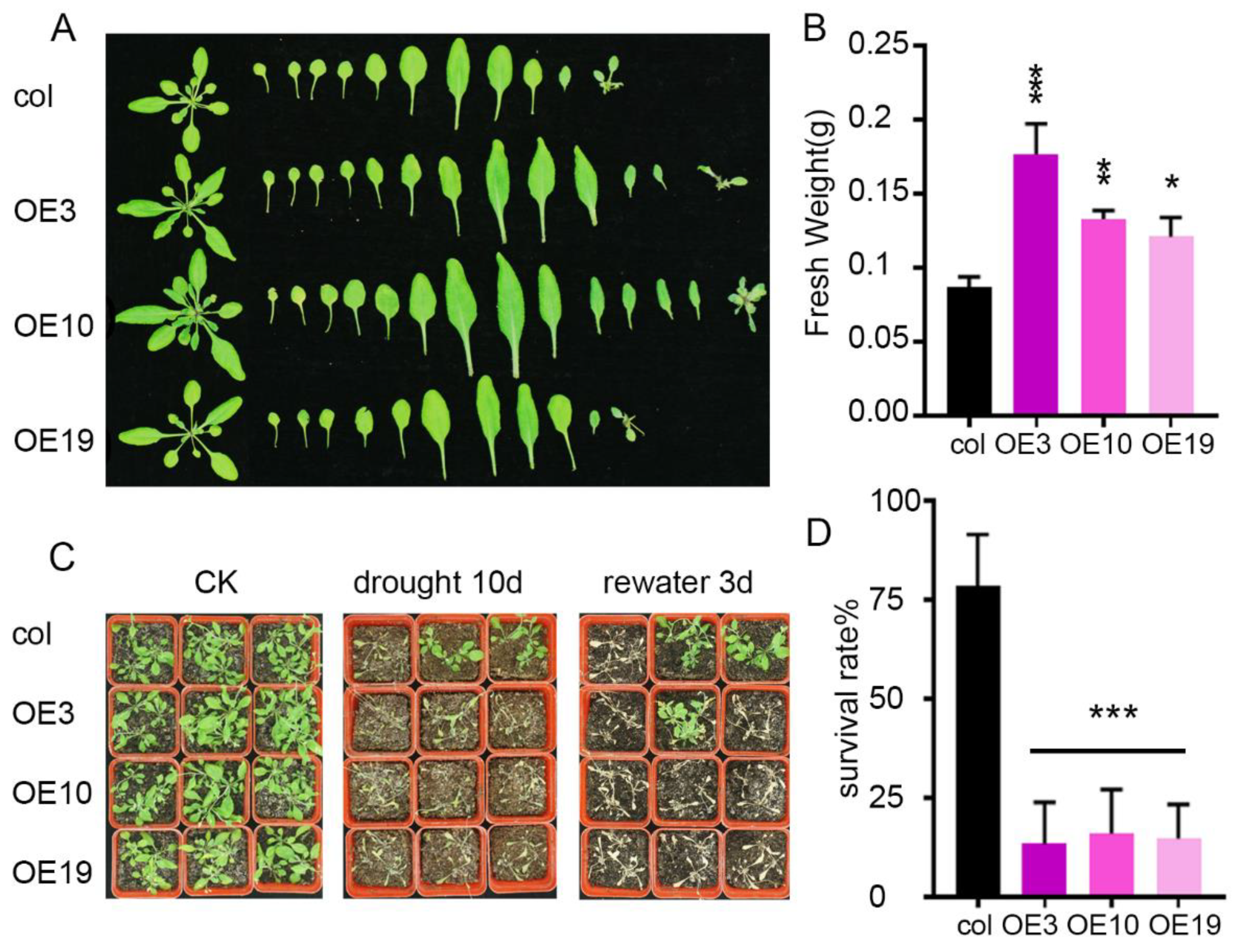
2.3. Overexpression of ZmLBD5 Decreased Drought Tolerance in Transgenic Arabidopsis
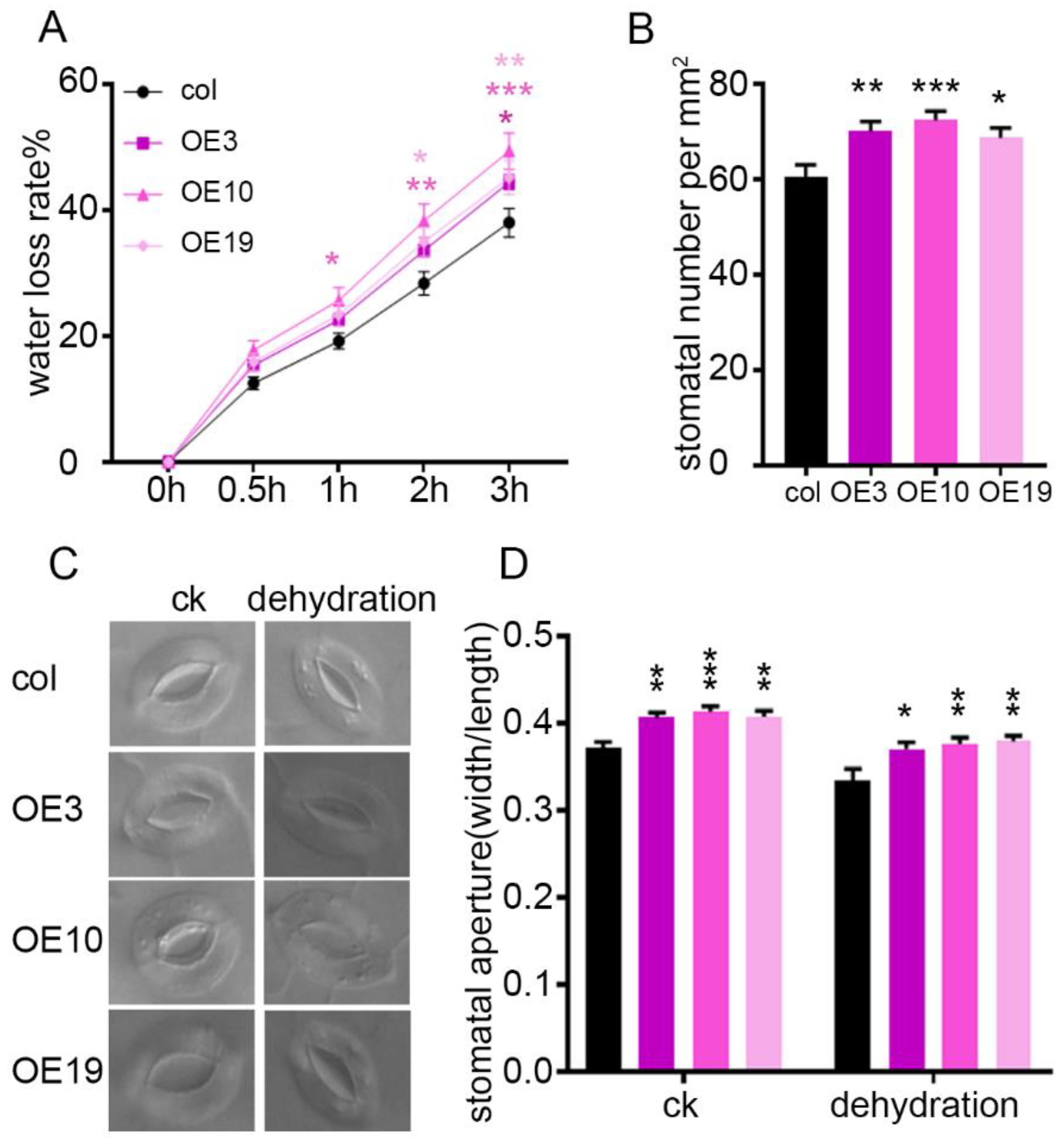
2.4. ZmLBD5 Increased the Water Loss Rate by Enhancing the Stomatal Density and Aperture
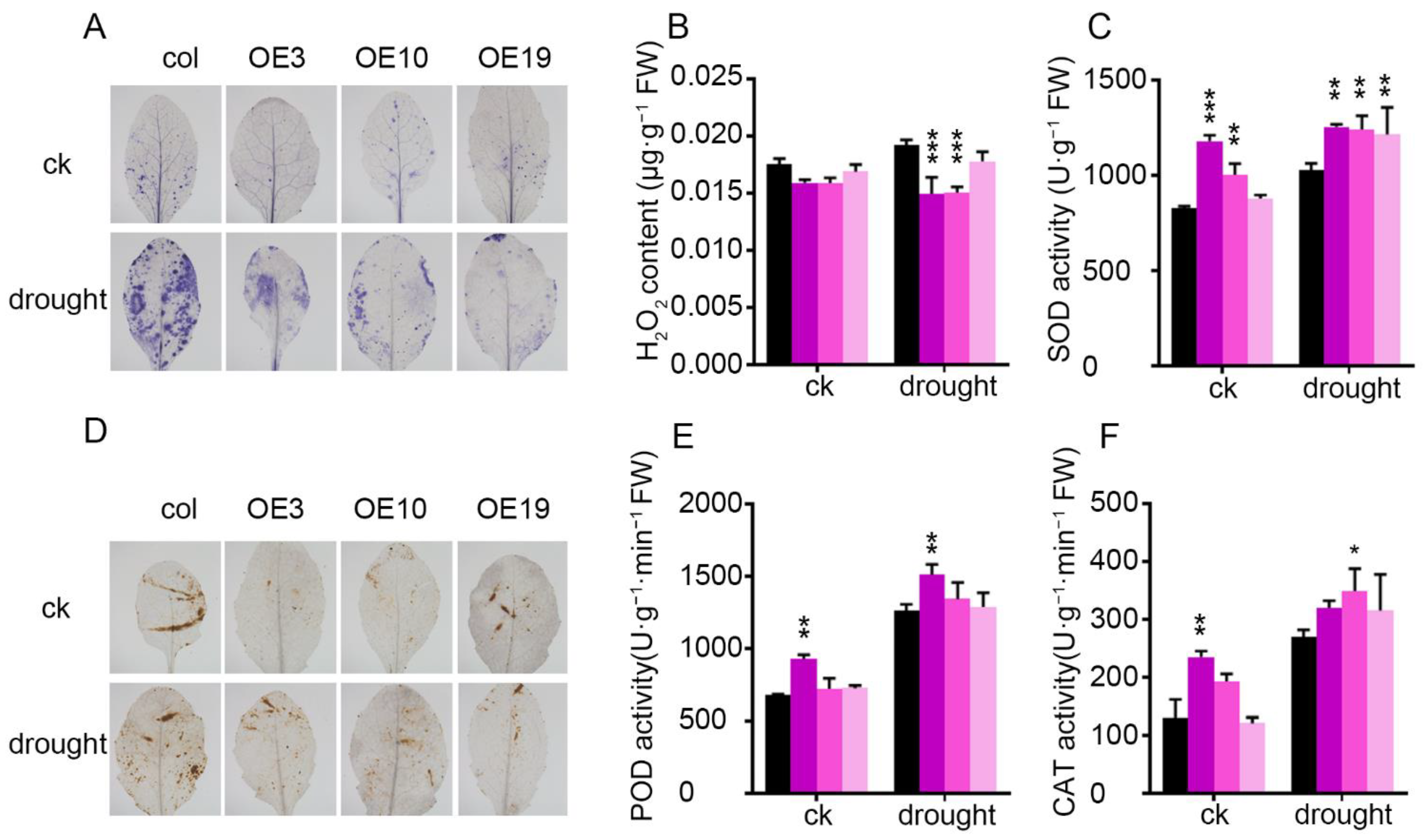
2.5. Overexpression of ZmLBD5 Improved Antioxidant Enzyme Activity and Blocked ROS Accumulation in Arabidopsis
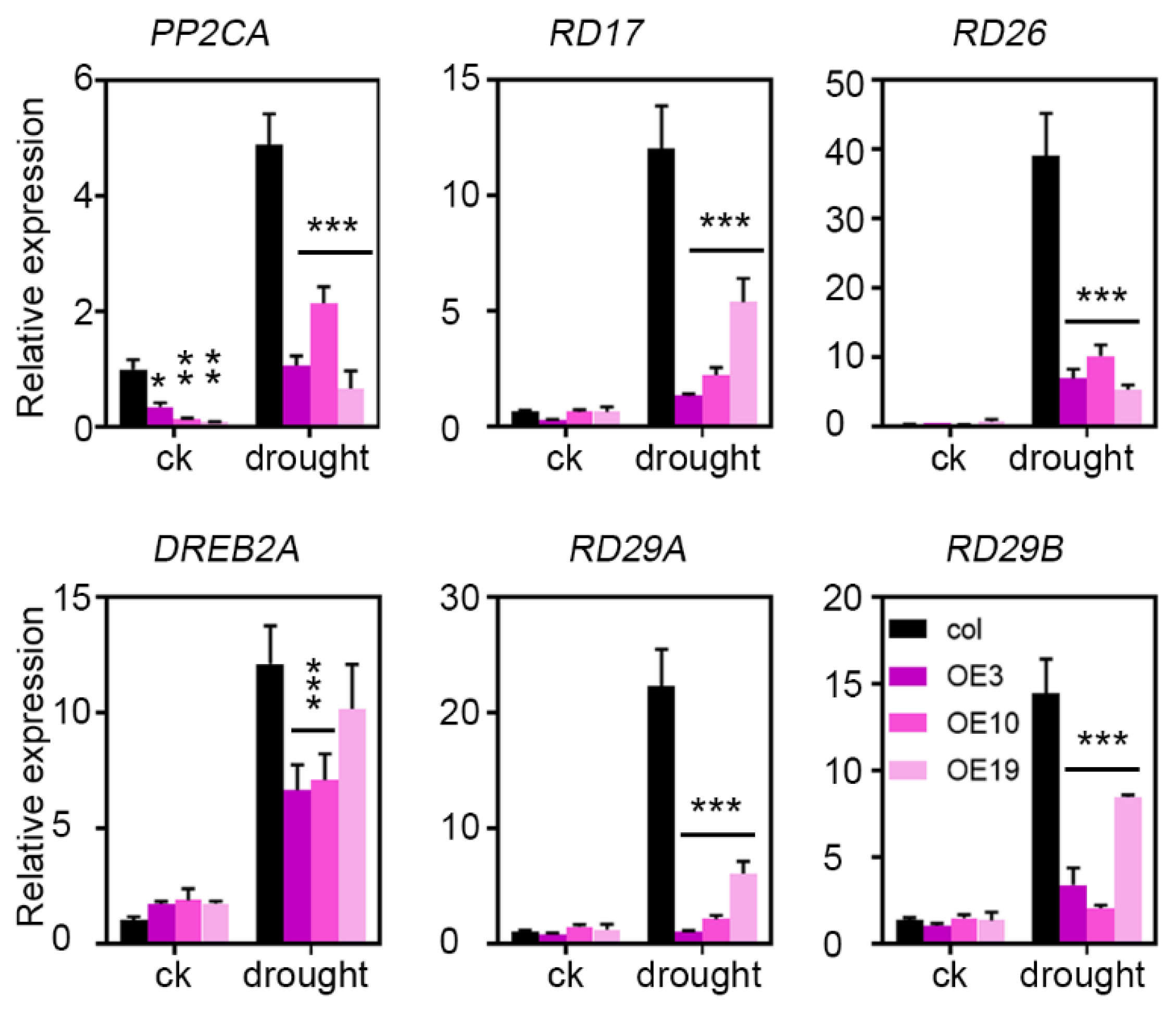
2.6. ZmLBD5 Negatively Regulates Drought-Related Genes’ Expression in Transgenic Arabidopsis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials and Growth Conditions
5.2. Sequence Analysis
5.3. Subcellular Localization
5.4. RNA Extraction and Quantitative RT-qPCR Analysis
5.5. Generation of Transgenic Plants and Phenotypic Analysis
5.6. Water Loss Measurement
5.7. Stomatal Density and Stomatal Aperture
5.8. ROS Measurements
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Blum, A. Drought resistance, water-use efficiency, and yield potential—are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 2005, 56, 1159–1168. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef]
- Yang, L.; Jake, F.; Hui, W.; Xinzhi, N.; Pingsheng, J.; Robert, L.; Robert, K.; Brian, S.; Baozhu, G. Stress sensitivity is associated with differential accumulation of reactive oxygen and nitrogen species in maize genotypes with contrasting levels of drought tolerance. Int. J. Mol. Sci. 2015, 16, 24791–24819. [Google Scholar] [CrossRef]
- Xiong, H.; Yu, J.; Miao, J.; Li, J.; Zhang, H.; Wang, X.; Liu, P.; Zhao, Y.; Jiang, C.; Yin, Z.; et al. Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol. 2018, 178, 451–467. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Majer, C.; Xu, C.; Berendzen, K.W.; Hochholdinger, F. Molecular interactions of rootless concerning crown and semial roots, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philosphical Trans. Rayal Soc. B 2012, 367, 1542–1551. [Google Scholar] [CrossRef]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB domain proteins: Beyond lateral organ boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, M.; Lee, M.R.; Park, S.K.; Kim, J. Lateral organ boundaries domain (LBD) 10 interacts with sidecar pollen/LBD27 to control pollen development in Arabidopsis. Plant J. 2015, 81, 794–809. [Google Scholar] [CrossRef]
- Berckmans, B.; Vassileva, V.; Schmid, S.P.C.; Maes, S.; Veylder, L.D. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 2011, 23, 3671–3683. [Google Scholar] [CrossRef]
- Evans, M.M.S. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies lateral organ boundaries domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenetics Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Zhang, S.Z.; Zheng, C.C. Genomewide analysis of lateral organ boundaries domain gene family in zea mays. J. Genet. 2014, 93, 79–91. [Google Scholar] [CrossRef]
- Guo, B.J.; Wang, J.; Lin, S.; Tian, Z.; Zhou, K.; Luan, H.Y.; Lyu, C.; Zhang, X.Z.; Xu, R.G. A genome-wide analysis of the asymmetric leaves2/lateral organ boundaries(AS2/LOB) gene family in barley (Hordeum vulgare L.). Biomed. Biotechnol. 2016, 17, 763–774. [Google Scholar] [CrossRef]
- Gupta, K.; Gupta, S. Molecular and in silico characterization of tomato LBD transcription factors reveals their role in fruit development and stress responses. Plant Gene 2021, 27, 100309. [Google Scholar] [CrossRef]
- Grimplet, J.; Pimentel, D.; Agudelo-Romero, P.; Martinez-Zapater, J.M.; Fortes, A.M. The lateral organ boundaries domain gene family in grapevine: Genome-wide characterization and expression analyses during developmental processes and stress responses. Sci. Rep. 2017, 7, 15968. [Google Scholar] [CrossRef]
- Lu, Q.; Shao, F.; Macmillan, C.; Wilson, I.W.; van der Merwe, K.; Hussey, S.G.; Myburg, A.A.; Dong, X.; Qiu, D. Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018, 16, 124–136. [Google Scholar] [CrossRef]
- Liu, H.; Cao, M.; Chen, X.; Ye, M.; Zhao, P.; Nan, Y.; Li, W.; Zhang, C.; Kong, L.; Kong, N.; et al. Genome-wide analysis of the lateral organ boundaries domain (LBD) gene family in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 5360. [Google Scholar] [CrossRef]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C.P. Direct repression of KNOX loci by the asymmetric leaves1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef]
- Bortiri, E.; Chuck, G.; Vollbrecht, E.; Rocheford, T.; Martienssen, R.; Hake, S. ramosa2 encodes a lateral organ boundary domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 2006, 18, 574–585. [Google Scholar] [CrossRef]
- Taramino, G.; Sauer, M.; Stauffer, J.L., Jr.; Multani, D.; Niu, X.; Sakai, H.; Hochholdinger, F. The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 2007, 50, 649–659. [Google Scholar] [CrossRef]
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an auxin response factor in auxin signaling. Plant Cell 2005, 17, 1387–1396. [Google Scholar] [CrossRef]
- Goh, T.; Toyokura, K.; Yamaguchi, N.; Okamoto, Y.; Uehara, T.; Kaneko, S.; Takebayashi, Y.; Kasahara, H.; Ikeyama, Y.; Okushima, Y.; et al. Lateral root initiation requires the sequential induction of transcription factors LBD16 and PUCHI in Arabidopsis thaliana. New Phytol. 2019, 224, 749–760. [Google Scholar] [CrossRef]
- Ye, L.; Wang, X.; Lyu, M.; Siligato, R.; Eswaran, G.; Vainio, L.; Blomster, T.; Zhang, J.; Mähönen, A.P. Cytokinins initiate secondary growth in the Arabidopsis root through a set of LBD genes. Curr. Biol. 2021, 31, 3365–3373.e3367. [Google Scholar] [CrossRef]
- Bdeir, R.; Busov, V.; Yordanov, Y.; Gailing, O. Gene dosage effects and signatures of purifying selection in lateral organ boundaries domain (LBD) genes LBD1 and LBD18. Plant Syst. Evol. 2016, 302, 433–445. [Google Scholar] [CrossRef]
- Yordanov, Y.S.; Busov, V. Boundary genes in regulation and evolution of secondary growth. Plant Signal Behav. 2011, 6, 688–690. [Google Scholar] [CrossRef][Green Version]
- Jeon, E.; Young Kang, N.; Cho, C.; Joon Seo, P.; Chung Suh, M.; Kim, J. LBD14/ASL17 positively regulates lateral root formation and is involved in ABA response for root architecture in Arabidopsis. Plant Cell Physiol. 2017, 58, 2190–2201. [Google Scholar] [CrossRef]
- Jeon, B.W.; Kim, J. Role of LBD14 during ABA-mediated control of root system architecture in Arabidopsis. Plant Signal Behav. 2018, 13, e1507405. [Google Scholar] [CrossRef]
- Guo, Z.; Xu, H.; Lei, Q.; Du, J.; Li, C.; Wang, C.; Yang, Y.; Yang, Y.; Sun, X. The Arabidopsis transcription factor LBD15 mediates ABA signaling and tolerance of water-deficit stress by regulating ABI4 expression. Plant J. 2020, 104, 510–521. [Google Scholar] [CrossRef]
- Ma, W.; Wu, F.; Sheng, P.; Wang, X.; Zhang, Z.; Zhou, K.; Zhang, H.; Hu, J.; Lin, Q.; Cheng, Z.; et al. The LBD12-1 transcription factor suppresses apical meristem size by repressing argonaute 10 expression. Plant Physiol. 2017, 173, 801–811. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Mori, M.; Ichikawa, T.; Matsui, K.; Kuroda, H.; et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant 2010, 3, 125–142. [Google Scholar] [CrossRef]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef]
- Wang, P.; Song, C.P. Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol. 2008, 178, 703–718. [Google Scholar] [CrossRef]
- Miller, G. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Ma, B.; Hou, Q.; Wan, X. Phylogeny and functions of LOB domain proteins in plants. Intern. J. Mol. Sci. 2020, 21, 2278. [Google Scholar] [CrossRef]
- Zhang, X.; He, Y.; He, W.; Su, H.; Wang, Y.; Hong, G.; Xu, P. Structural and functional insights into the LBD family involved in abiotic stress and flavonoid synthases in Camellia sinensis. Sci. Rep. 2019, 9, 15651. [Google Scholar] [CrossRef]
- Xu, J.; Hu, P.; Tao, Y.; Song, P.; Gao, H.; Guan, Y. Genome-wide identification and characterization of the Lateral Organ Boundaries Domain (LBD) gene family in polyploid wheat and related species. PeerJ 2021, 9, e11811. [Google Scholar] [CrossRef]
- Zentella, R.; Zhang, Z.L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef]
- Ariel, F.; Diet, A.; Verdenaud, M.; Gruber, V.; Frugier, F.; Chan, R.; Crespi, M. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 2010, 22, 2171–2183. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, M.J.; Park, M.Y.; Han, K.H.; Kim, J. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol. Plant 2013, 6, 1722–1725. [Google Scholar] [CrossRef]
- Lee, H.W.; Kang, N.Y.; Pandey, S.K.; Cho, C.; Lee, S.H.; Kim, J. Dimerization in LBD16 and LBD18 transcription factors is critical for lateral root formation. Plant Physiol. 2017, 174, 301–311. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Kazan, K.; Manners, J.M. Lateral organ boundaries domain transcription factors: New roles in plant defense. Plant Signal. Behav. 2012, 7, 1702–1704. [Google Scholar] [CrossRef]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar] [CrossRef]
- Iwakawa, H.; Iwasaki, M.; Kojima, S.; Ueno, Y.; Soma, T.; Tanaka, H.; Semiarti, E.; Machida, Y.; Machida, C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007, 51, 173–184. [Google Scholar] [CrossRef]
- Huang, X.Y.; Chao, D.Y.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Song, J.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Abscisic acid and hydrogen peroxide are involved in drought priming-induced drought tolerance in wheat (Triticum aestivum L.). Plant Biol. 2020, 22, 1113–1122. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Dong, F.; Gao, J.; Galbraith, D.W.; Song, C.-P. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 2001, 126, 1438–1448. [Google Scholar] [CrossRef]
- Sergiev, I.; Alexieva, V.; Karanov, E.N.; Karanov, E.; Sergiev, L.M.; Karanova, E.; Alexieva, V. Effect of spermine, atrazine and combination between them on some endogenous protective systems and stress markers in plants. Comptes Rendus Acad. Bulg. Sci. 1997, 51, 121–124. [Google Scholar]
- Otter, P.A. Apoplastic peroxidases and lignification in needles of Norway Spruce (Picea abies L.). Plant Physiol. 1994, 106, 53–60. [Google Scholar]
- Chance, M.A. The assay of catalases and peroxidases. Methods Biochem. Anal. 1954, 1, 357–424. [Google Scholar]
- Fridovich, B.A. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar]

| Site Name | Sequence | Position | Strand | Function |
|---|---|---|---|---|
| ABRE | ACGTG | −1984 | - | Abscisic acid responsiveness |
| ABRE | CGTACGTGCA | −1730 | - | Abscisic acid responsiveness |
| ABRE | CACGTG | −1596 | + | Abscisic acid responsiveness |
| ABRE | ACGTG | −1595 | + | Abscisic acid responsiveness |
| ABRE | ACGTG | −1528 | + | Abscisic acid responsiveness |
| ABRE | ACGTG | −71 | - | Abscisic acid responsiveness |
| ABRE | CCACGTGG | −1597 | + | Abscisic acid responsiveness |
| DRE | GCCGAC | −1896 | - | Dehydration-responsive element |
| DRE | GCCGAC | −1495 | - | Dehydration-responsive element |
| DRE | ACCGAGA | −38 | + | Dehydration-responsive element |
| LTR | CCGAAA | −1635 | + | Low-temperature responsiveness |
| LTR | CCGAAA | −262 | + | Low-temperature responsiveness |
| MBS | CAACTG | −597 | - | MYB-binding site involved in drought-inducibility |
| MYBRS | CAACCA | −1566 | - | MYB recognition site |
| MYBRS | CAACTG | −597 | - | MYB recognition site |
| MYBRS | TAACCA | −593 | - | MYB recognition site |
| MYBRS | CAACCA | −518 | + | MYB recognition site |
| MYBRS | CAACCA | −100 | + | MYB recognition site |
| MYBRS | CAACCA | −96 | + | MYB recognition site |
| MYBRS | CCGTTG | −1844 | + | MYB recognition site |
| MYBRS | TAACCA | −593 | - | MYB recognition site |
| CCAAT-box | CAACGG | −1844 | - | MYBHv1-binding site |
| ARE | AAACCA | −1631 | + | Anaerobic induction |
| ARE | AAACCA | −259 | + | Anaerobic induction |
| G-box | CACGTC | −1984 | + | Light responsiveness |
| G-box | GCCACGTGGA | −1598 | + | Light responsiveness |
| G-box | CACGTG | −1596 | + | Light responsiveness |
| G-Box | CACGTG | −1596 | + | Light responsiveness |
| G-Box | CACGTT | −1529 | - | Light responsiveness |
| G-box | CACGTC | −71 | + | Light responsiveness |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, J.; Zhang, W.; Zheng, D.; Xiong, H.; Feng, X.; Zhang, X.; Wang, Q.; Wu, F.; Xu, J.; Lu, Y. ZmLBD5 Increases Drought Sensitivity by Suppressing ROS Accumulation in Arabidopsis. Plants 2022, 11, 1382. https://doi.org/10.3390/plants11101382
Xiong J, Zhang W, Zheng D, Xiong H, Feng X, Zhang X, Wang Q, Wu F, Xu J, Lu Y. ZmLBD5 Increases Drought Sensitivity by Suppressing ROS Accumulation in Arabidopsis. Plants. 2022; 11(10):1382. https://doi.org/10.3390/plants11101382
Chicago/Turabian StyleXiong, Jing, Weixiao Zhang, Dan Zheng, Hao Xiong, Xuanjun Feng, Xuemei Zhang, Qingjun Wang, Fengkai Wu, Jie Xu, and Yanli Lu. 2022. "ZmLBD5 Increases Drought Sensitivity by Suppressing ROS Accumulation in Arabidopsis" Plants 11, no. 10: 1382. https://doi.org/10.3390/plants11101382
APA StyleXiong, J., Zhang, W., Zheng, D., Xiong, H., Feng, X., Zhang, X., Wang, Q., Wu, F., Xu, J., & Lu, Y. (2022). ZmLBD5 Increases Drought Sensitivity by Suppressing ROS Accumulation in Arabidopsis. Plants, 11(10), 1382. https://doi.org/10.3390/plants11101382






