Genomic Analysis and Secondary Metabolites Production of the Endophytic Bacillus velezensis Bvel1: A Biocontrol Agent against Botrytis cinerea Causing Bunch Rot in Post-Harvest Table Grapes
Abstract
:1. Introduction
2. Results
2.1. Taxonomic Classification of the Bacterial Stain Bvel1
2.2. In Vitro Antagonistic Activity of Bacillus velezensis Bvel1 and Its Secreted Metabolites against B. cinerea
2.3. UHPLC-HRMS Analysis of Secreted Metabolites Produced by Strain Bvel1
2.4. Preventive and Curative Action of Strain Bvel1 against B. cinerea on Grape Berries
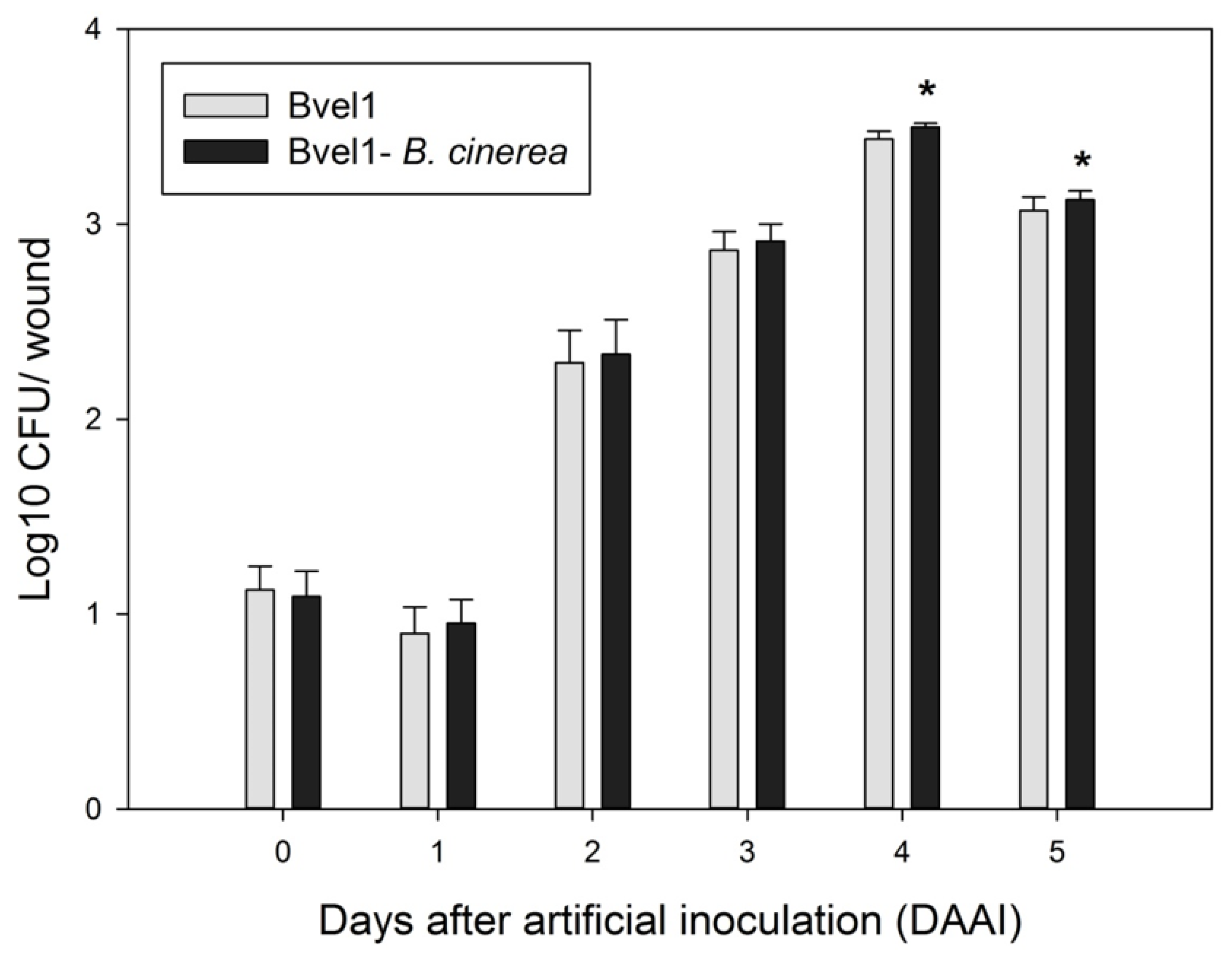
2.5. Bacillus velezensis Bvel1 Colonization in Wounded Berries
2.6. Genomic Insights into the Antifungal Activity of the Strain Bvel1
3. Discussion
4. Materials and Methods
4.1. Isolation of Endophytic Bacteria-Microorganisms and Culture Conditions
4.2. In Vitro Antagonistic Activity of Bacillus velezensis Bvel1 against B. cinerea
4.3. Extraction of Bacillus velezensis Bvel1 Secreted Agar-Diffusible Compounds
4.4. In Vitro Antagonistic Activity of Secreted Agar-Diffusible Compounds from Strain Bvel1 against B. cinera
4.5. TLC-Bioautography of Bvel1 Secreted Compounds
4.6. Orbitrap High Resolution Mass Spectrometry (UHPLC-HRMS) Analysis of Bvel1 Secreted Compounds
4.7. In Vivo Antagonistic Activity of Bvel1 against B. cinerea on Grape Berries
4.8. Colonization of the Strain Bvel1 on Wounded Grape Berries
4.9. Genome Sequencing
4.10. Functional Genome Analysis
4.11. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Poveda, J.; Barquero, M.; González-Andrés, F. Insight into the microbiological control strategies against Botrytis cinerea using systemic plant resistance activation. Agronomy 2020, 10, 1822. [Google Scholar] [CrossRef]
- Jiang, C.M.; Shi, J.L.; Liu, Y.L.; Zhu, C.Y. Inhibition of Aspergillus carbonarius and fungal contamination in table grapes using Bacillus subtilis. Food Control 2014, 35, 41–48. [Google Scholar] [CrossRef]
- Pertot, I.; Giovannini, O.; Benanchi, M.; Caffi, T.; Rossi, V.; Mugnai, L. Combining biocontrol agents with different mechanisms of action in a strategy to control Botrytis cinerea on grapevine. Crop Prot. 2017, 97, 85–93. [Google Scholar] [CrossRef]
- Fedele, G.; Brischetto, C.; Rossi, V. Biocontrol of Botrytis cinerea on grape berries as influenced by temperature and humidity. Front. Plant Sci. 2020, 11, 1232. [Google Scholar] [CrossRef]
- Fira, D.; Dimkic, I.; Beric, T.; Lozo, J.; Stankovic, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Cedeño, L.R.M.; Mosqueda, M.d.C.O.; Lara, P.D.L.; Cota, F.I.P.; Villalobos, S.d.L.S.; Santoyo, G. Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: Fundamentals, methods of application and future perspectives. Microbiol. Res. 2021, 242, 126612. [Google Scholar] [CrossRef] [PubMed]
- Keswani, C.; Singh, H.B.; García-Estrada, C.; Caradus, J.; He, Y.W.; Mezaache-Aichour, S.; Glare, T.R.; Borriss, R.; Sansinenea, E. Antimicrobial secondary metabolites from agriculturally important bacteria as next-generation pesticides. Appl. Microbiol. Biotechnol. 2020, 104, 1013–1034. [Google Scholar] [CrossRef]
- Arrebola, E.; Jacobs, R.; Korsten, L. Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against post-harvest fungal pathogens. J. Appl. Microbiol. 2010, 108, 386–395. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, L.; Zeriouh, H.; Romero, D.; Cubero, J.; de Vicente, A.; and Pérez-García, A. The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate and salicylic acid-dependent defence responses. Microb. Biotechnol. 2013, 6, 264–274. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shen, Q.; Zhang, R. Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signalling pathways. Mol. Plant Microbe Interact. 2018, 31, 560–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Héloir, M.C.; Zhang, X.; Geissler, M.; Trouvelot, S.; Jacquens, L.; Henkel, M.; Su, X.; Fang, X.; Wang, Q. Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol. Plant Pathol. 2019, 20, 1037–1050. [Google Scholar] [CrossRef] [Green Version]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [Green Version]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef]
- Romero, D.; De Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.-W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Perez-Garcia, A. The Iturin and Fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waewthongrak, W.; Leelasuphakul, W.; McCollum, G. Cyclic Lipopeptides from Bacillus subtilis ABS-S14 Elicit Defense-Related Gene Expression in Citrus Fruit. PLoS ONE 2014, 9, e109386. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Xue, Y.; Huang, Z.; Jiang, M.; Lu, F.; Bie, X.; Miao, S.; Lu, Z. Bacillomycin D inhibits growth of Rhizopus stolonifer and induces defense-related mechanism in cherry tomato. Appl. Microbiol. Biotechnol. 2019, 103, 7663–7674. [Google Scholar] [CrossRef]
- Farzand, A.; Moosa, A.; Zubair, M.; Khan, A.R.; Massawe, V.C.; Tahir, H.A.S.; Sheikh, T.M.M.; Ayaz, M.; Gao, X. Suppression of Sclerotinia sclerotiorum by the induction of systemic resistance and regulation of antioxidant pathways in tomato using fengycin produced by Bacillus amyloliquefaciens FZB42. Biomolecules 2019, 9, 613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Ren, J.; Li, P.; Feng, S.; Dong, P.; Ren, M. Potential of microbial endophytes to enhance the resistance to postharvest diseases of fruit and vegetables. J. Sci. Food Agric. 2021, 101, 1744–1757. [Google Scholar] [CrossRef] [PubMed]
- Lastochkina, O.; Pusenkova, L.; Garshina, D.; Yuldashev, R.; Shpirnaya, I.; Kasnak, C.; Palamutoglu, R.; Mardanshin, I.; Garipova, S.; Sobhani, M.; et al. The effect of endophytic bacteria Bacillus subtilis and salicylic acid on some resistance and quality traits of stored Solanum tuberosum L. Tubers infected with Fusarium dry rot. Plants 2020, 9, 738. [Google Scholar] [CrossRef] [PubMed]
- Chaouachi, M.; Marzouk, T.; Jallouli, S.; Elkahoui, S.; Gentzbittel, L.; Ben, C.; Dj´ebali, N. Activity assessment of tomato endophytic bacteria bioactive compounds for the postharvest biocontrol of Botrytis cinerea. Post. Biol. Technol. 2021, 172, 111389. [Google Scholar] [CrossRef]
- Damasceno, C.L.; Duarte, E.A.A.; dos Santos, L.B.P.R.; de Oliveira, T.A.S.; de Jesus, F.N.; de Oliveira, L.M.; Góes-Neto, A.; Soares, A.C.F. Postharvest biocontrol of anthracnose in bananas by endophytic and soil rhizosphere bacteria associated with sisal (Agave sisalana) in Brazil. Biol. Control 2020, 137, 104016. [Google Scholar] [CrossRef]
- Fan, B.; Blom, J.; Klenk, H.P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “Operational Group, B. amyloliquefaciens” within the B. subtilis species complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [Green Version]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–667. [Google Scholar] [CrossRef]
- Luo, C.; Zhou, H.; Zou, J.; Wang, X.; Zhang, R.; Xiang, Y.; Chen, Z. Bacillomycin L and surfactin contribute synergistically to the phenotypic features of Bacillus subtilis 916 and the biocontrol of rice sheath blight induced by Rhizoctonia solani. Appl. Microbiol. Biotechnol. 2015, 99, 1897–1910. [Google Scholar] [CrossRef] [PubMed]
- Janek, T.; Drzymała, K.; Dobrowolski, A. In vitro efficacy of the lipopeptide biosurfactant surfactin-C15 and its complexes with divalent counterions to inhibit Candida albicans biofilm and hyphal formation. Biofouling 2020, 36, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, C.; Liang, J.; Wu, L.; Gao, W.; Jiang, J. Iturin A extracted from Bacillus subtilis WL2 Affects Phytophthora infestans via cell structure disruption, oxidative stress, and energy supply dysfunction. Front. Microbiol. 2020, 11, 2205. [Google Scholar]
- Parker, J.B.; Walsh, C.T. Action and timing of BacC and BacD in the late stages of biosynthesis of the dipeptide antibiotic bacilysin. Biochemistry 2013, 52, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Im, S.M.; Yu, N.H.; Joen, H.W.; Kim, S.O.; Park, H.W.; Park, A.R.; Kim, J.C. Biological control of tomato bacterial wilt by oxydifficidin and difficidin-producing Bacillus methylotrophicus DR-08. Pest. Biochem. Physiol. 2020, 163, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Cheon, H.I.; Yeo, M.S.; Kim, K.M.; Kang, J.S.; Pyo, J. Determination of siderophore from Bacillus mojavensis using liquid chromatography quadrupole time-of-flight tandem mass spectrometry. J. Life Sci. 2019, 29, 19. [Google Scholar]
- Javvadi, S.G.; Cescutti, P.; Rizzo, R.; Lonzarich, V.; Navarini, L.; Licastro, D.; Venturi, V. The spent culture supernatant of Pseudomonas syringae contains azelaic acid. BMC Microbiol. 2018, 18, 199. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Liu, X.H.; Wu, M.B.; Ge, S. Molecular insights into the antifungal mechanism of bacilysin. J. Mol. Model. 2018, 24, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, C.A.; Bowman, M.J.; Rooney, A.P. Iturinic lipopeptide diversity in the Bacillus subtilis species group—Important antifungals for plant disease biocontrol applications. Front. Microbiol. 2019, 10, 1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Zhang, L.; Goodwin, P.H.; Xia, M.; Zhang, J.; Wang, Q.; Liang, J.; Sun, R.; Wu, C.; Yang, L. Isolation, identification, and complete genome assembly of an endophytic Bacillus velezensis YB-130, potential biocontrol agent against Fusarium graminearum. Front. Microbiol. 2020, 11, 598285. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Kloepper, J.W.; Pare, P.W. Bacterial volatiles induced systemic resistance in Arabidopsis. Plant Physiol. 2004, 134, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Raymaekers, K.; Ponet, L.; Holtappels, D.; Berckmans, B.; Cammue, B.P.A. Screening for novel biocontrol agents applicable in plant disease management—A review. Biol. Control 2020, 144, 104240. [Google Scholar] [CrossRef]
- Kohl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [Green Version]
- Zeriouh, H.; Romero, D.; García-Gutiérrez, L.; Cazorla, F.M.; de Vicente, A.; Pérez-García, A. The Iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol. Plant Microb. Interact. 2011, 24, 1540–1552. [Google Scholar] [CrossRef] [Green Version]
- Zeriouh, H.; de Vicente, A.; Pérez-García, A.; Romero, D. Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 2014, 16, 2196–2211. [Google Scholar] [CrossRef]
- Fan, H.; Zhang, Z.; Li, Y.; Zhang, X.; Duan, Y.; Wang, Q. Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front. Microbiol. 2017, 8, 1973. [Google Scholar] [CrossRef]
- Yamamoto, S.; Shiraishi, S.; Suzuki, S. Are cyclic lipopeptides produced by Bacillus amyloliquefaciens S13-3 responsible for the plant defence response in strawberry against Colletotrichum gloeosporioides? Lett. Appl. Microbiol. 2015, 60, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Farace, G.; Fernandez, O.; Jacquens, L.; Coutte, F.; Krier, F.; Jacques, P.; Clément, C.; Barka, E.A.; Jacquard, C.; Dorey, S. Cyclic lipopeptides from Bacillus subtilis activate distinct patterns of defence responses in grapevine. Mol. Plant Pathol. 2015, 16, 177–187. [Google Scholar] [CrossRef]
- Park, K.; Park, Y.-S.; Ahamed, J.; Dutta, S.; Ryu, H.; Lee, S.-H.; Balaraju, K.; Manir, M.; Moon, S.-S. Elicitation of induced systemic resistance of chili pepper by iturin A analogs derived from Bacillus vallismortis EXTN-1. Can. J. Plant Sci. 2016, 96, 564–570. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.; Tonelli, M.L.; Figueredo, M.S.; Ibáñez, F.; Fabra, A. The lipopeptide surfactin triggers induced systemic resistance and priming state responses in Arachis hypogaea L. Eur. J. Plant Pathol. 2018, 152, 845–851. [Google Scholar] [CrossRef]
- Bacon, C.W.; Hinton, D.M.; Mitchell, T.R.; Snook, M.E.; Olubajo, B. Characterization of endophytic strains of Bacillus mojavensis and their production of surfactin isomers. Biol. Control 2012, 62, 1–9. [Google Scholar] [CrossRef]
- Pršic, J.; Ongena, M. Elicitors of plant immunity triggered by beneficial bacteria. Front. Plant Sci. 2020, 11, 1675. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Balti, R.; Neves, A.R. Bioprotective mechanisms of lactic acid bacteria against fungal spoilage of food. Curr. Opin. Biotechnol. 2019, 56, 138–146. [Google Scholar] [CrossRef]
- Cecchini, N.M.; Roychoudhry, S.; Speed, D.J.; Steffes, K.; Tambe, A.; Zodrow, K.; Konstantinoff, K.; Jung, H.W.; Engle, N.L.; Tschaplinski, T.J.; et al. Underground azelaic acid–conferred resistance to Pseudomonas syringae in Arabidopsis. Mol. Plant Microbe Interact. 2019, 32, 86–94. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Li, Y.; Zhang, Y.Y.; Qiao, H.T.; He, J.T.; Yuan, Q.; Chen, X.; Fan, J. High-cell-density culture enhances the antimicrobial and freshness effects of Bacillus subtilis S1702 on table grapes (Vitis vinifera cv. Kyoho). Food Chem. 2019, 286, 541–549. [Google Scholar] [CrossRef]
- Pellegrini, M.; Pagnani, G.; Bernardi, M.; Mattedi, A.; Spera, D.M.; Del Gallo, M. Cell-free supernatants of plant growth-promoting bacteria: A review of their use as biostimulant and microbial biocontrol agents in sustainable. Sustainability 2020, 12, 9917. [Google Scholar] [CrossRef]
- Dimopoulou, A.; Theologidis, I.; Liebmann, B.; Kalantidis, K.; Vassilakos, N.; Skandalis, N. Bacillus amyloliquefaciens MBI600 differentially induces tomato defense signaling pathways depending on plant part and dose of application. Sci. Rep. 2019, 9, 19120. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Hu, J. Plipastatin A1 produced by a marine sediment-derived Bacillus amyloliquefaciens SH-B74 contributes to the control of gray mold disease in tomato. 3 Biotech 2018, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Calvo, H.; Mendiara, I.; Arias, E.; Blanco, D.; Venturini, M.E. The role of iturin A from B. amyloliquefaciens BUZ-14 in the inhibition of the most common postharvest fruit rots. Food Microbiol. 2019, 82, 62–69. [Google Scholar] [CrossRef] [Green Version]
- Touré, Y.; Ongena, M.; Jacques, P.; Guiro, A.; Thonart, P. Role of lipopeptides produced by Bacillus subtilis GA1 in the reduction of grey mould disease caused by Botrytis cinerea on apple. J. Appl. Microbiol. 2004, 96, 1151–1160. [Google Scholar] [CrossRef]
- Debois, D.; Jourdan, E.; Smargiasso, N.; Thonart, P.; De Pauw, E.; Ongena, M. Spatiotemporal monitoring of the antibiome secreted by Bacillus biofilms on plant roots using MALDI mass spectrometry imaging. Anal. Chem. 2014, 86, 4431–4438. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Lamshöft, M.; Zühlke, S.; Spiteller, M. An endophytic fungus from Hypericum perforatum that produces hypericin. J. Nat. Prod. 2008, 71, 159–162. [Google Scholar] [CrossRef]
- Cheng, X.; Ji, X.; Li, J.; Qi, W.; Qiao, K. Characterization of antagonistic Bacillus methylotrophicus isolated from rhizosphere and its biocontrol effects on maize stalk rot. Phytopathology 2019, 109, 571–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertrand, S.; Schumpp, O.; Bohni, N.; Bujard, A.; Azzollini, A.; Monod, M.; Gindro, K.; Wolfender, J.-L. Detection of metabolite induction in fungal co-cultures on solid media by high-throughput differential ultra-high pressure liquid chromatography–time-of-flight mass spectrometry fingerprinting. J. Chromatogr. A 2013, 1292, 219–228. [Google Scholar] [CrossRef]
- Cawoy, H.; Debois, D.; Franzil, L.; De Pauw, E.; Thonart, P.; Ongena, M. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microb. Biotechnol. 2015, 8, 281–295. [Google Scholar] [CrossRef]
- Asari, S.; Ongena, M.; Debois, D.; De Pauw, E.; Chen, K.; Bejai, S.; Meijer, J. Insights into the molecular basis of biocontrol of Brassica pathogens by Bacillus amyloliquefaciens UCMB5113 lipopeptides. Ann. Bot. 2017, 120, 551–562. [Google Scholar] [CrossRef]
- Calvo, H.; Marco, P.; Blanco, D.; Oria, R.; Venturini, M.E. Potential of new strain Bacillus amyloliquefaciens BUZ-14 as a biocontrol agent of postharvest fruit diseases. Food Microbiol. 2017, 63, 101–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Townsend, G.K.; Heuberger, J.W. Methods for estimating losses caused by diseases in fungicide experiments. Plant Dis. Rep. 1943, 27, 340–343. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressive mauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 25, 5. [Google Scholar]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG database update: Focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 2014, 42, D490–D495. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Zhu, Z.; Fu, L.; Niu, B.; Li, W. WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genom. 2011, 12, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
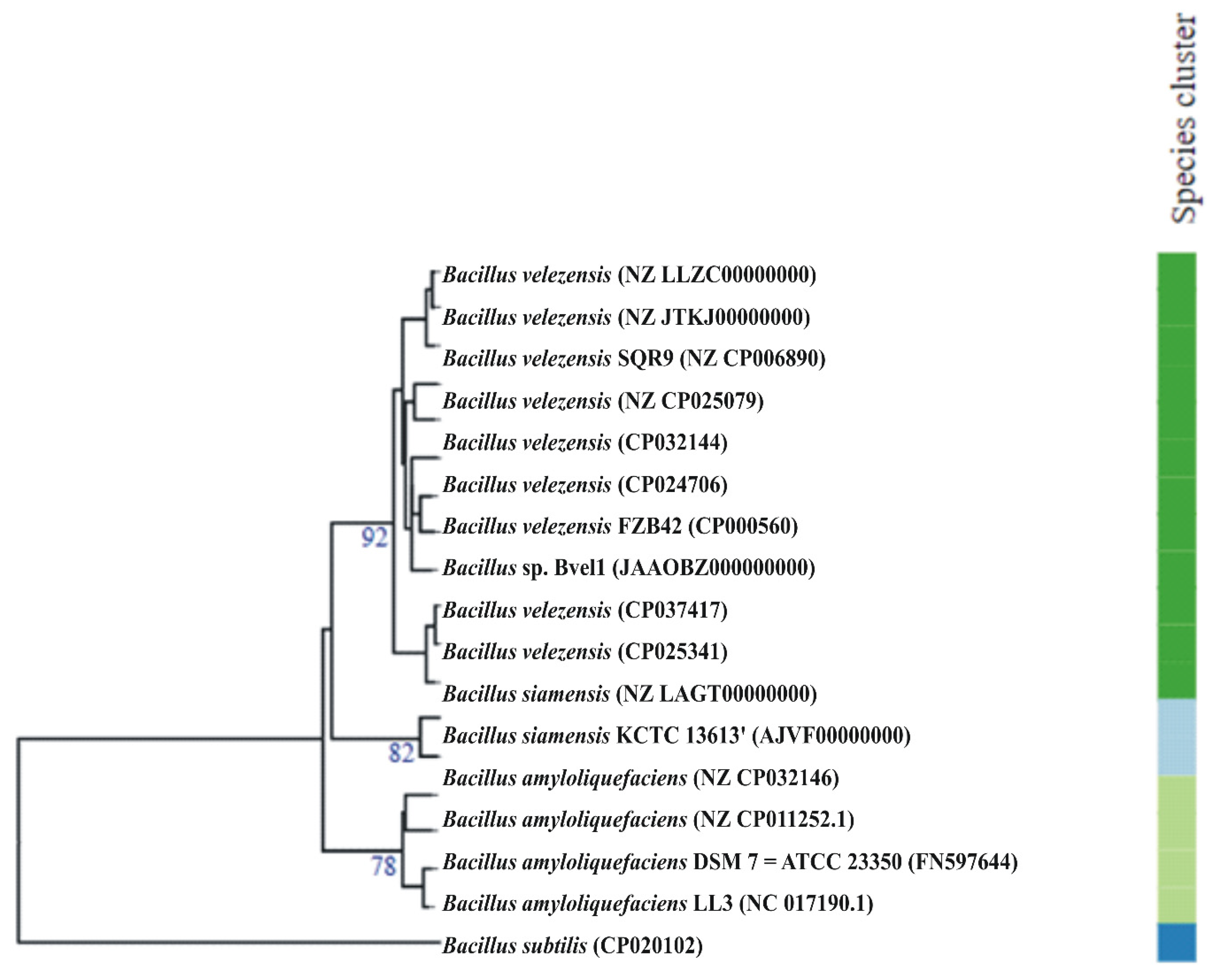
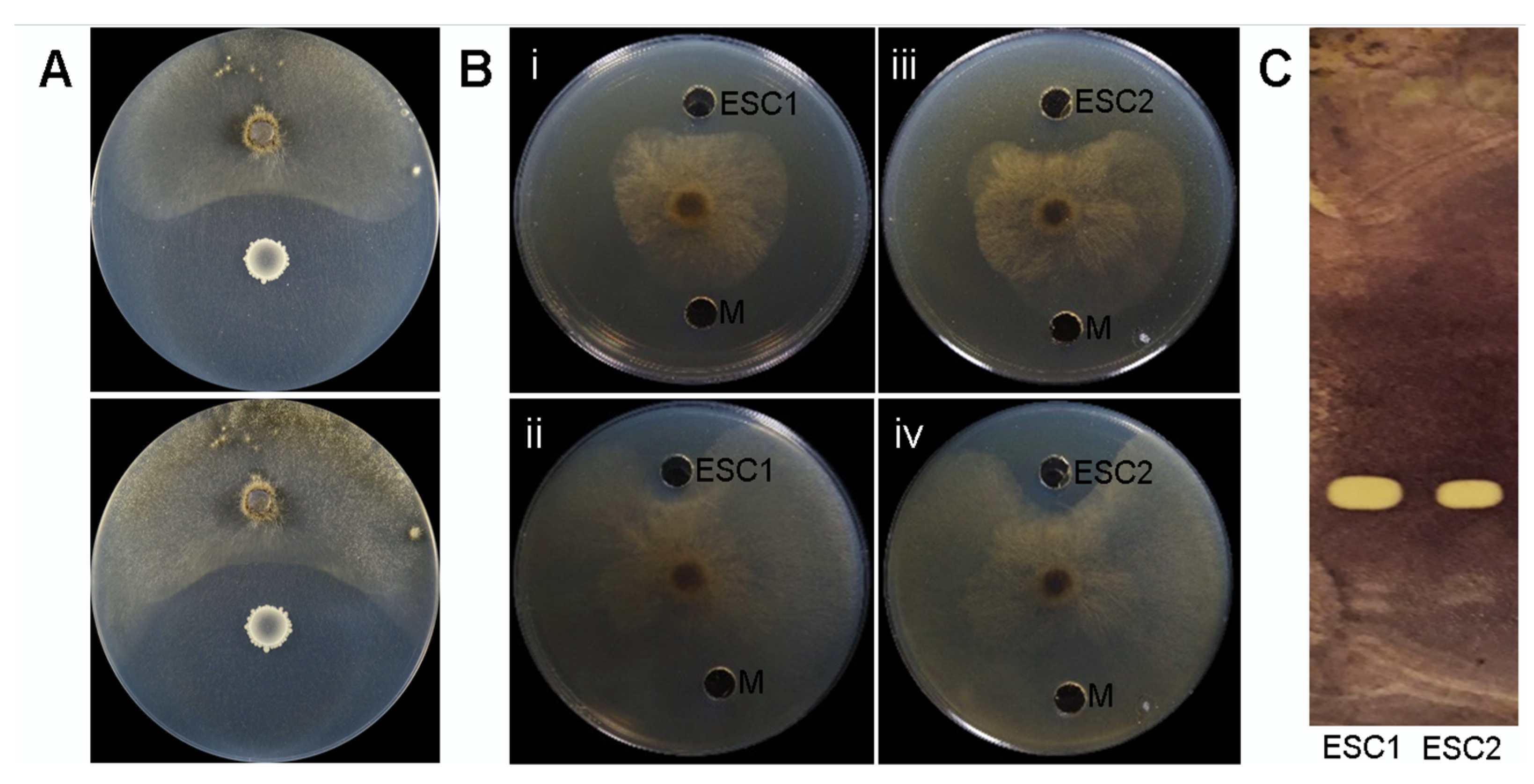
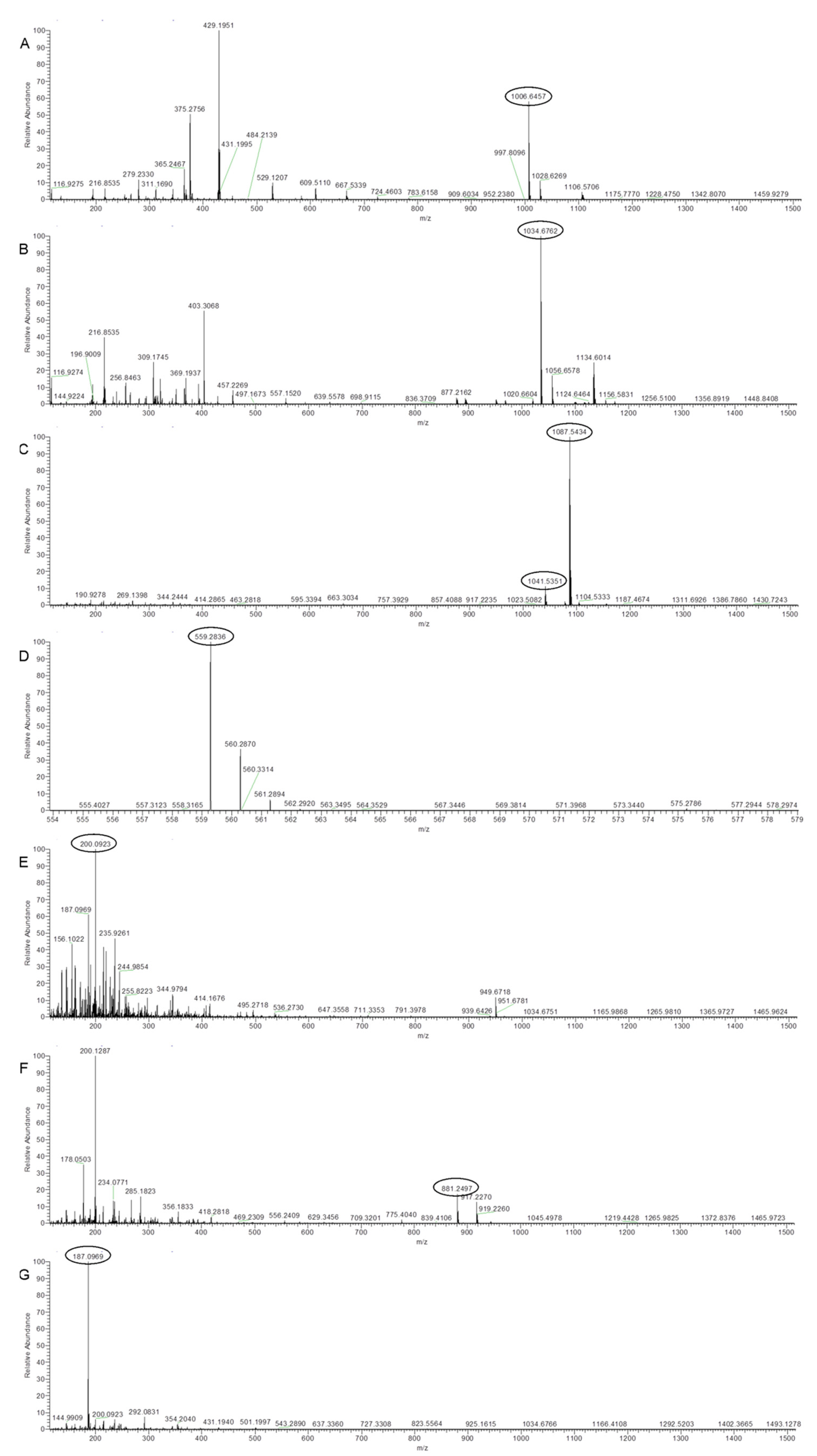

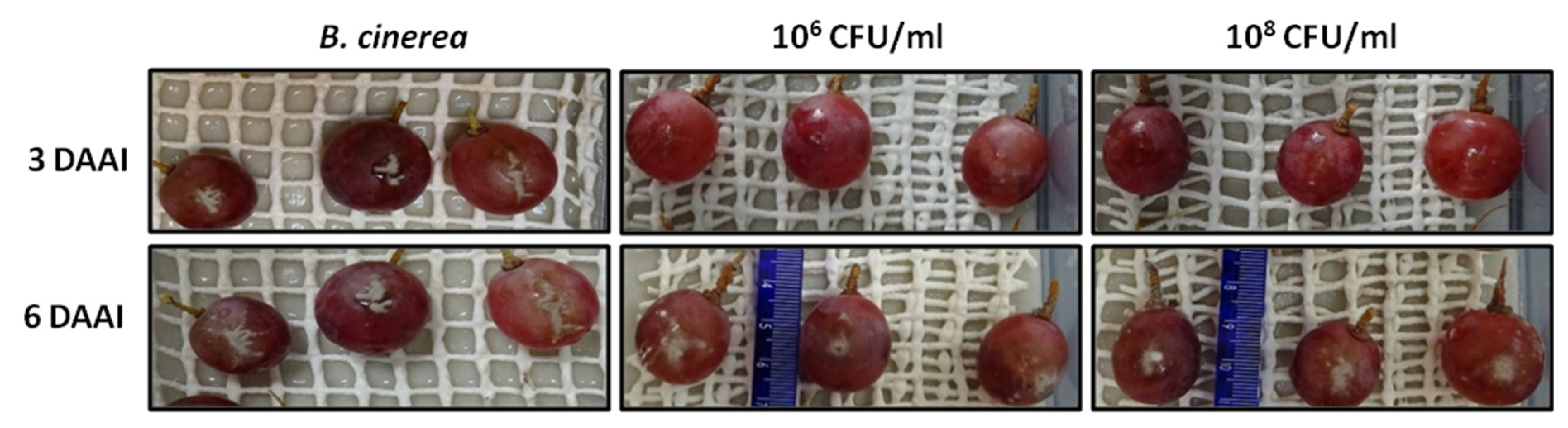
| Bacterial Strains | OrthoANI (%) | dDDH (%) |
|---|---|---|
| Bacillus velezensis Bvel1 | 100.00 | 100.00 |
| B. amyloliquefaciens DSM 7 T | 93.94 | 55.10 |
| B. amyloliquefaciens LL3 | 93.86 | 54.50 |
| B. amyloliquefaciens YP6 | 94.03 | 55.10 |
| B. velezensis Y2 | 97.70 | 80.00 |
| B. velezensis FZB42 | 97.70 | 80.10 |
| B. velezensis QST713 | 97.62 | 79.20 |
| B. velezensis Lzh-a42 | 97.82 | 80.00 |
| B. velezensis LB002 | 98.70 | 99.84 |
| B. velezensis SQR9 | 97.70 | 79.40 |
| B. velezensis NRRL B-41580 T | 97.75 | 79.50 |
| Antibiotic Compounds | Molecular Formula | Experimental m/z | RT (min) | Δm (ppm) | Adduct | References |
|---|---|---|---|---|---|---|
| Surfactin A-C13 | C51H89N7O13 | 1006.6457 | 22,699 | 2.22 | [M − H]− | [26] |
| Surfactin A-C15 | C53H93N7O13 | 1034.6762 | 23,945 | 1.39 | [M − H]− | [27] |
| Iturin A2-C14 | C48H74N12O14 | 1041.5351 | 14,310 | −1.22 | [M − H]− | [28] |
| L-dihydroanticapsin | C9H15NO4 | 200.0923 | 11,395 | 2.83 | [M − H]− | [29] |
| Oxydifficidin | C31H45O7P | 559.2836 | 16,950 | 3.01 | [M − H]− | [30] |
| Bacillibactin | C39H42N6O18 | 881.2497 | 11,880 | 2.85 | [M − H]− | [31] |
| Azelaic acid | C9H16O4 | 187.0969 | 11,012 | 2.22 | [M − H]− | [32] |
| Cluster Number | Synthetase | Most Similar Known Cluster | MIBiG ID (% of Genes Show Similarity) | Predicted Size (bp) |
|---|---|---|---|---|
| 1 | NPRS, TransATPKS | Rhizocticin | BGC0000926_c1 (22%) | 77.624 |
| 2 | NRPS | Surfactin | BGC0000433_c1 (82%) | 65.407 |
| 3 | PKS-like | Butirosin | BGC0000693_c1 (7%) | 41.224 |
| 4 | Terpene | - | - | 20.740 |
| 5 | Lanthipeptide-class-II | - | - | 28.889 |
| 6 | TransATPKS | Macrolactin | BGC0000181_c1 (100%) | 86.366 |
| 7 | TransATPKS-NRPS | Bacillaene | BGC0001089_c1 100(%) | 109.176 |
| 8 | NPRS, TransATPKS-NRPS | Fengycin | BGC0001095_c1 (93%) | 114.939 |
| 9 | Terpene | - | - | 41.246 |
| 10 | T3PKS | - | - | 20.739 |
| 11 | TransATPKS | Difficidin | BGC0000176_c1 (100%) | 106.166 |
| 12 | NPRS | Bacillibactin | BGC0000309_c1 (100%) | 51.789 |
| 13 | NPRS | Bacilycin | BGC0001184_c1 (100%) | 41.418 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nifakos, K.; Tsalgatidou, P.C.; Thomloudi, E.-E.; Skagia, A.; Kotopoulis, D.; Baira, E.; Delis, C.; Papadimitriou, K.; Markellou, E.; Venieraki, A.; et al. Genomic Analysis and Secondary Metabolites Production of the Endophytic Bacillus velezensis Bvel1: A Biocontrol Agent against Botrytis cinerea Causing Bunch Rot in Post-Harvest Table Grapes. Plants 2021, 10, 1716. https://doi.org/10.3390/plants10081716
Nifakos K, Tsalgatidou PC, Thomloudi E-E, Skagia A, Kotopoulis D, Baira E, Delis C, Papadimitriou K, Markellou E, Venieraki A, et al. Genomic Analysis and Secondary Metabolites Production of the Endophytic Bacillus velezensis Bvel1: A Biocontrol Agent against Botrytis cinerea Causing Bunch Rot in Post-Harvest Table Grapes. Plants. 2021; 10(8):1716. https://doi.org/10.3390/plants10081716
Chicago/Turabian StyleNifakos, Kallimachos, Polina C. Tsalgatidou, Eirini-Evangelia Thomloudi, Aggeliki Skagia, Dimitrios Kotopoulis, Eirini Baira, Costas Delis, Konstantinos Papadimitriou, Emilia Markellou, Anastasia Venieraki, and et al. 2021. "Genomic Analysis and Secondary Metabolites Production of the Endophytic Bacillus velezensis Bvel1: A Biocontrol Agent against Botrytis cinerea Causing Bunch Rot in Post-Harvest Table Grapes" Plants 10, no. 8: 1716. https://doi.org/10.3390/plants10081716
APA StyleNifakos, K., Tsalgatidou, P. C., Thomloudi, E.-E., Skagia, A., Kotopoulis, D., Baira, E., Delis, C., Papadimitriou, K., Markellou, E., Venieraki, A., & Katinakis, P. (2021). Genomic Analysis and Secondary Metabolites Production of the Endophytic Bacillus velezensis Bvel1: A Biocontrol Agent against Botrytis cinerea Causing Bunch Rot in Post-Harvest Table Grapes. Plants, 10(8), 1716. https://doi.org/10.3390/plants10081716









