Salinity Effects on Gene Expression, Morphological, and Physio-Biochemical Responses of Stevia rebaudiana Bertoni In Vitro
Abstract
:1. Introduction
2. Results
2.1. Effect of NaCl on Morphological Responses of Stevia Rebaudiana Bertoni In Vitro
2.2. Effect of NaCl on Physiological Responses of Stevia Rebaudiana Bertoni In Vitro
2.3. Effect of NaCl on Gene Expression and Biochemical Response of Stevia Rebaudiana Bertoni In Vitro
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Mode of Excision
4.3. Callus Induction, Shoots Regeneration and Rooting
4.4. Estimation of Photosynthetic Pigments and Proline Accumulation
4.5. Isozymes Electrophoresis
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sreedhar, R.V.; Venkatachalam, L.; Thimmaraju, R.; Bhagyalakshmi, N.; Narayan, M.S.; Ravishankar, G.A. Direct organogenesis from leaf explants of Stevia rebaudiana and cultivation in bioreactor. Biol. Plant. 2008, 52, 355–360. [Google Scholar]
- Ibrahim, I.A.; Nasr, M.I.; Mohammed, B.R.; EL-Zifzafi, M.M. Plant growth regulators affecting in vitro cultivation of Stevia rebaudiana. Sugar Tech. 2008, 3, 248–253. [Google Scholar] [CrossRef]
- Lyakhovkin, A.G.; Long, T.D.; Titov, D.A.; Anh, M.P. Cultivation and Utilization of Stevia (Stevia rebaudiana Bertoni); Agricultural Publishing House: Hanoi, Vietnam, 1993; pp. 1–44. [Google Scholar]
- Abbas, M.S.; Dobeie, A.M.; Azzam, C.R.; Soliman, A.S. Identification of salt-tolerant Genotypes among Egyptian and Nigerian Peanut (Arachis hypogaea L.) Using Biochemical and Molecular Tools. In Mitigating Environmental Stresses for Agricultural Sustainability in Egypt; Springer International Publishing: New York, NY, USA, 2021; pp. 437–469. [Google Scholar] [CrossRef]
- ElSayed, A.I.; Boulila, M.; Rafudeen, M.S.; Sengupta, S.; Rady, M.M. Melatonin regulatory mechanisms and phylogenetic analyses implying new sequences of melatonin biosynthesis-related genes extracted from peanut under salinity stress. Plants 2020, 9, 854. [Google Scholar] [CrossRef]
- Abo-Doma, A.; Azzam, C.R. Hunting of some differentially expressed genes under salt stress in wheat. Egypt. J. Plant Breed. 2007, 11, 233–244. [Google Scholar]
- Azzam, C.R.; Edris, S.; Mansour, A.A. Changes in wheat P5CS gene expression in response to salt stress in wheat. Egypt. J. Genet. Cytol. 2009, 38, 375–386. [Google Scholar]
- Rady, M.M.; Kuşvuran, A.; Alharby, H.F.; Alzahrani, Y.; Kuşvuran, S. Pretreatment with proline or an organic bio-stimulant induces salt tolerance in wheat plants by improving antioxidant redox state and enzymatic activities and reducing the oxidative stress. J. Plant Growth Regul. 2019, 38, 449–462. [Google Scholar] [CrossRef]
- Rehman, H.; Alharby, H.F.; Bamagoos, A.A.; Abdelhamid, M.T.; Rady, M.M. Sequenced application of glutathione as an antioxidant with organic biostimulant improves physiological and metabolic adaptation to salinity in wheat. Plant Physiol. Biochem. 2021, 158, 43–52. [Google Scholar] [CrossRef]
- Azzam, C.R.; Abd El Naby, Z.M.; Mohamed, N.A. Salt Tolerance Associated With Molecular Markers In Alfalfa. J. Biosci. Appl. Res. 2019, 5, 416–428. [Google Scholar] [CrossRef]
- Zaki, S.S.; Rady, M.M. Moringa oleifera leaf extract improves growth, physiochemical attributes, antioxidant defence system, and yields of salt-stressed Phaseolus vulgaris L. plants. Int. J. Chem Tech Res. 2015, 8, 120–134. [Google Scholar]
- Rady, M.O.A.; Semida, W.M.; Abd El-Mageed, T.A.; Hemida, K.A.; Rady, M.M. Up-regulation of antioxidative defense systems by glycine betaine foliar application in onion plants confers tolerance to salinity stress. Sci. Hortic. 2018, 240, 614–622. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Semida, W.M.; Rady, M.M.; Mohamed, G.F.; Hemida, K.A.; Alhammad, B.A.; Hassan, M.M.; Shami, A. Sequenced Antioxidants Application Rectifies Ion Imbalance and Strengthens Antioxidant Systems in Salt-stressed Cucumber. Plants 2020, 9, 1783. [Google Scholar] [CrossRef]
- Taha, R.S.; Seleiman, M.F.; Alotaibi, M.; Alhammad, B.A.; Rady, M.M.; Mahdi, A.H.A. Exogenous potassium treatments elevate salt tolerance and performances of Glycine max by boosting antioxidant defense system under actual saline field conditions. Agronomy 2020, 10, 1741. [Google Scholar] [CrossRef]
- Flowers, T.J. Improving crop salt tolerance. Am. J. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Hernandez, J.A.; Jimenez, A.; Mullineaux, P.; Sevilla, F. Tolerance of pea (Pisum sativum L.) to long term salt stress is associated with induction of antioxidant defenses. Plant Cell Enviorn. 2000, 23, 853–862. [Google Scholar]
- Khan, M.H.; Panda, S.K. Induction of Oxidative Stress in Roots of Oryza sativa L. in Response to Salt Stress. Biol. Plant. 2002, 45, 625–627. [Google Scholar] [CrossRef]
- Queirós, F.; Fidalgo, F.; Santos, I.; Salema, R.R. In vitro selection of salt tolerant cell lines in Solanum tuberosum L. Biol. Plant. 2007, 51, 728–734. [Google Scholar] [CrossRef]
- Gerami, M.; Majidian, P.; Ghorbanpour, A.; Alipour, Z. Stevia rebaudiana Bertoni responses to salt stress and chitosan elicitor. Physiol. Mol. Biol. Plants 2020, 26, 965–974. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Ann. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, H.J.; Lips, S.H. Cell suspension cultures of Solanum tuberosum L. as a model system for N and salinity response. Effect of salinity on NO3- uptake and PM-ATPase activity. J. Plant Physiol. 1997, 150, 103–109. [Google Scholar] [CrossRef]
- Al-Taweel, S.K.; Azzam, C.R.; Khaled, K.A.; Abdel-Aziz, R.M. Improvement of stevia (stevia rebaudiana Bertoni) and steviol glycoside through traditional breeding and biotechnological approaches. Sabrao J. Breed. Genet. 2021, 53, 88–111. [Google Scholar]
- Taware, A.S.; Harke, S.N.; Mukadam, D.S.; Chavan, A.M.; Taware, S.D. Effect of different extracts of callus and plantlets of Stevia rebaudiana (Bertoni) on seed germination of some agricultural crops. Afr. J. Biotechnol. 2010, 9, 6675–6683. [Google Scholar] [CrossRef]
- Janarthanam, B.; Gopalakrishnan, M.; Lakshmi, M.S.G.; Sekar, T. Plant regeneration from leaf-derived callus of Stevia rebaudiana Bertoni. Plant Tissue Cult. Biotechnol. 2009, 19, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Patel, R.M.; Shah, R.R. Regeneration of Stevia Plant through Callus Culture. Indian J. Pharm. Sci. 2009, 2, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Sheng, R.L.; Yuan, H.Y.; Huang, S.Z. Influences of Na2CO3 stress on physiological metabolisms of different alkali tolerant varieties of Stevia rebaudiana. Agric Sci. Technol. 2011, 12, 1837–1841. [Google Scholar]
- Hasegawa, P.M.; Bressan, R.A.; Nelson, D.E.; Samaras, Y.; Rhodes, D. Tissue culture in the improvement of salt tolerance in plants. In Monographs on Theoretical and Applied Genetics: Breeding Plants with Resistance to Problem Soils; Springer: Berlin/Heidelberg, Germany, 1994; pp. 83–125. [Google Scholar]
- Tal, M. In vitro selection for salt tolerance in crop plants: Theoretical and practical considerations. Vitro Cell. Dev. Biol. 1994, 30, 175–180. [Google Scholar]
- Gu, R.; Liu, Q.; Pie, D.; Jiang, X. Understanding saline and osmotic tolerance of Populus euphratica suspended cells. Plant Cell Tissue Organ Cult. 2004, 78, 261–265. [Google Scholar]
- Chen, R.; Gyokusen, K.; Saito, A. Selection, regeneration and protein profile characteristics of NaCl-tolerant callus of Robinia pseudoacaia L. J. Res. 2001, 6, 43–48. [Google Scholar] [CrossRef]
- Jaiswal, R.; Singh, N.P. Plant Regeneration from NaCl-Tolerant Callus/Cell Lines of Chickpea. ICPN 2001, 8, 21–23. [Google Scholar]
- Mubarak, M.H.; Belal, A.H.; El-Dein, T.N.; El-Sarag, E.I. In vitro response growth Stevia rebaudiana to salinity and drought. Sinai J. Appl. Sci. 2012, 1, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Zeng, J.; Chen, A.; Li, D.; Yi, B.; Wu, W. Effects of salt stress on the growth, physiological responses, and glycoside contents of Stevia rebaudiana Bertoni. J. Agric. Food Chem. 2013, 61, 5720–5726. [Google Scholar] [CrossRef]
- Pandey, M.; Chikara, S.K. In vitro Regeneration and Effect of Abiotic Stress on Physiology and Biochemical Content of Stevia Rebaudiana ‘Bertoni’. J. Plant Sci. Res. 2014, 1, 113. [Google Scholar]
- Rathore, S.; Singh, N.; Singh, S.K. Influence of NaCl on biochemical parameters of two cultivars of Stevia rebaudiana regenerated in vitro. J. Stress Physiol. Biochem. 2014, 10, 287–296. [Google Scholar]
- Tanksley, S.D.; Ortan, T. Iosymes in plant genetics and breeding. In Isozymes in Plant Genetics and Breeding, 1st ed.; Elsevier: Amsterdam, The Netherlands, 1983; Volume 1A. [Google Scholar]
- Zhang, M.; Fang, Y.; Ji, Y.; Jiang, Z.; Wang, L. Effects of salt stress on ion content, antioxidant enzymes and protein profile in different tissues of Broussonetia papyrifera. S. Afr. J. Bot. 2013, 85, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kalir, A.; Omri, G.; Poljakoff-Mayber, A. Peroxidase and catalase activity in leaves of Halimione portulacoides exposed to salinity. Physiol. Plant. 1984, 238–244. [Google Scholar] [CrossRef]
- Aly, A.A.H.; Maraei, R.W.; Aldrussi, I. Changes in peroxidase and polyphenol oxidase activity and transcript levels of related genes in two Egyptian bread wheat cultivars (Triticum aestivum L.) affected by gamma irradiation and salinity stress. Bangladesh J. Bot. 2019, 48, 177–186. [Google Scholar] [CrossRef]
- Cai, C.; Wu, S.; Niu, E.; Cheng, C.; Guo, W. Identification of genes related to salt stress tolerance using intron-length polymorphic markers, association mapping and virus-induced gene silencing in cotton. Sci. Rep. 2017, 7, 528. [Google Scholar] [CrossRef] [Green Version]
- Latef, A.A.H.A.; Omer, A.M.; Badawy, A.A.; Osman, M.S.; Ragaey, M.M. Strategy of salt tolerance and interactive impact of Azotobacter chroococcum and/or Alcaligenes faecalis inoculation on canola (Brassica napus L.) plants grown in saline soil. Plants 2021, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Fallah, F.; Nokhasi, F.; Ghaheri, M.; Kahrizi, D.; Beheshti Ale Agha, A.; Ghorbani, T.; Kazemi, E.; Ansarypour, Z. Effect of salinity on gene expression, morphological and biochemical characteristics of stevia rebaudiana Bertoni under in vitro conditions. Cell. Mol. Biol. 2017, 63, 102–106. [Google Scholar] [CrossRef]
- Abdol-Qados, A.M.S. Effect of salt stress on plant growth and metabolism of bean plant Vicia faba (L.). J. Saudi Soc. Agric. Sci. 2011, 10, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, K.; Srivastava, A. Assessment of salinity tolerance of Vinga mungovar. Pu-19 using ex vitro and in vitro methods. Asian J. Biotechnol. 2010, 2, 73–85. [Google Scholar] [CrossRef] [Green Version]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Motos, J.R.; Díaz-Vivancos, P.; Álvarez, S.; Fernández-García, N.; Sánchez-Blanco, M.J.; Hernández, J.A. Physiological and biochemical mechanisms of the ornamental Eugenia myrtifolia L. plants for coping with NaCl stress and recovery. Planta 2015, 242, 829–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acosta-Motos, J.R.; Díaz-Vivancos, P.; Álvarez, S.; Fernández-García, N.; Sánchez-Blanco, M.J.; Hernández, J.A. NaCl-induced physiological and biochemical adaptive mechanisms in the ornamental Myrtus communis L. plants. J. Plant Physiol. 2015, 183, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Sadek, H.B. Identification of Fingerprints and Marker-Assisted Selection for Stress Tolerance in Some Accessions of Stevia rebaudiana. Master’s Thesis, Genetics Department, Faculty of Agriculture, Ain Shams University, Cairo, Egypt, 2003. [Google Scholar]
- El-Zifzafi, M.A. Physiological Studies on Stevia rebaudiana Bertoni through Tissue Cultures Techniques and Its Suitability for Desert Regions. Ph.D. Thesis, Faculty of Agriculture, Cairo University, Cairo, Egypt, 2003. [Google Scholar]
- Hossain, M.A.; Shamim, A.H.M.; Jahan, T.A.; Hasan, M.N. Micropropagation of Stevia. Int. J. Sustain. Crop Prod. 2008, 3, 1–9. [Google Scholar]
- Pratibha, G.; Satyawati, S.; Sanjay, S. Callusing in Stevia rebaudiana (Natural Sweetener) for Steviol Glycoside Production. Int. J. Agric. Biol. Sci. 2010, 4, 893–897. [Google Scholar]
- Tiwari, J.K.; Munshi, A.D.; Kumar, R.; Pandey, R.N.; Arora, A.; Bhat, J.S.; Sureja, A.K. Effect of salt stress on cucumber: Na+- K+ ratio, osmolyte concentration, phenols, and chlorophyll content. Acta Physiol. Plant 2010, 32, 103–114. [Google Scholar] [CrossRef]
- Cantabella, C.; Piqueras, A.; Acosta-Motos, J.R.; Bernal-Vicente, A.; Hernandez, J.A.; Diaz-Vivancos, P. Salt-tolerance mechanisms induced in Stevia rebaudiana Bertoni: Effects on mineral nutrition, antioxidative metabolism and steviol glycoside content. Plant Physiol. Biotechnol. 2017, 115, 484–496. [Google Scholar] [CrossRef]
- Guan, Z.; Su, Y.; Teng, N.; Chen, S.; Sun, H.; Li, C.; Chen, F. Morphological, physiological and structural responses of two species of Artemisia to NaCl stress. Sci. World J. 2013, 2013, 10. [Google Scholar] [CrossRef]
- Darko, E.; Végh, B.; Khalil, R.; Marˇcek, T.; Szalai, G.; Pál, M.; Janda, T. Metabolic responses of wheat seedlings to osmotic stress induced by various osmolytes under iso-osmotic conditions. PLoS ONE 2019, 14, e0226151. [Google Scholar] [CrossRef]
- Duarte, B.; Santos, D.; Marques, J.C.; Cacador, I. Ecophysiological adaptations of two halophytes to salt stress: Photosynthesis, PS II photochemistry and antioxidant feedback-Implications for resilience in climate change. Plant Physiol. Biochem. 2013, 67, 178–188. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Erturk, U.; Ivritepe, S.N.; Erlikaya, C.Y.; Bor, M.; Ozdemir, F.; Turkan, I. Responses of the cherry rootstock to salinity in vitro. Biol. Plant. 2007, 51, 597–600. [Google Scholar] [CrossRef]
- Sherif, F.E. In vitro NaCl tolerance of Artemisia dracunculus. Int. J. Med. Arom. Plants 2012, 2, 549–557. [Google Scholar]
- Jamil, M.; Bashir, S.; Anwar, S.; Bibi, S.; Bangash, A.; Ullah, F.; Shikrha, E. Effect of salinity on physiological and biochemical characteristics of different varieties of rice. Pak. J. Bot. 2012, 44, 7–13. [Google Scholar]
- Khawale, R.N.; Singh, S.K.; Patel, V.B.; Singh, S.P. Changes due to in vitro sodium chloride induced salinity in grape (Vitis vinifera L.). Indian J. Plant Physiol. 2003, 28, 378–382. [Google Scholar]
- El-Sabrout, M.B. Micropropagation and salt tolerance evaluation in some grape cultivars. II-Salt tolerance evaluation. J. Agric. Sci. Mansura Univ. 2003, 28, 6833–6855. [Google Scholar]
- Santos, C.V. Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves among the amino acids, proline is the main effector in this response. Sci. Hortic. 2004, 103, 93–99. [Google Scholar] [CrossRef]
- Tavakkoli, E.; Rengasamy, P.S.R.; McDonald, G.K. High concentrations of Na+ and Cl− ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J. Exp. Bot. 2010, 61, 4449–4459. [Google Scholar] [CrossRef]
- Poonlaphdecha, J.; Maraval, L.; Roques, S.; Audebert, A.; Boulanger, R.; Bry, X.; Gunata, Z. Effect of timing and duration of salt treatment during growth of a fragrant rice on variety on yield and 2- acetyl-1- pyrroline, proline and GABA levels. J. Agric. Food Chem. 2012, 60, 3824–3830. [Google Scholar] [CrossRef]
- Szabados, L.; Savoure, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Dar, M.I.; Naikoo, M.I.; Rehman, F.; Naushin, F.; Khan, F.A. Proline Accumulation in Plants: Roles in Stress Tolerance and Plant Development. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Springer: New Delhi, India, 2016; pp. 155–166. [Google Scholar]
- Errabii, T.; Gandonou, C.B.; Essalmani, H.; Abrini, J.; Idaomar, M.; Senhaji, N.S. Effects of NaCl and mannitol induced stress on sugarcane (Saccharum sp.) callus cultures. Acta Physiol. Plant. 2007, 29, 95–102. [Google Scholar] [CrossRef]
- Fayek, M.A.; Shabaan, E.A.; Zayed, N.S.; El-Obeidy, A.A.; Taha, R.A. Effect of salt stress on chemical and physiological contents Jojoba (Simmondsia chinensis (Link) chneider) using in vitro culture. World J. Agric. Sci. 2010, 6, 446–450. [Google Scholar]
- Zhang, X.; Yin, H.; Chen, S.; He, J.; Guo, S. Changes in antioxidant enzyme activity and transcript levels of related genes in Limonium sinense Kuntze seedlings under NaCl stress. J. Chem. 2014, 2014, 6. [Google Scholar] [CrossRef] [Green Version]
- Arzani, A.; Ashraf, A. Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit. Rev. Plant Sci. 2016, 35, 146–189. [Google Scholar] [CrossRef]
- Abdel-Hameid, N.R.; Elanany, M.A.M.; Haiba, A.A.A.; AbdEl-Hady, E.A.A. Relative mutagencity of some food preservatives on plant cells. Aust. J. Basic Appl. Sci. 2011, 5, 2817–2826. [Google Scholar]
- Azzam, C.R.; Abd-Elnaby, Z.M.; Salem, A.K. Influence of Agro-Ecological Conditions on Gene Expression, Yield and Yield Components of the Mono-Cut (Fahl) Type of Berseem. Egypt. J. Plant Breed. 2012, 16, 135–159. [Google Scholar] [CrossRef]
- Ksouri, R.; Megdiche, W.; Falleh, H.; Trabelsi, N.; Boulaaba, M.; Smaoui, A.; Abdelly, C. Influence of biological, environmental and technical factors on phenolic content and antioxidant activities of Tunisian halophytes. C. R. Biol. 2008, 331, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Megdiche, W.; Ben Amor, N.; Debez, A.; Hessini, K.; Ksouri, R.; Abdelly, C. Physiological and biochemical traits involved in the genotypic variability to salt tolerance of Tunisian Cakile Maritime. Afr. J. Ecol. 2009, 47, 774–783. [Google Scholar] [CrossRef]
- Naffeti, M.; Sriti, J.; Hamdaoui, G.; Kchouk, E.M.; Marzouk, B. Salinity impact on fruit yield, essential oil composition and antioxidant activities of Coriandrum sativum fruit extracts. Food Chem. 2011, 124, 221–225. [Google Scholar] [CrossRef]
- Radi, A.A.; Farghaly, F.A.; Hamada, A.M. Physiological and biochemical responses of salt-tolerant and salt-sensitive wheat and bean cultivars to salinity. J. Biol. Earth Sci. 2013, 3, 72–88. [Google Scholar]
- Sairam, R.K.; Chandrasekar, V.; Srivastava, G.C. Comparison of hexaploid and tetraploid wheat cultivars in their responses to water stress. Biol. Plant 2001, 44, 89–94. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Liu, H.S.; Meng, F.T.; Zhang, S.T.; Zhang, Z.H.; Kang, G.Z. The effect of drought stress on physiological characters of leaves and seed filling characteristics of the new wheat cultivar Yamai 36 during the late developmental stage. Sci. Agric. Sin. 2000, 33, 94–96. [Google Scholar]
- Biruk, A.; Kazlovskaya, Z. Prospects for using of isozyme markers in identification of stevia cultivars. Sodinink. Ir Daržininkyste. 2008, 27, 359–364. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol. Plant. 1962, 15, 473–495. [Google Scholar] [CrossRef]
- Constan, D.; Patel, R.; Keegstra, K.; Jarvis, P. An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 2004, 38, 93–106. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water stress studies. Plant Soil. 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Iglesias, L.; Lima, H.; Simon, J.P. Isozyme identification of zygotic and nucellar seedlings in citrus. J. Hered. 1974, 65, 81–84. [Google Scholar] [CrossRef]
- Stegemann, H.; Burgermeister, W.; Franksen, H.; Krogerrecklenfor, E. Manual of gel electrophoresis, and isoelectric focusing with the apparatus PANTA-PHOR. Inst. Biochem. Messeweg. 1985, 11, D-3300. [Google Scholar]
- Rahnama, H.; Ebrahimzadeh, H. Antioxidant Isozymes Activities in Potato Plants (Solanum tuberosum L.) Under Salt Stress. J. Sci. Islamic Repub. Iran 2006, 17, 225–230. [Google Scholar]
- Baaziz, M.; Aissam, F.; Brakez, Z.; Bendiab, K.; El Hadrami, I. Electrophoretic patterns of acid soluble proteins and active isoforms of peroxidase and polyphenoloxidase typifying calli and somatic embryos of two reputed date palm cultivars in Morocco. Euphytica 1994, 76, 159–168. [Google Scholar] [CrossRef]
- Jonathan, F.W.; Weeden, N.F. Visualization and interpretation of plant isozymes. In Isozymes in Plant Biology; Soltis, D.E., Soltis, P.S., Eds.; Chpman and Hall: London, UK, 1990; pp. 5–45. [Google Scholar] [CrossRef]
- Snedecor, G.W.; Cochran, W.G. Statistical Methods, 7th ed.; The Iowa State University Press: Ames, IA, USA, 1980; p. 593. [Google Scholar]

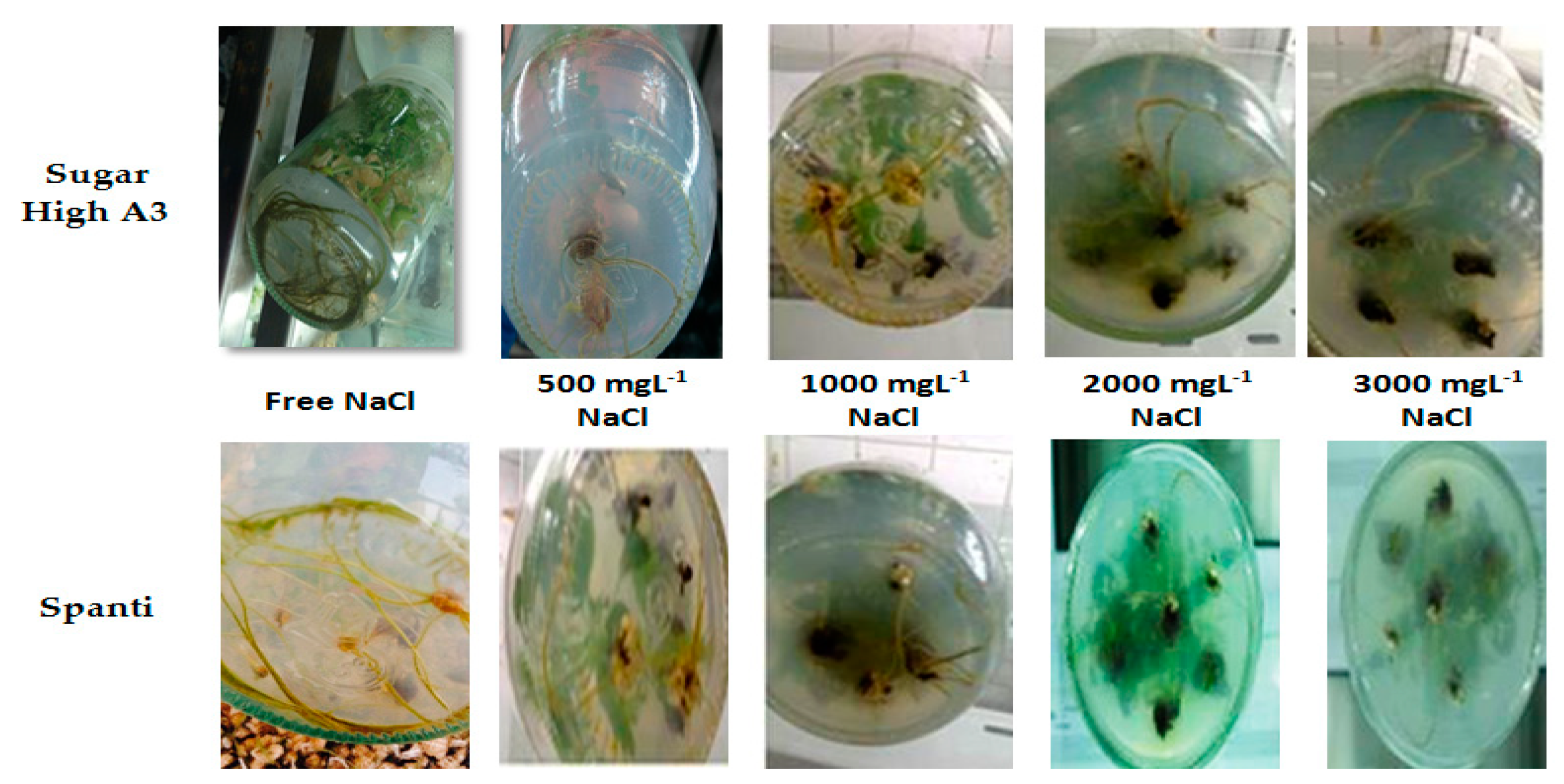

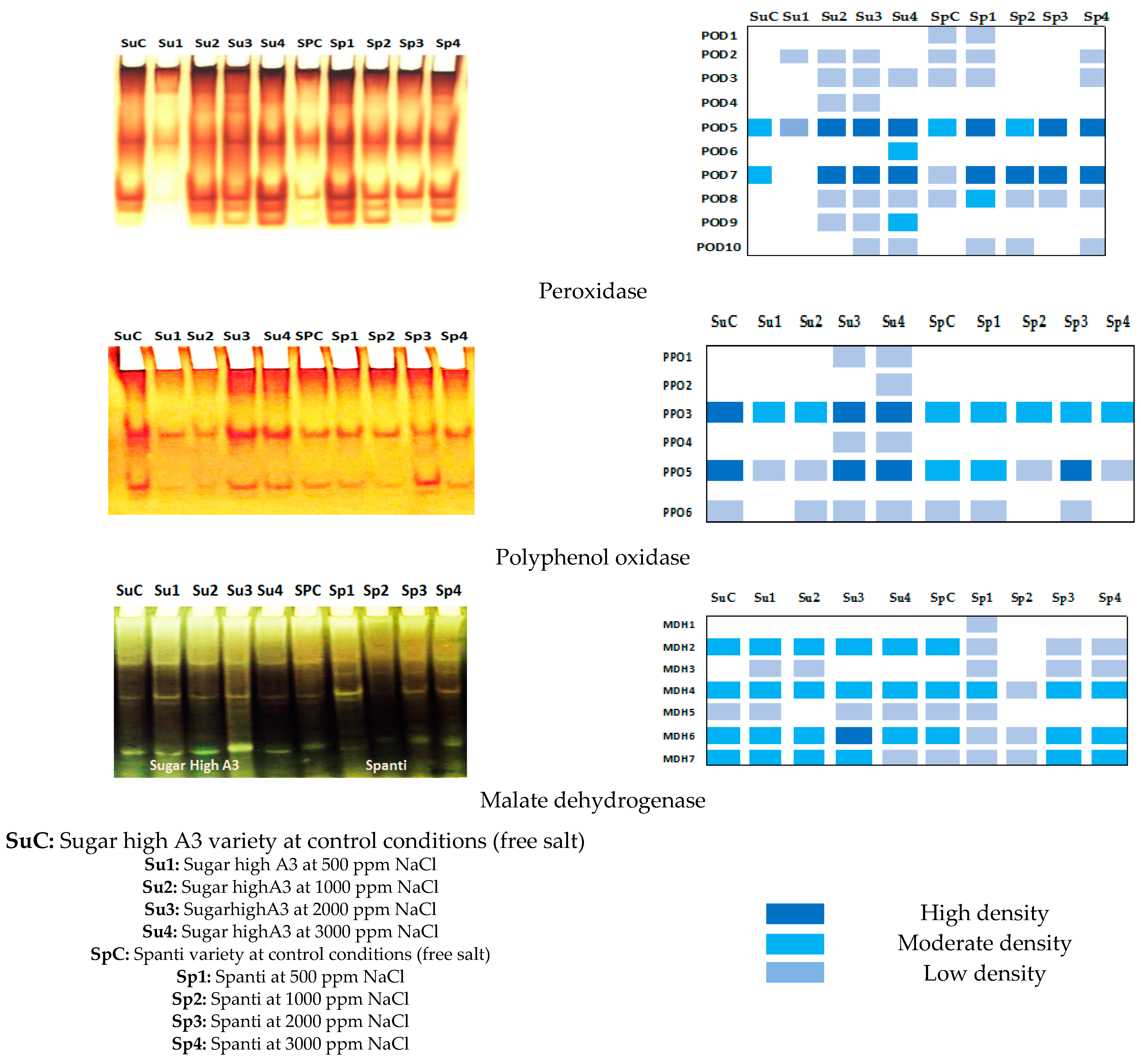
| Parameters | NaCl mg L−1 Concentrations | Stevia Varieties | Means | |
|---|---|---|---|---|
| Sugar High A3 | Spanti | |||
| Survival callus% | Control | 62.50 a | 62.00 a | 62.25 a |
| 500 | 53.09 b | 50.60 b,c | 51.85 b | |
| 1000 | 52.30 b | 43.66 d | 47.98 c | |
| 2000 | 48.05 c | 36.17 e | 42.11 d | |
| 3000 | 44.20 d | 32.65 f | 38.43 e | |
| Means | 52.13 a | 45.01 b | ||
| Regeneration% | Control | 42.11 a | 34.93 b,c | 38.52 a |
| 500 | 35.83 a,b | 30.16 c | 32.99 b | |
| 1000 | 36.03 a,b | 23.12 d,e | 29.57 c | |
| 2000 | 35.61 b,c | 20.14 e | 27.88 c,d | |
| 3000 | 29.81 c | 18.09 e | 23.95 d | |
| Means | 35.90 a | 25.30 b | ||
| Plantlet length (cm) | Control | 5.15 b | 4.86 b | 5.01 a,b |
| 500 | 5.66 a | 4.44 b | 5.05 a | |
| 1000 | 4.73 b | 4.23 b | 4.48 a,b | |
| 2000 | 4.26 b | 3.74 b | 4.00 b | |
| 3000 | 4.02 b | 3.13 b | 3.58 b | |
| Means | 4.76 a | 4.08 b | ||
| No. of leaf plantlets−1 | Control | 9.61 a | 8.76 ab | 9.19 a |
| 500 | 8.83 a,b | 7.51 a,c | 8.17 a,b | |
| 1000 | 7.56 a,b,c | 7.02 b,c,d | 7.29 b | |
| 2000 | 7.15 b,c,d | 5.53 d | 6.34 c | |
| 3000 | 6.11 c,d | 5.31 d | 5.71 c | |
| Means | 7.85 a | 6.82 b | ||
| No. of multiple shoots | Control | 15.60 a | 12.50 b,c,d | 14.05 a |
| 500 | 14.42 a,b | 10.60 d,e,f | 12.51 b | |
| 1000 | 13.27 a,b | 8.36 e,f,g | 10.82 b,c | |
| 2000 | 12.87 b,c | 7.68 f,g | 10.28 c | |
| 3000 | 10.23 c,d,e | 6.68 g | 8.46 d | |
| Means | 13.22 a | 9.16 b | ||
| Root length (cm) | Control | 7.55 a | 6.57 c,d | 7.06 a |
| 500 | 7.22 a,b | 6.23 c,d | 6.73 a | |
| 1000 | 6.71 b,c | 6.23 d,e | 6.47 b | |
| 2000 | 6.17 c,d,e | 5.21 f | 5.49 c | |
| 3000 | 6.06 e | 5.85 f | 5.96 c | |
| Means | 6.66 a | 6.02 b | ||
| No. of root plantlets−1 | Control | 7.26 a | 6.27 a,b | 6.77 a |
| 500 | 7.31 a | 5.05 c | 6.18 a,b | |
| 1000 | 6.38 a,b | 5.11 c | 5.75 b | |
| 2000 | 5.26 b,c | 3.36 d | 4.31 c | |
| 3000 | 5.01 c | 3.76 d | 4.39 c | |
| Means | 6.24 a | 4.96 b | ||
| Peroxidase Groups | Relative Mobility | Stevia Varieties | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sugar High A3 | Spanti | ||||||||||
| SuC | Su1 | Su2 | Su3 | Su4 | SpC | Sp1 | Sp2 | Sp3 | Sp4 | ||
| POD1 | 0.18 | 0 | 0 | 0 | 0 | 0 | 1− | 1− | 0 | 0 | 0 |
| POD2 | 0.23 | 0 | 1− | 1− | 1− | 0 | 1− | 1− | 0 | 0 | 1− |
| POD3 | 0.29 | 0 | 0 | 1− | 1− | 1− | 1− | 1− | 0 | 0 | 1− |
| POD4 | 0.34 | 0 | 0 | 1− | 1− | 0 | 0 | 0 | 0 | 0 | 0 |
| POD5 | 0.52 | 1+ | 1− | 1++ | 1++ | 1++ | 1+ | 1++ | 1+ | 1++ | 1++ |
| POD6 | 0.57 | 0 | 0 | 0 | 0 | 1− | 0 | 0 | 0 | 0 | 0 |
| POD7 | 0.83 | 1+ | 0 | 1++ | 1++ | 1++ | 1− | 1++ | 1++ | 1++ | 1++ |
| POD8 | 0.87 | 0 | 0 | 1− | 1− | 1− | 1− | 1+ | 1− | 1− | 1− |
| POD9 | 0.91 | 0 | 0 | 1− | 1− | 1+ | 0 | 0 | 0 | 0 | 0 |
| POD10 | 0.98 | 0 | 0 | 0 | 1− | 1− | 0 | 1− | 1− | 0 | 1− |
| Polyphenol Oxidase Groups | Relative Mobility | Stevia Varieties | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sugar High A3 | Spanti | ||||||||||
| SuC | Su1 | Su2 | Su3 | Su4 | SpC | Sp1 | Sp2 | Sp3 | Sp4 | ||
| PPO 1 | 0.38 | 0 | 0 | 0 | 1− | 1− | 0 | 0 | 0 | 0 | 0 |
| PPO 2 | 0.42 | 0 | 0 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | 0 |
| PPO 3 | 0.58 | 1++ | 1+ | 1+ | 1++ | 1++ | 1+ | 1+ | 1+ | 1+ | 1+ |
| PPO 4 | 0.62 | 0 | 0 | 0 | 1− | 1− | 0 | 0 | 0 | 0 | 0 |
| PPO 5 | 0.85 | 1++ | 1− | 1− | 1++ | 1++ | 1+ | 1+ | 1− | 1++ | 1− |
| PPO 6 | 0.88 | 1− | 0 | 1− | 1− | 1− | 1− | 1− | 0 | 1− | 0 |
| Malate Dehydrogenase Groups | Relative Mobility | Stevia Varieties | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sugar High A3 | Spanti | ||||||||||
| SuC | Su1 | Su2 | Su3 | Su4 | SpC | Sp1 | Sp2 | Sp3 | Sp4 | ||
| MDH 1 | 0.29 | 0 | 0 | 0 | 0 | 0 | 0 | 1− | 0 | 0 | 0 |
| MDH 2 | 0.33 | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1− | 0 | 1− | 1− |
| MDH 3 | 0.40 | 0 | 1− | 1− | 0 | 0 | 0 | 1− | 0 | 1− | 1− |
| MDH 4 | 0.52 | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1− | 1+ | 1+ |
| MDH 5 | 0.55 | 1− | 1− | 0 | 1− | 1− | 1− | 1− | 0 | 0 | 0 |
| MDH 6 | 0.78 | 1+ | 1+ | 1+ | 1++ | 1+ | 1+ | 1− | 1− | 1+ | 1+ |
| MDH 7 | 0.83 | 1+ | 1+ | 1+ | 1+ | 1− | 1− | 1− | 1− | 1+ | 1+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzam, C.R.; Al-Taweel, S.K.; Abdel-Aziz, R.M.; Rabea, K.M.; Abou-Sreea, A.I.B.; Rady, M.M.; Ali, E.F. Salinity Effects on Gene Expression, Morphological, and Physio-Biochemical Responses of Stevia rebaudiana Bertoni In Vitro. Plants 2021, 10, 820. https://doi.org/10.3390/plants10040820
Azzam CR, Al-Taweel SK, Abdel-Aziz RM, Rabea KM, Abou-Sreea AIB, Rady MM, Ali EF. Salinity Effects on Gene Expression, Morphological, and Physio-Biochemical Responses of Stevia rebaudiana Bertoni In Vitro. Plants. 2021; 10(4):820. https://doi.org/10.3390/plants10040820
Chicago/Turabian StyleAzzam, Clara R., Sudad K. Al-Taweel, Ranya M. Abdel-Aziz, Karim M. Rabea, Alaa I. B. Abou-Sreea, Mostafa M. Rady, and Esmat F. Ali. 2021. "Salinity Effects on Gene Expression, Morphological, and Physio-Biochemical Responses of Stevia rebaudiana Bertoni In Vitro" Plants 10, no. 4: 820. https://doi.org/10.3390/plants10040820






