B Chromosomes in Genus Sorghum (Poaceae)
Abstract
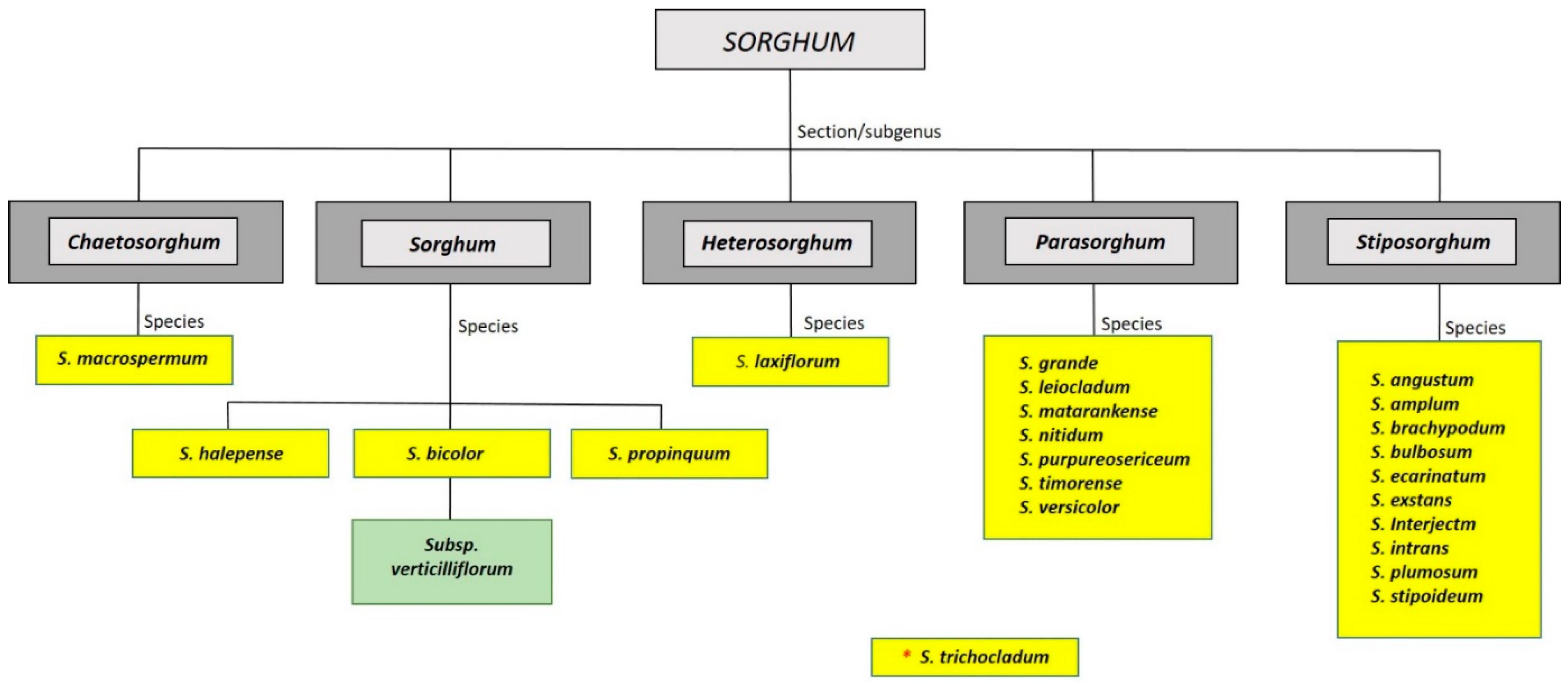
:1. Introduction
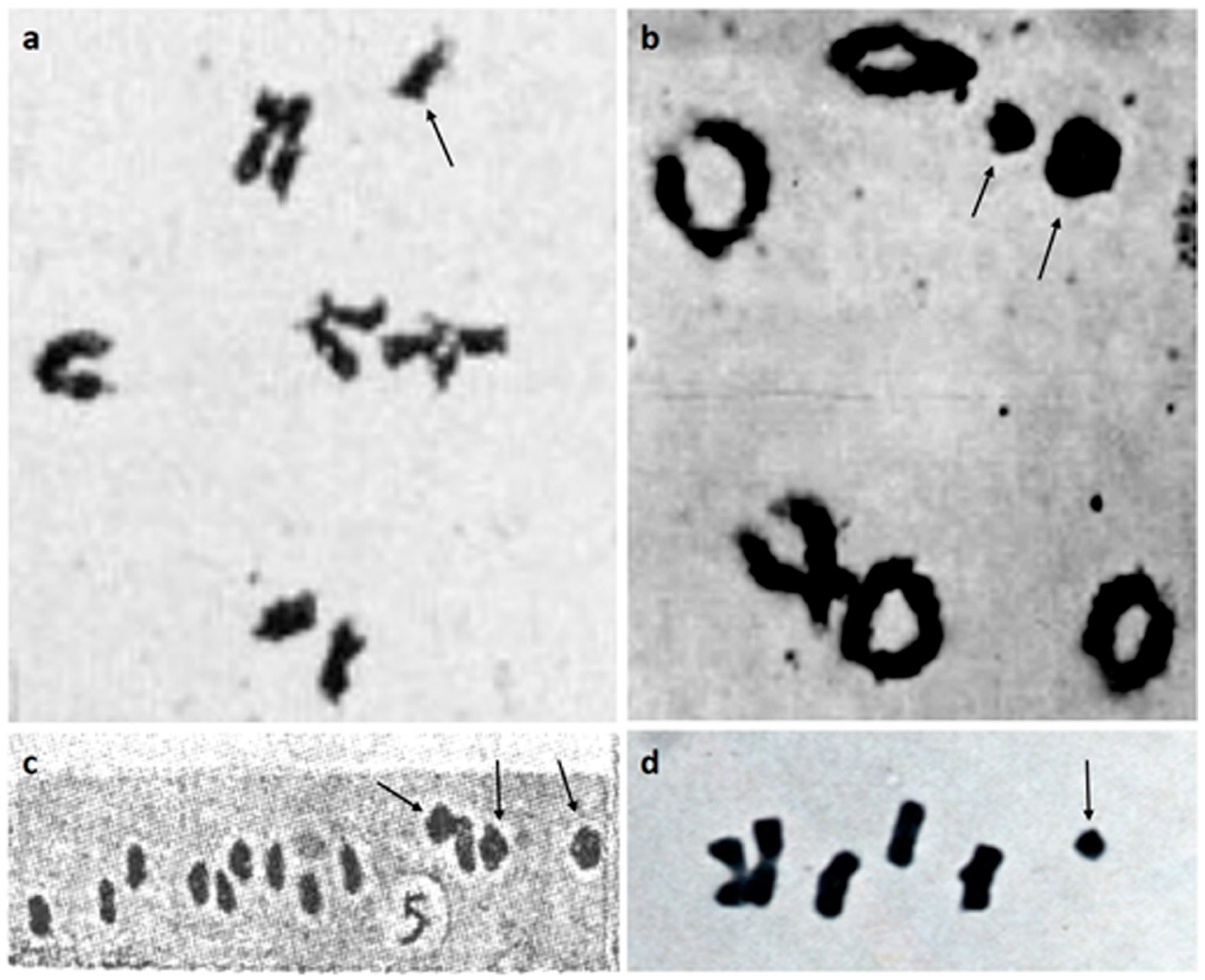
2. B Chromosomes in the Genus Sorghum
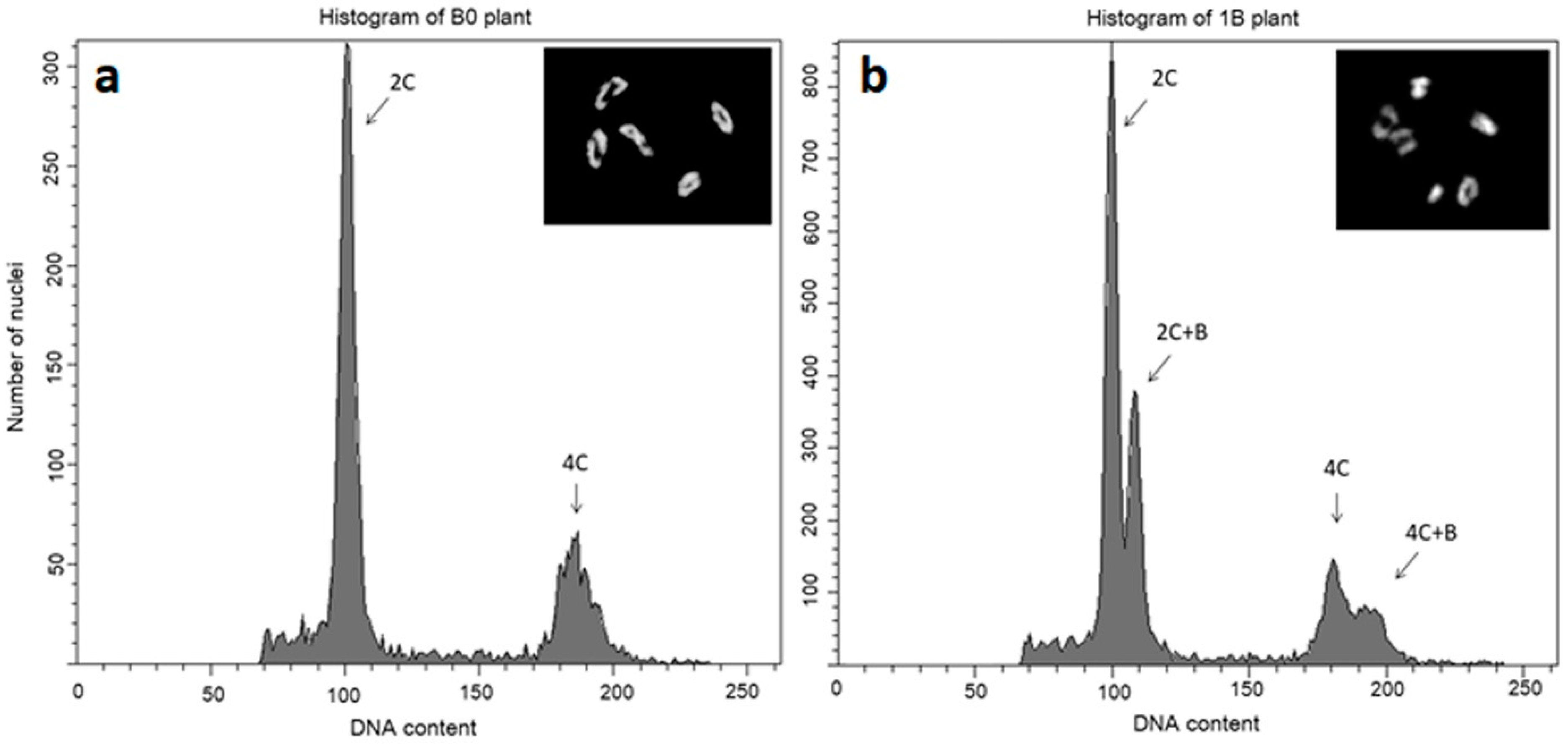
3. Elimination and Maintenance of B Chromosomes
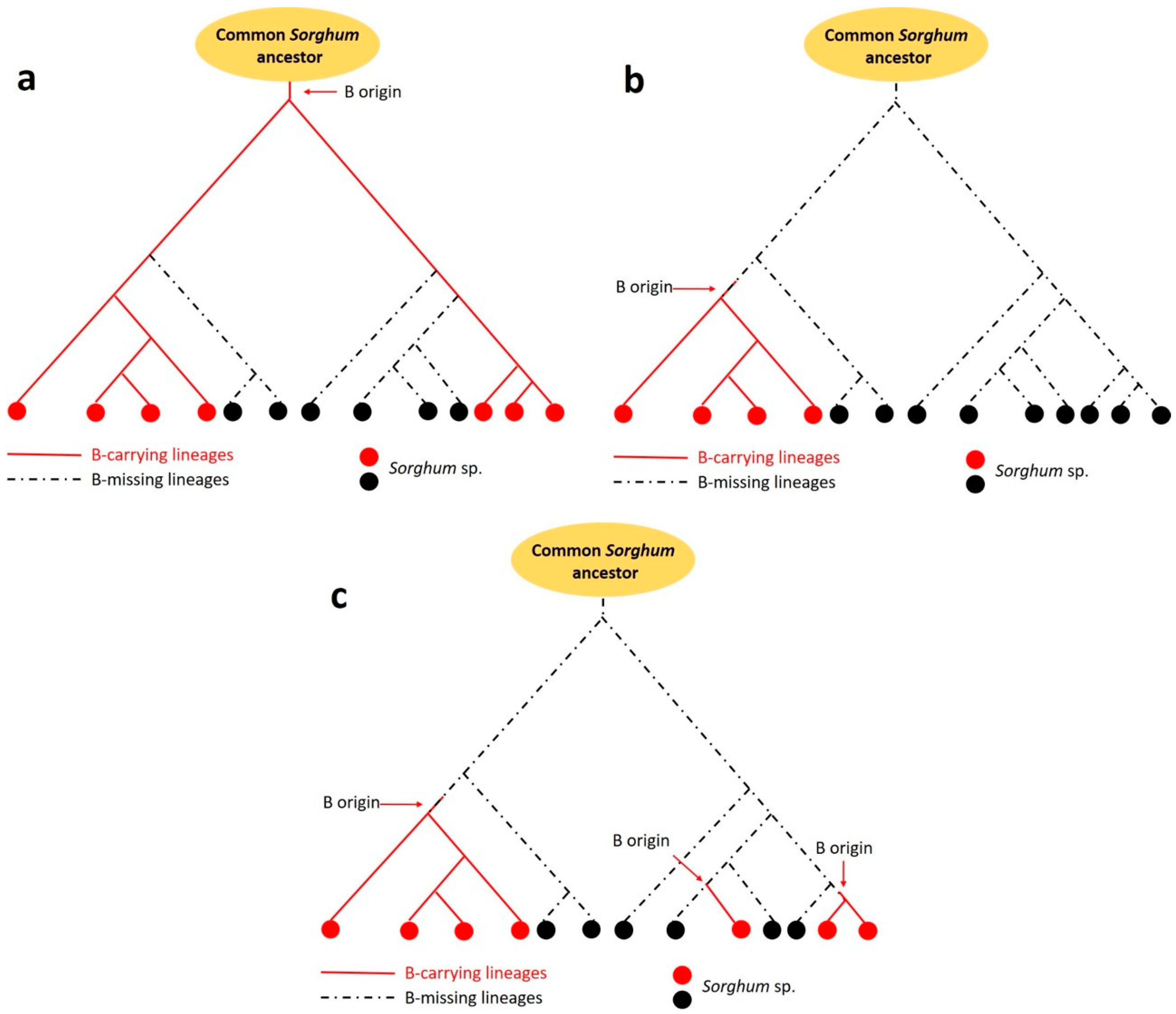
4. Did B Chromosomes Emerge Several Times in the Genus Sorghum?
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- D’Ambrosio, U.; Alonso-Lifante, M.P.; Barros, K.; Kovařík, A.; Mas de Xaxars, G.; Garcia, S. B-chrom: A database on B-chromosomes of plants, animals and fungi. New Phytol. 2017, 216, 635–642. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.N. B chromosomes in plants. New Phytol. 1995, 131, 411–434. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Sharbel, T.F.; Baukeboom, L.W. B-chromosome evolution. Phil. Trans. R. Soc. Lond. B 2000, 355, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Baukeboom, L.W. Bewildering Bs: An impression of the 1st B-chromosome conference. Heredity 1994, 73, 328–336. [Google Scholar] [CrossRef] [Green Version]
- Werren, J.H.; Stouthamer, R. PSR (paternal sex ratio) chromosomes: The ultimate selfish genetic elements. Genetica 2003, 117, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Carlson, W.R. A procedure for localizing genetic factors controlling mitotic nondisjunction in the B chromosome of maize. Chromosoma 1973, 42, 127–136. [Google Scholar] [CrossRef]
- Alfenito, M.R.; Birchler, J.A. Molecular characterization of a maize B chromosome centric sequence. Genetics 1993, 135, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Stark, E.A.; Connerton, I.; Bennet, S.T.; Barnes, S.R.; Parker, J.S.; Forster, J.W. Molecular analysis of the structure of the maize B-chromosome. Chromosome Res. 1996, 4, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Lamb, J.C.; Vega, J.M.; Dawe, R.K.; Birchler, J.A.; Jiang, J. Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 2005, 17, 1412–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandery, M.J.; Forster, J.W.; Blunden, R.; Jones, R.N. Identification of a family of repeated sequences on the rye B chromosome. Genome 1990, 33, 908–913. [Google Scholar] [CrossRef]
- Klemme, S.; Banaei-Moghaddam, A.M.; Macas, J.; Wicker, T.; Novák, P.; Houben, A. High-copy sequences reveal distinct evolution of the rye B chromosome. New Phytol. 2013, 199, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Banaei-Moghaddam, A.M.; Schubert, V.; Kumke, K.; Wei, O.; Klemme, S.; Nagaki, K.; Macas, J.; González-Sánchez, M.; Heredia, V.; Gómez-Revilla, D.; et al. Nondisjunction in favor of a chromosome: The mechanism of rye B chromosome drive during pollen mitosis. Plant Cell 2012, 24, 4124–4134. [Google Scholar] [CrossRef] [Green Version]
- Blunden, R.; Wilkes, T.J.; Forster, J.W.; Jimenez, M.M.; Sandery, M.J.; Karp, A.; Jones, R.N. Identification of the E3900 family, a second family of rye B chromosome specific repeated sequences. Genome 1993, 36, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.E. Mirrored genome size distributions in monocot and eudicot plants. Acta Biotheor. 2001, 49, 43–51. [Google Scholar] [CrossRef]
- Trivers, R.; Burt, A.; Palestis, B.G. B chromosomes and genome size in flowering plants. Genome 2004, 47, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Palestis, B.G.; Trivers, R.; Burt, A.; Jones, R.N. The distribution of B chromosomes across species. Cytogenet. Genome Res. 2004, 106, 151–158. [Google Scholar] [CrossRef]
- Levin, D.A.; Palestis, B.G.; Jones, R.N.; Trivers, R. Phyletic hotspots for B chromosomes in angiosperms. Evolution 2005, 59, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N. B-Chromosome Drive. Am. Nat. 1991, 137, 430–442. [Google Scholar] [CrossRef]
- Müntzing, A. Cytological studies of extra fragment chromosomes in rye; the mechanism of non-disjunction at the pollen mitosis. Hereditas 1946, 32, 97–119. [Google Scholar] [CrossRef]
- Roman, H. Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 1947, 32, 391–409. [Google Scholar]
- Carlson, W.R.; Chou, T.S. B chromosome nondisjunction in corn: Control by factors near the centromere. Genetics 1981, 97, 379–389. [Google Scholar]
- Han, F.; Lamb, J.C.; Yu, W.; Gao, Z.; Birchler, J.A. Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. Plant Cell 2007, 19, 524–533. [Google Scholar] [CrossRef] [Green Version]
- Roman, H. Directed fertilization in maize. Proc. Natl. Acad. Sci. USA 1948, 34, 36–42. [Google Scholar] [CrossRef] [Green Version]
- Carlson, W.R. Factors affecting preferential fertilization in maize. Genetics 1969, 62, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Rusche, M.L.; Mogensen, H.L.; Shi, L.; Keim, P.; Rougier, M.; Chaboud, A.; Dumas, C. B chromosome behavior in maize pollen as determined by a molecular probe. Genetics 1997, 147, 1915–1921. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, N. A cytological study on 8-chromosome rye. Cytologia 1934, 6, 68–77. [Google Scholar] [CrossRef] [Green Version]
- Nur, U. A mitotically unstable supernumerary chromosome with an accumulation mechanism in a grasshopper. Chromosoma 1963, 14, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Kayano, H. Accumulation of B chromosomes in the germ line of Locusta migratoria. Heredity 1971, 27, 119–123. [Google Scholar] [CrossRef] [Green Version]
- Kayano, H. Cytogenetic studies in Lilium callosum. III. Preferential segregation of a supernumerary chromosome in EMCs. Proc. Jpn. Acad. 1957, 33, 553–558. [Google Scholar] [CrossRef] [Green Version]
- Nur, U. A supernumerary chromosome with an accumulation mechanism in the lecanoid genetic system. Chromosoma 1962, 13, 249–271. [Google Scholar] [CrossRef]
- Raman, V.S.; Krishnaswami, D. Accessory chromosomes in sorghum nitidum Pers. J. Indian Bot. Soc. 1960, 39, 278–280. [Google Scholar]
- Zuk, J. The additional heterochromatic chromosome and its influence on sex chromosome pairing in Rumex. Heredity 1969, 24, 69–74. [Google Scholar] [CrossRef]
- Holmes, D.S.; Bougourd, S.M. B-chromosome selection in Allium schoenoprasum II. Experimental populations. Heredity 1991, 67, 117–122. [Google Scholar] [CrossRef] [Green Version]
- Bakkali, M.; Cabrero, J.; López-León, M. The B chromosome polymorphism of the grasshopper Eyprepocnemis plorans in North Africa. I. B variants and frequency. Heredity 1999, 83, 428–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Marco Ferro, D.A.; Moreira-Filho, O.; Bertollo, L.A.C. B chromosome polymorphism in the fish, Astyanax scabripinnis. Genetica 2003, 119, 147–153. [Google Scholar] [CrossRef]
- Plowman, A.; Bougourd, S. Selectively advantageous effects of B chromosomes on germination behaviour in Allium schoenoprasum L. Heredity 1994, 72, 587–593. [Google Scholar] [CrossRef]
- Dherawattana, A.; Sadanaga, K. Cytogenetics of a crown rust-resistant hexaploid oat with 42 + 2 fragment chromosomes1. Crop Sci. 1973, 13, 591–594. [Google Scholar] [CrossRef]
- Han, Y.; Liu, X.; Benny, U.; Kistler, H.C.; VanEtten, H.D. Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J. 2001, 25, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Martis, M.M.; Klemme, S.; Banaei-Moghaddam, A.M.; Blattner, F.R.; Macas, J.; Schmutzer, T.; Scholz, U.; Gundlach, H.; Wicker, T.; Simkova, H.; et al. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 2012, 109, 13343–13346. [Google Scholar] [CrossRef] [Green Version]
- Page, B.T.; Wanous, M.K.; Birchler, J.A. Characterization of a maize chromosome 4 centromeric sequence: Evidence for an evolutionary relationship with the B chromosome centromere. Genetics 2001, 159, 291–302. [Google Scholar]
- Cheng, Y.M.; Lin, B.Y. Cloning and characterization of maize B chromosome sequences derived from microdissection. Genetics 2003, 164, 299–310. [Google Scholar] [PubMed]
- Banaei-Moghaddam, A.M.; Meier, K.; Karimi-Ashtiyani, R.; Houben, A. Formation and expression of pseudogenes on the B chromosome of rye. Plant Cell 2013, 25, 2536–2544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houben, A.; Banaei-Moghaddam, A.M.; Klemme, S.; Timmis, J.N. Evolution and biology of supernumerary B chromosomes. Cell. Mol. Life Sci. 2014, 71, 467–478. [Google Scholar] [CrossRef]
- Makunin, A.I.; Kichigin, I.G.; Larkin, D.M.; O’Brien, P.C.; Ferguson-Smith, M.A.; Yang, F.; Proskuryakova, A.A.; Vorobieva, N.V.; Chernyaeva, E.N.; O’Brien, S.J.; et al. Contrasting origin of B chromosomes in two cervids (Siberian roe deer and grey brocket deer) unravelled by chromosome-specific DNA sequencing. BMC Genom. 2016, 17, 618. [Google Scholar] [CrossRef] [Green Version]
- Miao, V.P.; Covert, S.F.; Van Etten, H.D. A fungal gene for antibiotic resistance on a dispensable (“B”) chromosome. Science 1991, 254, 1773–1776. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Conte, M.A.; Fantinatti, B.E.A.; Cabral-de-Mello, D.C.; Carvalho, R.F.; Vicari, M.R.; Kocher, T.D.; Martins, C. Origin and evolution of B chromosomes in the cichlid fish Astatotilapia latifasciata based on integrated genomic analyses. Mol. Biol. Evol. 2014, 31, 2061–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Gabriel, T.S.; Martis, M.M.; Gursinsky, T.; Schubert, V.; Vrána, J.; Doležel, J.; Grundlach, H.; Altschmied, L.; Scholz, U.; et al. Rye B chromosomes encode a functional Argonaute-like protein with in vitro slicer activities similar to its A chromosome paralog. New Phytol. 2017, 213, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Domínguez, B.; Ruiz-Ruano, F.; Cabrero, J.; Corral, J.M.; López-León, M.D.; Sharbel, T.F.; Camacho, J.P.M. Protein-coding genes in B chromosomes of the grasshopper Eyprepocnemis plorans. Sci. Rep. 2017, 7, 45200. [Google Scholar]
- Yoshida, K.; Terai, Y.; Mizoiri, S.; Aibara, M.; Nishihara, H.; Watanabe, M.; Kuroiwa, A.; Hirai, H.; Hirai, Y.; Matsuda, Y.; et al. B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 2011, 7, e100220. [Google Scholar] [CrossRef]
- Makunin, A.I.; Romanenko, S.A.; Beklemisheva, V.R.; Perelman, P.L.; Druzhkova, A.S.; Petrova, K.O.; Prokopov, D.Y.; Chernyaeva, E.N.; Johnson, J.L.; Kukekova, A.V.; et al. Sequencing of supernumerary chromosomes of red fox and raccoon dog confirms a non-random gene acquisition by B chromosomes. Genes 2018, 9, 405. [Google Scholar] [CrossRef] [Green Version]
- Huskins, C.L.; Smith, S.G. A cytological study of the genus Sorghum PERS. II. The meiotic chromosomes. J. Genet. 1934, 28, 387–395. [Google Scholar] [CrossRef]
- Wu, T. B-chromosomes in Sorghum stipoideum. Heredity 1992, 68, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Janaki-Ammal, E. Chromosome Diminution in a Plant. Nature 1940, 146, 839–840. [Google Scholar] [CrossRef]
- Darlington, C.D.; Thomas, P.T. Morbid mitosis and the activity of inert chromosomes in sorghum. Proc. R. Soc. Lond. B 1941, 130, 127–150. [Google Scholar]
- Raman, V.S.; Meenakshi, K.; Thangam, M.S.; Sivagnanam, L. The cytological behaviour of B-chromosomes in Sorghum halepense. Abst. Madras Agric. J. 1964, 51, 72–73. [Google Scholar]
- Raman, V.S.; Thangam, M.S. Paternal transmission of accessory chromosomes in a species of Eu-Sorghum. Sci. Cult. 1965, 31, 150–151. [Google Scholar]
- Wu, T.P.; Pi, C.P. Accessory chromosome in Sorghum nitidum Pers. Taiwania 1975, 20, 147–161. [Google Scholar]
- Wu, T.P. B chromosomes in Sorghum purpureo-sericeum. Proc. Natl. Sci. Counc. B 1984, 8, 198–209. [Google Scholar]
- D’cruz, R.; Deshmukh, J.N. Behaviour of B chromosomes in Sorghum purpureosericeum. Poona Agric. Col. Mag. 1960, 51, 30–34. [Google Scholar]
- Wu, T.P. Pachytene morphology of Sorghum nitidum chromosome complement. Cytologia 1978, 43, 433–440. [Google Scholar] [CrossRef] [Green Version]
- Friebe, B. Nucleolar activity of B-chromosomes in Allium cernuum (Alliaceae). Plant Syst. Evol. 1989, 163, 87–92. [Google Scholar] [CrossRef]
- Fröst, S.; Östergren, G. Crepis pannonica and Crepis conyzaefolia—Two more species having accessory chromosomes. Hereditas 1959, 45, 211–214. [Google Scholar] [CrossRef]
- Raman, V.S.; Meenakshi, K.; Thangam, M.S. Accessory chromosomes and their meiotic behaviour in hybrids of grain sorghum and Johnson grass. Cytologia 1976, 41, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Durkin, S.G.; Glover, T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007, 41, 169–192. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ma, L.; Yang, F.; Fei, S.; Li, L. 45S rDNA regions are chromosome fragile sites expressed as gaps in vitro on metaphase chromosomes of root-tip meristematic cells in Lolium spp. PLoS ONE 2008, 3, e2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustamante, F.O.; Rocha, L.C.; Torres, G.A.; Davide, L.C.; Mittelmann, A.; Techio, V.H. Distribution of rDNA in diploid and polyploid Lolium multiflorum Lam. and fragile sites in 45S rDNA regions. Crop Sci. 2014, 54, 617–625. [Google Scholar] [CrossRef] [Green Version]
- Lan, H.; Chen, C.L.; Miao, Y.; Yu, C.X.; Guo, W.W.; Xu, Q.; Deng, X.X. Fragile sites of ‘Valencia’ sweet orange (Citrus sinensis) chromosomes are related with active 45s rDNA. PLoS ONE 2016, 11, e0151512. [Google Scholar] [CrossRef]
- Karafiátová, M.; Bednářová, M.; Said, M.; Čížková, J.; Holušová, K.; Blavet, N.; Bartoš, J. B chromosome of Sorghum purpureosericeum reveals the first pieces of its sequence. J. Exp. Bot. 2020, 72, 1606–1616. [Google Scholar] [CrossRef]
- Ruban, A.; Schmutzer, T.; Wu, D.D.; Fuchs, J.; Boudichevskaia, A.; Rubtsova, M.; Pistrick, K.; Melzer, M.; Himmelbach, A.; Schubert, V.; et al. Supernumerary B chromosomes of Aegilops speltoides undergo precise elimination in roots early in embryo development. Nat. Commun. 2020, 11, 2764. [Google Scholar] [CrossRef]
- Jones, R.N.; Gonzáles-Sanchez, M.; González-García, M.; Vega, J.M.; Puertas, M.J. Chromosome with their life of their own. Cytogenet. Genome Res. 2008, 120, 265–280. [Google Scholar] [CrossRef]
- Wu, D.; Ruban, A.; Fuchs, J.; Macas, J.; Novák, P.; Vaio, M.; Zhou, Y.; Houben, A. Nondisjunction and unequal spindle organization accompany the drive of Aegilops speltoides B chromosomes. New Phytol. 2019, 223, 1340–1352. [Google Scholar] [CrossRef]
- Eickbush, D.G.; Eickbush, T.H.; Werren, J.H. Molecular characterization of repetitive DNA sequences from a B chromosome. Chromosoma 1991, 101, 575–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houben, A.; Verlin, D.; Leach, C.R.; Timmis, J.N. The genomic complexity of micro B chromosomes of Brachycome dichromosomatica. Chromosoma 2001, 110, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.C.; Meyer, J.M.; Corcoran, B.; Kato, A.; Han, F.; Birchler, J.A. Distinct chromosomal distributions of highly repetitive sequences in maize. Chromosome Res. 2007, 15, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Franks, T.K.; Houben, A.; Leach, C.R.; Timmis, J.N. The molecular organisation of a B chromosome tandem repeat sequence from Brachycome dichromosomatica. Chromosoma 1996, 105, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Garber, E.D. Cytotaxonomic studies in the genus Sorghum. Univ. Calif. Publ. Bot. 1950, 23, 283–361. [Google Scholar]
- Lazarides, M.; Hacker, J.B.; Andrew, M.H. Taxonomy, cytology and ecology of indigenous Australian sorghums (Sorghum Moench: Andropogoneae: Poaceae). Aust. Syst. Bot. 1991, 4, 591–635. [Google Scholar] [CrossRef]
- Ananda, G.; Myrans, H.; Norton, S.L.; Gleadow, R.; Furtado, A.; Henry, R.J. Wild Sorghum as a promising resource for crop improvement. Front. Plant Sci. 2020, 11, 1108. [Google Scholar] [CrossRef]
- Sun, Y.; Skinner, D.Z.; Liang, G.H.; Hulbert, S.H. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theor. Appl. Genetics 1994, 89, 26–32. [Google Scholar] [CrossRef]
- Ng’uni, D.; Geleta, M.; Fatih, M.; Bryngelsson, T. Phylogenetic analysis of the genus Sorghum based on combined sequence data from cpDNA regions and ITS generate well-supported trees with two major lineages. Ann. Bot. 2010, 105, 471–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Liu, H.; Wen, J.; Peterson, P.M. Infrageneric phylogeny and temporal divergence of Sorghum (Andropogoneae, Poaceae) based on low-copy nuclear and plastid sequences. PLoS ONE 2014, 9, e104933. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, J.S.; Ramachandran, D.; Henderson, A.; Freeman, J.; Carlise, M.; Harris, A.; Willison-Headley, Z. Phylogenetic reconstruction using four low-copy nuclear loci strongly supports a polyphyletic origin of the genus Sorghum. Ann. Bot. 2015, 116, 291–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, S.F.; Lin, Y.P.; Lin, B.Y. Characterization of AFLP sequences from regions of maize B chromosome defined by 12 B-10L translocations. Genetics 2005, 169, 375–388. [Google Scholar] [CrossRef] [Green Version]
- McAllister, B.F.; Werren, J.H. Hybrid origin of a B chromosome (PSR) in the parasitic wasp Nasonia vitripennis. Chromosoma 1997, 106, 243–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-León, M.D.; Neves, N.; Schwarzacher, T.; Heslop-Harrison, J.S.; Hewitt, G.M.; Camacho, J.P.M. Possible origin of a B chromosome deduced from its DNA composition using double FISH technique. Chromosome Res. 1994, 2, 87–92. [Google Scholar] [CrossRef]
- Sharbel, T.F.; Green, D.M.; Houben, A. B-chromosome origin in the endemic New Zealand frog Leiopelma hochstetteri through sex chromosome devolution. Genome 1998, 41, 14–22. [Google Scholar] [CrossRef]
- Donald, T.; Leach, C.; Clough, A.; Timmis, J.N. Ribosomal RNA genes and the B chromosome of Brachycome dichromosomatica. Heredity 1995, 74, 556–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamilena, M.; Garrido-Ramos, M.; Rejón, M.R.; Rejón, C.R.; Parker, J.S. Characterization of repeated sequences from microdissected B chromosomes of Crepis capillaris. Chromosoma 1995, 104, 113–120. [Google Scholar] [CrossRef]
- Dhar, M.K.; Friebe, B.; Koul, A.K.; Gill, B.S. Origin of an apparent B chromosome by mutation, chromosome fragmentation and specific DNA sequence amplification. Chromosoma 2002, 111, 332–340. [Google Scholar] [CrossRef]
- Fantinatti, B.E.; Mazzuchelli, J.; Valente, G.T.; Cabral-de-Mello, D.C.; Martins, C. Genomic content and new insights on the origin of the B chromosome of the cichlid fish Astatotilapia latifasciata. Genetica 2011, 139, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Green, D.M. Muller’s Ratchet and the evolution of supernumerary chromosomes. Genome 1990, 33, 818–824. [Google Scholar] [CrossRef]
- Magoon, M.L.; Shambulingappa, K.G. Karyomorphology of Sorghum propinquum and its bearing on the origin of 40-chromosome sorghum. Chromosoma 1961, 12, 460–465. [Google Scholar] [CrossRef]
- Barnaud, A.; Deu, M.; Garine, E.; Chantereau, J.; Bolteu, J.; Koïda, E.O.; McKey, D.; Joly, H.I. A weed–crop complex in sorghum: The dynamics of genetic diversity in a traditional farming system. Am. J. Bot. 2009, 96, 1869–1879. [Google Scholar] [CrossRef] [Green Version]
- Sagnard, F.; Deu, M.; Dembélé, D.; Leblois, R.; Touré, L.; Diakité, M.; Calatayud, C.; Vaksmann, M.; Bouchet, S.; Mallé, Y.; et al. Genetic diversity, structure, gene flow and evolutionary relationships within the Sorghum bicolor wild–weedy–crop complex in a western African region. Theor. Appl. Genet. 2011, 123, 1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapre, A.B.; Deshpande, D.S. Origin of B chromosomes in Coix L. through spontaneous interspecific hybridization. J. Hered. 1987, 78, 191–196. [Google Scholar] [CrossRef]
- Schartl, M.; Nanda, I.; Schlupp, I.; Wilde, B.; Epplen, J.T.; Schmid, M.; Parzefall, J. Incorporation of subgenomic amounts of DNA as compensation for mutational load in a gynogenetic fish. Nature 1995, 373, 68–71. [Google Scholar] [CrossRef]
- Perfectti, F.; Werren, J.H. The interspecific origin of B chromosomes: Experimental evidence. Evolution 2001, 55, 1069–1073. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bednářová, M.; Karafiátová, M.; Hřibová, E.; Bartoš, J. B Chromosomes in Genus Sorghum (Poaceae). Plants 2021, 10, 505. https://doi.org/10.3390/plants10030505
Bednářová M, Karafiátová M, Hřibová E, Bartoš J. B Chromosomes in Genus Sorghum (Poaceae). Plants. 2021; 10(3):505. https://doi.org/10.3390/plants10030505
Chicago/Turabian StyleBednářová, Martina, Miroslava Karafiátová, Eva Hřibová, and Jan Bartoš. 2021. "B Chromosomes in Genus Sorghum (Poaceae)" Plants 10, no. 3: 505. https://doi.org/10.3390/plants10030505
APA StyleBednářová, M., Karafiátová, M., Hřibová, E., & Bartoš, J. (2021). B Chromosomes in Genus Sorghum (Poaceae). Plants, 10(3), 505. https://doi.org/10.3390/plants10030505





