Small Non-Coding RNAs at the Crossroads of Regulatory Pathways Controlling Somatic Embryogenesis in Seed Plants
Abstract
1. Introduction
2. Small RNA Expression Profiles in Embryonic Tissues
3. Induction and Early Somatic Embryogenesis
3.1. Plant Growth Regulators and Stress Signaling Associated miRNAs
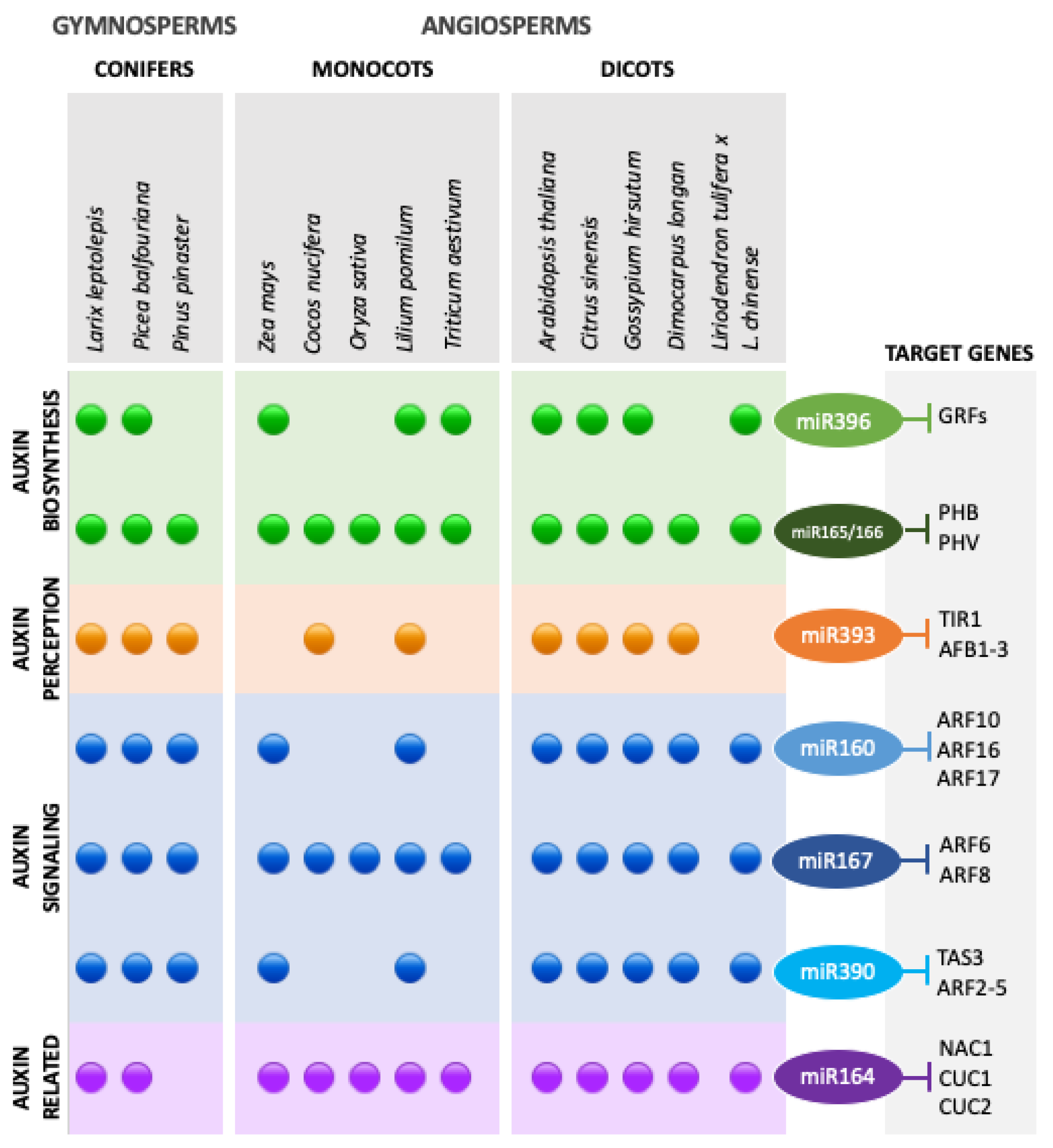
3.2. Auxins and miRNA–ARF Interactions
4. Late Somatic Embryogenesis
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Cho, S.H.; Coruh, C.; Axtell, M.J. miR156 and miR390 Regulate tasiRNA Accumulation and Developmental Timing in Physcomitrella patens. Plant Cell 2012, 24, 4837–4849. [Google Scholar] [CrossRef]
- Tsuzuki, M.; Futagami, K.; Shimamura, M.; Inoue, C.; Kunimoto, K.; Oogami, T.; Tomita, Y.; Inoue, K.; Kohchi, T.; Yamaoka, S.; et al. An Early Arising Role of the MicroRNA156/529-SPL Module in Reproductive Development Revealed by the Liverwort Marchantia polymorpha. Curr. Biol. 2019, 29, 3307–3314. [Google Scholar] [CrossRef]
- Perdiguero, P.; Rodrigues, A.S.; Chaves, I.; Costa, B.; Alves, A.; de María, N.; Vélez, M.D.; Díaz-Sala, C.; Cervera, M.T.; Miguel, C.M. Comprehensive analysis of the isomiRome in the vegetative organs of the conifer Pinus pinaster under contrasting water availability. Plant. Cell Environ. 2020, 44, 706–728. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, A.; Kellner, M.J.; Mosiolek, M.; Schon, M.A.; Nodine, M.D. “MicroRNA Dynamics and Functions During Arabidopsis Embryogenesis. Plant Cell 2019, 31, 2929–2946. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J. Classification and Comparison of Small RNAs from Plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, F.; Vaucheret, H.; Rajagopalan, R.; Lepers, C.; Gasciolli, V.; Mallory, A.; Hilbert, J.L.; Bastel, D.P.; Crété, P. Endogenous trans-acting siRNAs regulate the accumulation of arabidopsis mRNAs. Mol. Cell 2004, 16, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B. MicroRNAs in Control of Plant Development. J. Cell. Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. Micrornas as potential targets for abiotic stress tolerance in plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef]
- Alves, A.; Rodrigues, A.S.; Miguel, C. microRNAs in Plant Embryogenesis. In Plant microRNAs. Concepts and Strategies in Plant Sciences, 1st ed.; Miguel, C., Dalmay, T., Chaves, I., Eds.; Springer: Cham, Switzerland, 2020; pp. 99–120. [Google Scholar]
- ten Hove, C.A.; Lu, K.J.; Weijers, D. Building a plant: Cell fate specification in the early arabidopsis embryo. Development 2015, 142, 420–430. [Google Scholar] [CrossRef]
- Cairney, J.; Pullman, G.S. The cellular and molecular biology of conifer embryogenesis. New Phytol. 2007, 176, 511–536. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, Z.; Shen, K.; Zhao, P.; Sun, M.X. Cell lineage-specific transcriptome analysis for interpreting cell fate specification of proembryos. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Armenta-Medina, A.; Lepe-Soltero, D.; Xiang, D.; Datla, R. Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote. Dev. Biol. 2017, 431, 145–151. [Google Scholar] [CrossRef]
- von Arnold, S.; Clapham, D.; Abrahamsson, M. Embryology in conifers. In Advances in Botanical Research; Cánovas, F.M., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 89, pp. 157–184. [Google Scholar]
- Hofmann, F.; Schon, M.A.; Nodine, M.D. The embryonic transcriptome of Arabidopsis thaliana. Plant Reprod. 2019, 32, 77–91. [Google Scholar] [CrossRef]
- Rodrigues, A.S.; Miguel, C.M. The pivotal role of small non-coding RNAs in the regulation of seed development. Plant Cell Rep. 2017, 36, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.S.; Chaves, I.; Costa, B.V.; Lin, Y.C.; Lopes, S.; Milhinhos, A.; van de Peer, Y.; Miguel, C.M. Small RNA profiling in Pinus pinaster reveals the transcriptome of developing seeds and highlights differences between zygotic and somatic embryos. Sci. Rep. 2019, 9, 11327. [Google Scholar] [CrossRef]
- Liu, Y.; El-Kassaby, Y.A.; Liu, E.K. Global Analysis of Small RNA Dynamics during Seed Development of Picea glauca and Arabidopsis thaliana Populations Reveals Insights on their Evolutionary Trajectories. Front. Plant Sci. 2017, 8, 1719. [Google Scholar] [CrossRef]
- Källman, T.; Chen, J.; Gyllenstrand, N.; Lagercrantz, U. A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol. 2013, 162, 741–754. [Google Scholar] [CrossRef]
- Liu, Y.; El-Kassaby, Y.A. Landscape of fluid sets of hairpin-derived 21-/24-nt-long small RNAs at seed set uncovers special epigenetic features in Picea glauca. Genome Biol. Evol. 2017, 9, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Papareddy, R.; Páldi, K.; Paulraj; Kao, S.P.; Nodine, M. Chromatin Regulates Bipartite-Classified Small RNA Expression to Maintain Epigenome Homeostasis in Arabidopsis. Genome Biol. 2020, 21, 251. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, S.; Han, S.; Li, X.; Tong, Z.; Qi, L. Deciphering small noncoding RNAs during the transition from dormant embryo to germinated embryo in larches (Larix leptolepis). PLoS ONE 2013, 8, e81452. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Deng, C.; Xia, Y.; Kong, H.; Zhang, S.; Wang, J. Identification of novel miRNAs and miRNA expression profiling in embryogenic tissues of Picea balfouriana treated by 6-benzylaminopurine. PLoS ONE 2017, 12, e0176112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Han, S.; Wu, T.; Li, X.; Li, W.; Qi, L. Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta 2012, 236, 647–657. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, J.; Zhang, M.; Li, X.; Luo, K.; Lu, Z.; Zhang, C.; Jin, B. Identification and characterization of microrna expression in Ginkgo biloba L. Leaves. Int. J. Doc. Anal. Recognit. 2015, 11, 76. [Google Scholar] [CrossRef]
- Chu, Z.; Chen, J.; Xu, H.; Dong, Z.; Chen, F.; Cui, D. Identification and comparative analysis of microRNA in wheat (Triticum aestivum L.) callus derived from mature and immature embryos during In vitro culture. Front. Plant Sci. 2017, 30, 1302. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Juárez-González, V.T.; López-Ruiz, B.A.; Baldrich, P.; Luján-Soto, E.; Meyers, B.C.; Dinkova, T.D. The explant developmental stage profoundly impacts small RNA-mediated regulation at the dedifferentiation step of maize somatic embryogenesis. Sci. Rep. 2019, 9, 14511. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, T.; Li, L.; Han, S.; Li, X.; Zhang, S.; Qi, L. Dynamic expression of small RNA populations in larch (Larix leptolepis). Planta 2013, 237, 89–101. [Google Scholar] [CrossRef]
- Zhang, J.W.; Wu, T.; Han, S.Y.; Qi, L.W.; Zhang, S.G. Expression Analysis of 12 miRNA Families Specific to Conifers during Somatic Embryogenesis of Larch. Forest Res. 2012, 25, 411–418. [Google Scholar]
- Xu, X.; Chen, X.; Chen, Y.; Zhang, Q.; Su, L.; Chen, X.; Chen, Y.; Zhang, Z.; Lin, Y.; Lai, Z. Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour. Sci. Rep. 2020, 10, 4626. [Google Scholar] [CrossRef]
- Zhang, J.; Xue, B.; Gai, M.; Song, S.; Jia, N.; Sun, H. Small RNA and transcriptome sequencing reveal a potential miRNA-mediated interaction network that functions during somatic embryogenesis in Lilium pumilum DC. Fisch. Front. Plant Sci. 2017, 8, 566. [Google Scholar] [CrossRef]
- Lin, Y.; Lai, Z. Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.). PLoS ONE 2013, 8, e60337. [Google Scholar] [CrossRef]
- Chen, Y.; Si, L.; Chen, X.; Zhang, S.; Xu, X.; Zhang, Z.; Chen, Y.; XuHan, X.; Lin, Y.; Lai, Z. Genome-wide identification and characterization of long non-coding RNAs involved in the early somatic embryogenesis in Dimocarpus longan Lour. BMC Genomics 2018, 19, 805. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Xu, L.; Wang, Y.; Huang, D.; Yu, R.; Limera, C.; Gong, Y.; Liu, L. Genome-Wide Identification of Embryogenesis-Associated microRNAs in Radish (Raphanus sativus L.) by High-Throughput Sequencing. Plant Mol. Biol. Rep. 2014, 32, 900–915. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C.; Wang, A. Molecular regulation of plant somatic embryogenesis. Vitr. Cell. Dev. Biol. Plant 2013, 49, 631–642. [Google Scholar] [CrossRef]
- Fehér, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta Gene Regul. Mech 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Fehér, A.; Pasternak, T.P.; Dudits, D. Transition of somatic plant cells to an embryogenic state. Plant Cell Tiss Org. 2003, 74, 201–228. [Google Scholar] [CrossRef]
- Fehér, A. Callus, Dedifferentiation, Totipotency, Somatic Embryogenesis: What These Terms Mean in the Era of Molecular Plant Biology? Front. Plant Sci. 2019, 10, 536. [Google Scholar] [CrossRef] [PubMed]
- Elhiti, M.; Tahir, M.; Gulden, R.H.; Khamiss, K.; Stasolla, C. Modulation of embryo-forming capacity in culture through the expression of Brassica genes involved in the regulation of the shoot apical meristem. J. Exp. Bot. 2010, 61, 4069–4085. [Google Scholar] [CrossRef]
- Gaj, M.D. Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul. 2004, 43, 27–47. [Google Scholar] [CrossRef]
- Wójcikowska, B.; Gaj, M.D. Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep. 2017, 36, 843–858. [Google Scholar] [CrossRef]
- Wójcik, A.M.; Nodine, M.D.; Gaj, M.D. miR160 and miR166/165 Contribute to the LEC2-Mediated Auxin Response Involved in the Somatic Embryogenesis Induction in Arabidopsis. Front. Plant Sci 2017, 8, 2024. [Google Scholar] [CrossRef]
- Horstman, A.; Li, M.; Heidman, I.; Weeman, M.; Chen, B.; Muino, J.S.; Angenent, G.C.; Boutilier, K. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef]
- Méndez-Hernández, H.A.; Ledezma-Rodriguez, M.; Avilez-Montalvo, R.N.; Juárez-Gómez, Y.L.; Skeete, A.; Avilez-Montavo, J.; De-la-Peña, C.; Loyola-Vargas, V. Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 2019, 10, 77. [Google Scholar] [CrossRef]
- Wójcik, A.M.; Wójcikowska, B.; Gaj, M.D. Current perspectives on the auxin-mediated genetic network that controls the induction of somatic embryogenesis in plants. Int. J. Mol. Sci 2020, 21, 1333. [Google Scholar] [CrossRef]
- Vashisht, D.; Nodine, M.D. MicroRNA functions in plant embryos. Biochem. Soc. Trans. 2014, 42, 352–357. [Google Scholar] [CrossRef]
- Megraw, M.; Baev, V.; Rusinov, V.; Jensen, S.T.; Kalantidis, K.; Hatzigeorgiou, A.G. MicroRNA promoter element discovery in Arabidopsis. RNA 2006, 12, 1612–1619. [Google Scholar] [CrossRef]
- Zhao, X.; Li, L. “Comparative Analysis of MicroRNA Promoters in Arabidopsis and Rice. Genom Proteom. Bioinf. 2013, 11, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Szyrajew, K.; Bielewicz, D.; Dolata, J.; Wójcik, A.M.; Nowak, K.; Szczygiel-Sommer, A.; Szweykowska-Kulinska, Z.; Jarmolowski, A.; Gaj, M.D. MicroRNAs Are Intensively Regulated during Induction of Somatic Embryogenesis in Arabidopsis. Front. Plant Sci. 2017, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef] [PubMed]
- Gliwicka, M.; Nowak, K.; Balazadeh, S.; Mueller-Roeber, B.; Gaj, M.D. Extensive Modulation of the Transcription Factor Transcriptome during Somatic Embryogenesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e69261. [Google Scholar] [CrossRef]
- Wójcik, A.M.; Gaj, M.D. miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment. Planta 2016, 244, 231–243. [Google Scholar] [CrossRef]
- Long, J.M.; Liu, Z.; Wu, X.M.; Fang, Y.N.; Jia, H.H.; Xie, Z.Z.; Deng, X.X.; Guo, W.W. Genome-scale mRNA and small RNA transcriptomic insights into initiation of citrus apomixis. J. Exp. Bot. 2016, 67, 5743–5756. [Google Scholar] [CrossRef]
- von Arnold, S.; Clapham, D.; Egertsdotter, U.; Mo, L.H. Somatic embryogenesis in conifers—A case study of induction and development of somatic embryos in Picea abies. Plant Growth Regul. 1996, 20, 3–9. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Z.; Gong, P.; Zhang, J.; Ziaf, K.; Li, H.; Xiao, F.; Ye, Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 2011, 33, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ge, L.; Liang, R.; Li, W.; Ruan, K.; Lin, H.; Jin, Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 2009, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Zeng, J.; He, X.Q. MiR169 and its target PagHAP2-6 regulated by ABA are involved in poplar cambium dormancy. J. Plant Physiol. 2016, 198, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, L.; Fan, Y.; Wang, L. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 2015, 555, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010, 22, 3574–3588. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2415. [Google Scholar] [CrossRef]
- Mallory, A.C.; Dugas, D.V.; Bartel, D.P.; Bartel, B. MicroRNA Regulation of NAC-Domain Targets Is Required for Proper Formation and Separation of Adjacent Embryonic, Vegetative, and Floral Organs. Curr. Biol. 2004, 14, 1035–1046. [Google Scholar] [CrossRef]
- Guo, H.-S.; Xie, Q.; Fei, J.-F.; Chua, N.-H. MicroRNA Directs mRNA Cleavage of the Transcription Factor NAC1 to Downregulate Auxin Signals for Arabidopsis Lateral Root Development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. miRNA-based drought regulation in wheat. Funct. Integr. Genomics 2016, 16, 221–233. [Google Scholar] [CrossRef]
- Hernandez, Y.; Goswami, K.; Sanan-Mishra, N. Stress induced dynamic adjustment of conserved miR164:NAC module. Plant-Environ. Interact. 2020, 1, 134–151. [Google Scholar] [CrossRef]
- Li, W.F.; Shou-Gong, Z.; Han, S.Y.; Wu, T.; Zhang, J.H.; Qi, L.W. Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr. Plant Cell Tiss Org. 2013, 113, 131–136. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kief, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.M.; Miki, B.L.A.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.; Moronczyk, J.; Wójcik, A.; Gaj, M.D. AGL15 Controls the Embryogenic Reprogramming of Somatic Cells in Arabidopsis through the Histone Acetylation-Mediated Repression of the miRNA Biogenesis Genes. Int. J. Mol. Sci. 2020, 21, 6733. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, V.M. Involvement of plant hormones and plant growth regulators on in vitro somatic embryogenesis. Plant Growth Regul. 2005, 47, 91–110. [Google Scholar] [CrossRef]
- Li, S.G.; Li, W.F.; Han, S.Y.; Yang, W.H.; Qi, L.W. Stage-specific regulation of four HD-ZIP III transcription factors during polar pattern formation in Larix leptolepis somatic embryos. Gene 2013, 522, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Deng, C.; Zhu, T.; Ling, J.; Zhang, H.; Kong, L.; Zhang, S.; Wang, J.; Chen, X. Dynamics of physiological and miRNA changes after long-term proliferation in somatic embryogenesis of Picea balfouriana. Trees Struct. Funct. 2019, 33, 469–480. [Google Scholar] [CrossRef]
- Shen, Y.; Jiang, Z.; Lu, S.; Lin, H.; Gao, S.; Peng, H.; Yuan, G.; Xhang, Z.; Zhao, M.; Rong, T.; et al. Combined small RNA and degradome sequencing reveals microRNA regulation during immature maize embryo dedifferentiation. Biochem. Biophys. Res. Commun. 2013, 441, 425–430. [Google Scholar] [CrossRef]
- López-Ruiz, B.A.; Juárez-González, V.T.; Chávez-Hernández, E.C.; Dinkova, T.D. MicroRNA Expression and Regulation During Maize Somatic Embryogenesis. Methods Mol. Biol. 2018, 1815, 397–410. [Google Scholar]
- Sabana, A.A.; Antony, G.C.; Rahul, U.; Rajesh, M.K. In silico identification of microRNAs and their targets associated with coconut embryogenic calli. Agri Gene 2018, 7, 59–65. [Google Scholar] [CrossRef]
- Sinha, A.; Solanki, M.; Shukla, L.I. Evidences for differential expression of miR167d-5p, target, positional nucleotide preference, and its role in somatic and different stages of regenerating calli of Oryza sativa. Plant Cell Tiss Org. 2019, 136, 537–548. [Google Scholar] [CrossRef]
- Chen, C.J.; Liu, Q.; Zhang, Y.C.; Qu, L.H.; Chen, Y.Q.; Gautheret, D. Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biol. 2011, 8, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Liu, Y.B.; Zhou, C.; Li, X.M.; Zhang, X.S. The microRNA167 controls somatic embryogenesis in Arabidopsis through regulating its target genes ARF6 and ARF8. Plant Cel. Tiss Org. 2016, 124, 405–417. [Google Scholar] [CrossRef]
- Wu, X.M.; Liu, M.Y.; Ge, X.X.; Xu, Q.; Guo, W.W. Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta 2011, 233, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Mei, C.; Lian, S.; Yu, Y.-T.; Lu, K.; Wu, Z.; Wang, X.F.; Zhang, D.P. Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol. Biol. 2015, 88, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, L.; Yuan, D.; Lindsey, K.; Zhang, X. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J. Exp. Bot. 2013, 64, 1521–1536. [Google Scholar] [CrossRef]
- Hewezi, T.; Baum, T.J. Complex feedback regulations govern the expression of miRNA396 and its GRF target genes. Plant Signal. Behav. 2012, 7, 749–751. [Google Scholar] [CrossRef]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. mir390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef]
- Liu, P.P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chen, Y.Q. A new mechanism in plant engineering: The potential roles of microRNAs in molecular breeding for crop improvement. Biotechnol. Adv. 2010, 28, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Lai, Z.; Tian, Q.; Lin, L.; Lai, R.; Yang, M.; Zhang, D.; Chen, Y.; Zhang, Z. Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and -17 cleavage during somatic embryogenesis in Dimocarpus longan Lour. Front. Plant Sci. 2015, 6, 956. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Botor, M.; Moronczyk, J.; Wójcik, A.M.; Nodzynski, T.; Karcz, J.; Gaj, M.G. Trichostatin a triggers an embryogenic transition in arabidopsis explants via an auxin-related pathway. Front. Plant Sci. 2018, 9, 1353. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zheng, Y.; Ji, H.; Burnie, W.; Perry, S.E. Gene regulation by the AGL15 transcription factor reveals hormone interactions in somatic embryogenesis. Plant Physiol. 2016, 172, 2374–2387. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Singh, A.K.; Chaudhary, B. Target-mimicry based miRNA167-diminution ameliorates cotton somatic embryogenesis via transcriptional biases of auxin signaling associated miRNAs and genes. Plant Cell. Tiss Org. 2020, 141, 511–531. [Google Scholar] [CrossRef]
- Yao, X.; Chen, J.; Zhou, J.; Yu, H.; Ge, C.; Zhang, M.; Gao, X.; Dai, X.; Yang, Z.N.; Zhao, Y. An Essential Role for miRNA167 in Maternal Control of Embryonic and Seed Development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Park, C.; Guitérrez, C.L.; Wójcikowska, B.; Pencik, A.; Novak, O.; Chen, J.; Grunewald, W.; Dresselhaus, T.; Friml, J.; et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 2018, 4, 548–553. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-Directed Phasing during Trans-Acting siRNA Biogenesis in Plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Axtell, M.J.; Jan, C.; Rajagopalan, R.; Bartel, D.P. A Two-Hit Trigger for siRNA Biogenesis in Plants. Cell 2006, 127, 565–577. [Google Scholar] [CrossRef]
- Wu, X.M.; Kou, S.J.; Liu, Y.L.; Fang, Y.N.; Xu, Q.; Guo, W.W. Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA-and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol. J. 2015, 13, 383–394. [Google Scholar] [CrossRef]
- Ge, X.; Zhang, C.; Wang, Q.; Yang, Z.; Wang, Y.; Zhang, X.; Wu, Z.; Hou, Y.; Wu, J.; Li, F. ITRAQ protein profile differential analysis between somatic globular and cotyledonary embryos reveals stress, hormone, and respiration involved in increasing plantlet regeneration of Gossypium hirsutum L. J. Proteome Res. 2015, 14, 268–278. [Google Scholar] [CrossRef]
- Corredoira, E.; Merkle, S.A.; Martínez, M.T.; Toribio, M.; Canhoto, J.M.; Correia, S.I.; Ballester, A.; Vieitez, A.M. Non-Zygotic Embryogenesis in Hardwood Species. Crit. Rev. Plant Sci. 2019, 38, 29–97. [Google Scholar] [CrossRef]
- Zhang, L.; Li, W.; Xu, H.; Qi, L.; Han, S. Cloning and characterization of four differentially expressed cDNAs encoding NFYA homologs involved in responses to ABA during somatic embryogenesis in Japanese larch (Larix leptolepis). Plant Cell Tiss Org. 2014, 117, 293–304. [Google Scholar] [CrossRef]
- Willmann, M.R.; Mehalick, A.J.; Packer, R.L.; Jenik, P.D. MicroRNAs Regulate the Timing of Embryo Maturation in Arabidopsis. Plant Physiol. 2011, 155, 1871–1884. [Google Scholar] [CrossRef]
- Ma, C.; Burd, S.; Lers, A. MiR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef]
- Miguel, C.M.; Rupps, A.; Raschke, J.; Rodrigues, A.S.; Trontin, J.F. Impact of molecular studies on somatic embryogenesis development for implementation in conifer multi-varietal forestry. In Vegetative Propagation of Forest Trees; Park, Y.S., Bonga, J.M., Moon, H.K., Eds.; National Institute of Forest Science: Seoul, Korea, 2016; pp. 373–421. [Google Scholar]
- Siddiqui, Z.H.; Abbas, Z.K.; Ansari, M.W.; Khan, M.N. The role of miRNA in somatic embryogenesis. Genomics 2019, 111, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Liu, C.; Feng, M.; Liu, Y.; Wu, X.; Guo, W. miR156- SPL modules regulate induction of somatic embryogenesis in citrus callus. J. Exp Bot. 2018, 69, 2979–2993. [Google Scholar] [CrossRef]
- Wang, L.; Chong, K. The essential role of cytokinin signaling in root apical meristem formation during somatic embryogenesis. Front. Plant Sci. 2016, 6, 1196. [Google Scholar] [CrossRef]
- Luo, Y.C.; Zhou, H.; Chen, Y.L.; Yang, J.H.; Chen, Y.Q.; Qu, L.H. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 2006, 580, 5111–5116. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, J.; Han, S.; Yang, W.; Li, W.; Wei, H.; Li, X.; Qi, L. Four abiotic stress-induced miRNA families differentially regulated in the embryogenic and non-embryogenic callus tissues of Larix leptolepis. Biochem. Biophys. Res. Commun. 2010, 398, 355–360. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Arikit, S.; Meyers, B.C. Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 2015, 32, 2905–2918. [Google Scholar] [CrossRef]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef]
- Jeong, D.; Schmidt, A.A.; Rymarquis, L.A.; Park, S.; Ganssmann, M.; German, M.A.; Accerbi, M.; Zhai, J.; Fahlgren, N.; Fox, E.F.; et al. Parallel analysis of RNA ends enhances global investigation of microRNAs and target RNAs of Brachypodium distachyon. Genome Biol. 2013, 14, R145. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.A.; Bender, J. Reprogramming the epigenome during germline and seed development. Genome Biol. 2009, 10, 232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Song, Q.X.; Liu, Y.F.; Hu, Y.X.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Sarazin, V.; Duclercq, J.; Mendou, B.; Aubanelle, L.; Nicolas, Y.; Aono, M.; Pilard, S.; Guerineau, F.; Sangwan-Norreel, B.; Sangwan, R.S. Arabidopsis BNT1, an atypical TIR-NBS-LRR gene, acting as a regulator of the hormonal response to stress. Plant Sci. 2015, 239, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Lelu-Walter, M.A.; Klimaszewska, K.; Miguel, C.; Aronen, T.; Hargreaves, C.T.; Trontin, J.F. Somatic embryogenesis for more effective breeding and deployment of improved varieties in Pinus spp.: Bottlenecks and recent advances. In Somatic Embryogenesis: Fundamental Aspects and Applications; Springer: Cham, Switzerland, 2016; pp. 319–365. [Google Scholar]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Guo, Z.; Kuang, Z.; Wang, Y.; Zhao, Y.; Tao, Y.; Cheng, C.; Yang, J.; Lu, X.; Hao, C.; Wang, T.; et al. PmiREN: A comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020, 48, D1114–D1121. [Google Scholar] [CrossRef] [PubMed]
| Somatic Embryogenesis (SE)-Related miRNA Family | Described or Putative Targets | (Putative) Role in Embryogenesis | References | ||
|---|---|---|---|---|---|
| Gymnosperm-specific | miR1314 | Putative cellulose synthases (TIGR) | Embryo dormancy and germination | [28] | |
| Conifer- specific | miR946 miR947 miR951 miR1311 | unknown | Embryo dormancy and germination | [28] | |
| miR950 | NB-ARC | ||||
| miR1312 | GRF2, HB1 | ||||
| miR1313 | LRK1 | ||||
| miR1315 | receptor-like protein kinase | Embryogenic ability | [23] | ||
| miR1316 | LIP1, LIP2, TIR-NBS-LRR proteins | Notdetermined yet | [17,24,29] | ||
| miR3699 | unknown | Not determined yet | [17] | ||
| miR3701 | NBS-LRR proteins, cellulose synthase | Not determined yet | [17,29] | ||
| miR3702 miR3704 | unknown | Not determined yet | [24,29] | ||
| Angiosperm-specific | miR444 | MIKC-type MADS-box | SE induction | [27] | |
| miR827 | NLA and PHT5 | Regulate auxin metabolism in early SE | [30] | ||
| Monocot- specific (Poaceae- specific) | miR531 | Wpk4 protein kinase | Embryogenic callus and embryo development | [26] | |
| miR1139 | Myb1 | Embryogenic callus development | [26] | ||
| miR1878 | NBS-LRR resistance protein-like | ||||
| miR5049 | Photosystem 1 subunit 5 Hydrolase, mitochondrial | Embryogenic callus and embryo development | [26] | ||
| miR5067 | Wpk4 protein kinase | Not determined yet | [31] | ||
| Dicot- specific | miR158 | SPINDLY | Gibberellic acid responses | [4] | |
| miR163 | SAMT family members | Not determined yet | [32] | ||
| miR403 | AGO2, AGO3 | Embryo maturation | [4] | ||
| miR406 | Spliceosomal proteins | Early embryogenesis | [33] | ||
| Brassicaceae- specific | miR161 | EMB2654, ARF | Embryo maturation | [4,34] | |
| miR824 | AGAMOUS-LIKE16 | Embryo maturation | [4] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, A.; Cordeiro, D.; Correia, S.; Miguel, C. Small Non-Coding RNAs at the Crossroads of Regulatory Pathways Controlling Somatic Embryogenesis in Seed Plants. Plants 2021, 10, 504. https://doi.org/10.3390/plants10030504
Alves A, Cordeiro D, Correia S, Miguel C. Small Non-Coding RNAs at the Crossroads of Regulatory Pathways Controlling Somatic Embryogenesis in Seed Plants. Plants. 2021; 10(3):504. https://doi.org/10.3390/plants10030504
Chicago/Turabian StyleAlves, Ana, Daniela Cordeiro, Sandra Correia, and Célia Miguel. 2021. "Small Non-Coding RNAs at the Crossroads of Regulatory Pathways Controlling Somatic Embryogenesis in Seed Plants" Plants 10, no. 3: 504. https://doi.org/10.3390/plants10030504
APA StyleAlves, A., Cordeiro, D., Correia, S., & Miguel, C. (2021). Small Non-Coding RNAs at the Crossroads of Regulatory Pathways Controlling Somatic Embryogenesis in Seed Plants. Plants, 10(3), 504. https://doi.org/10.3390/plants10030504







