Carbon Assimilation, Isotope Discrimination, Proline and Lipid Peroxidation Contribution to Barley (Hordeum vulgare) Salinity Tolerance
Abstract
1. Introduction
2. Results
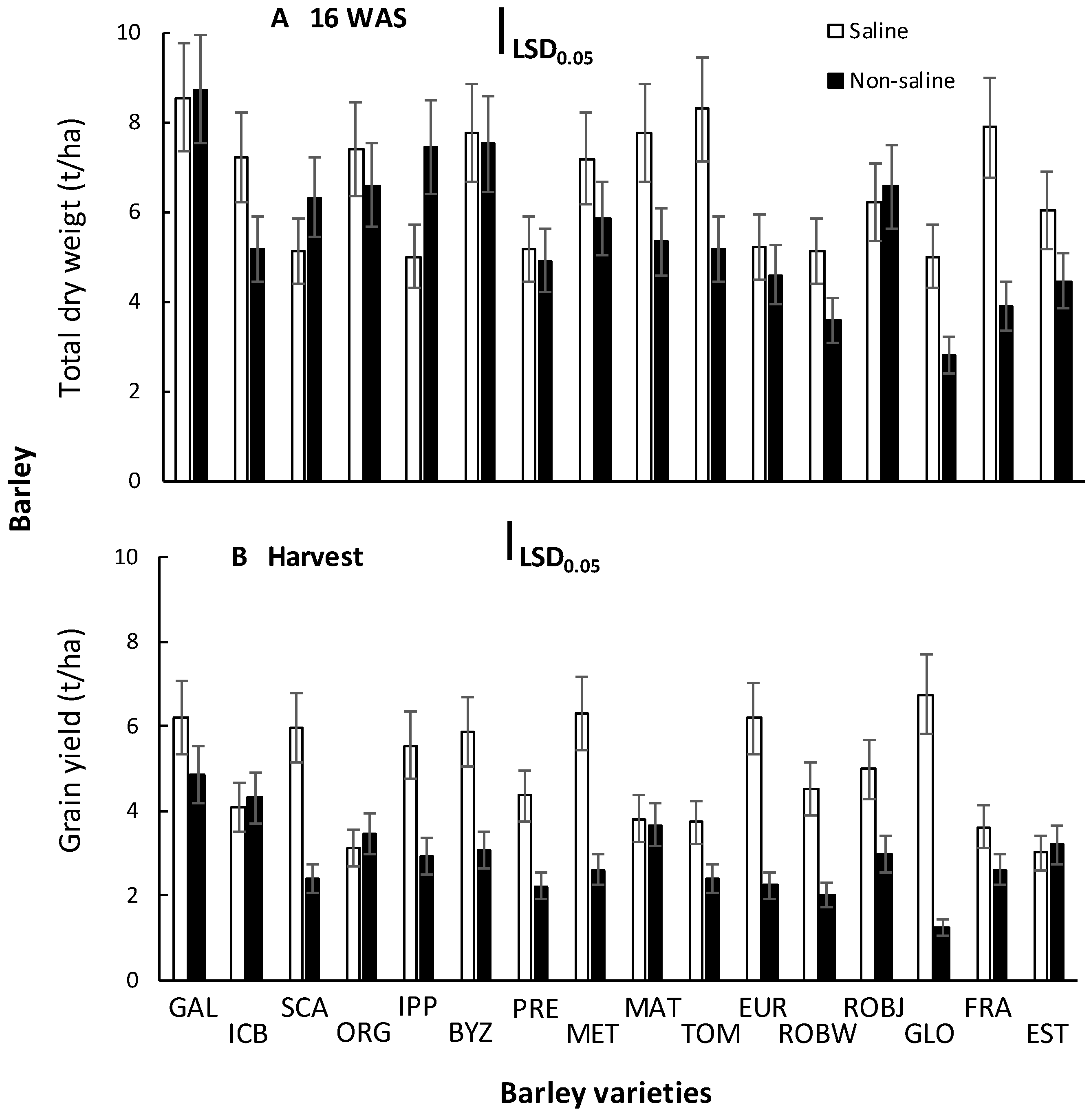
2.1. Barley Growth and Yield
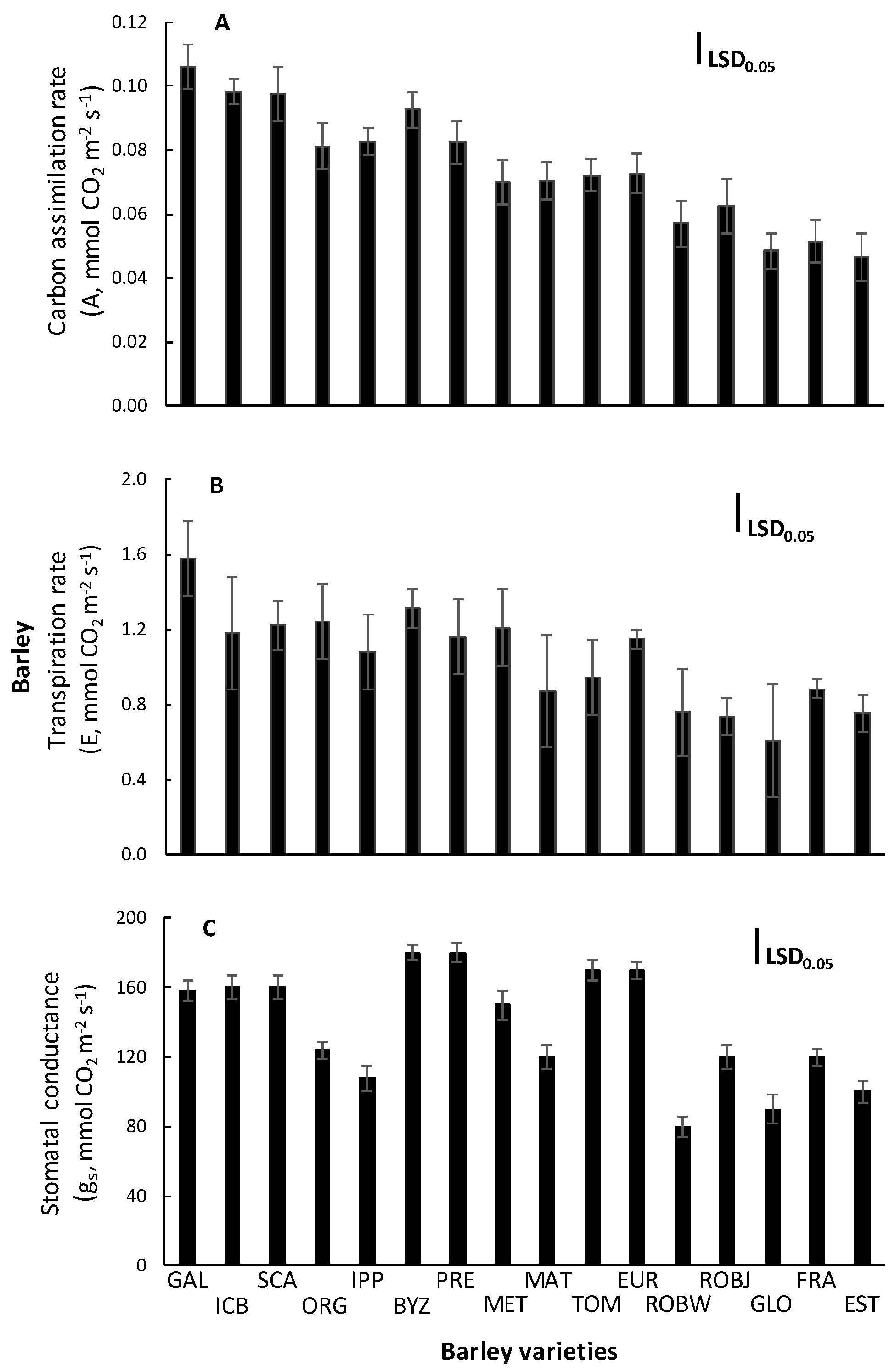
2.2. Gas Exchange Measurements
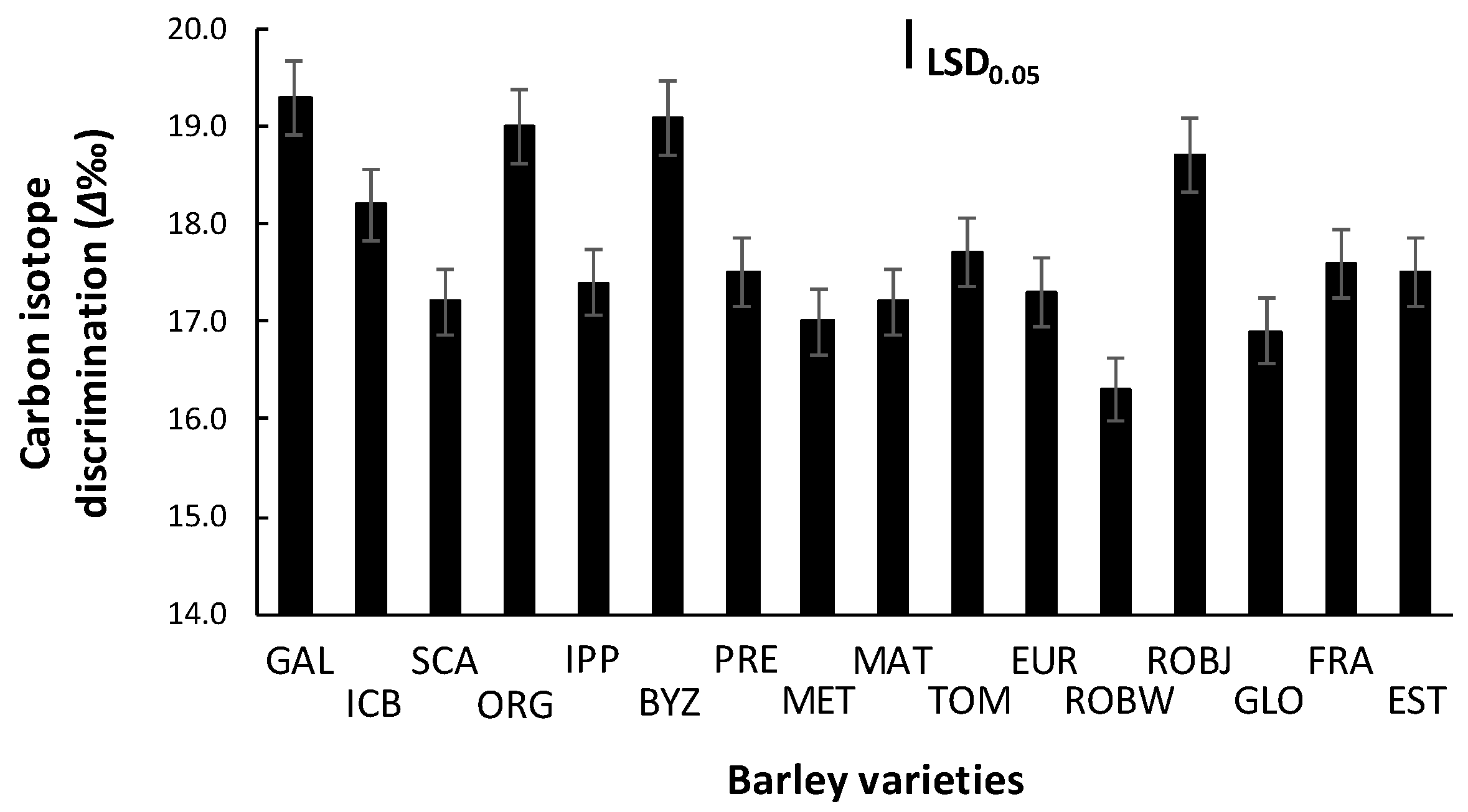
2.3. Carbon Isotope Discrimination (Δ)
2.4. Proline Determination
2.5. Lipid Peroxidation and Hydrogen Peroxide Assays
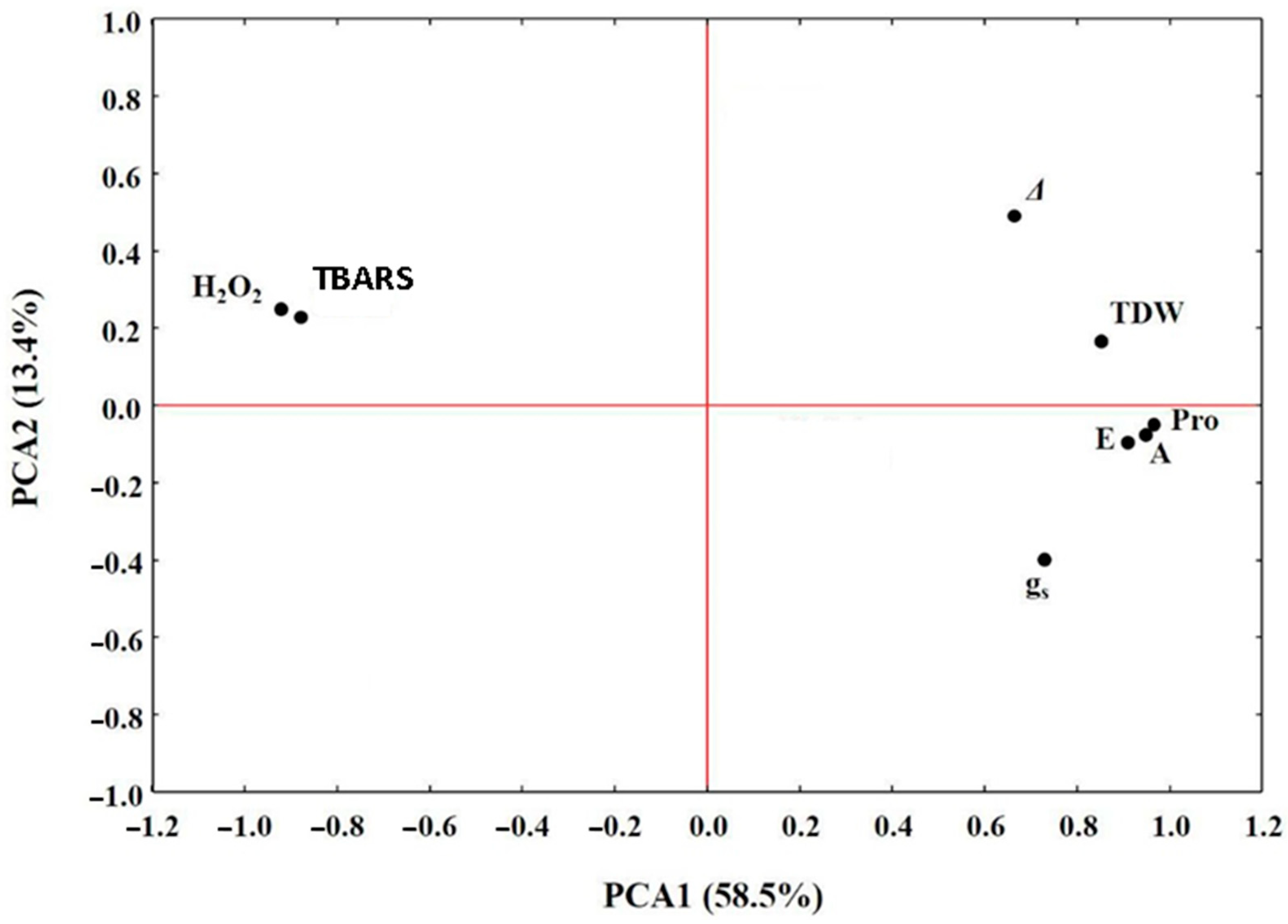
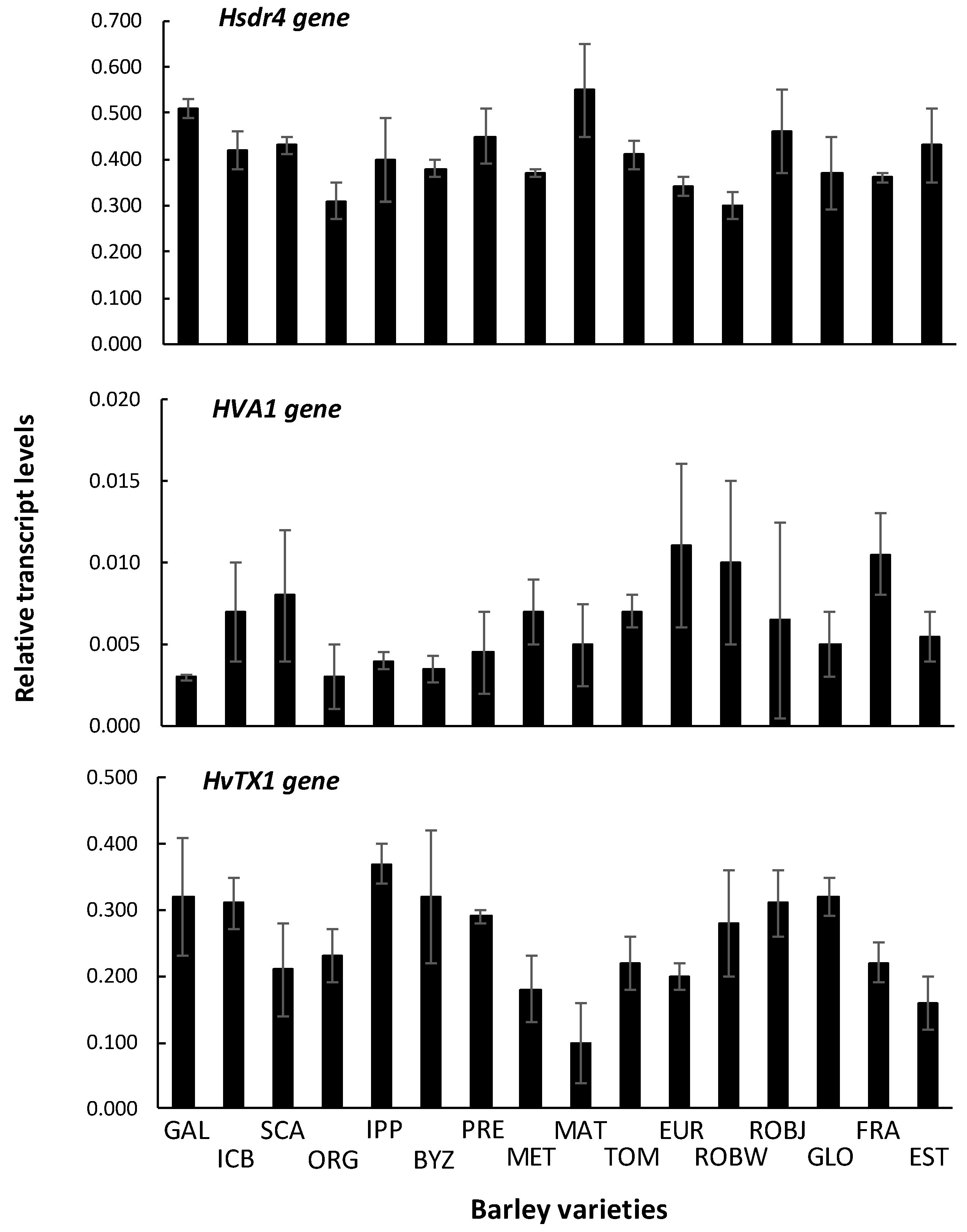
2.6. Gene Expression
3. Discussion
3.1. Barley Growth and Yield
3.2. Gas Exchange Measurements
3.3. Carbon Isotope Discrimination (Δ)
3.4. Proline Determination
3.5. Lipid Peroxidation and Hydrogen Peroxide Assays
3.6. Gene Expression
4. Materials and Methods
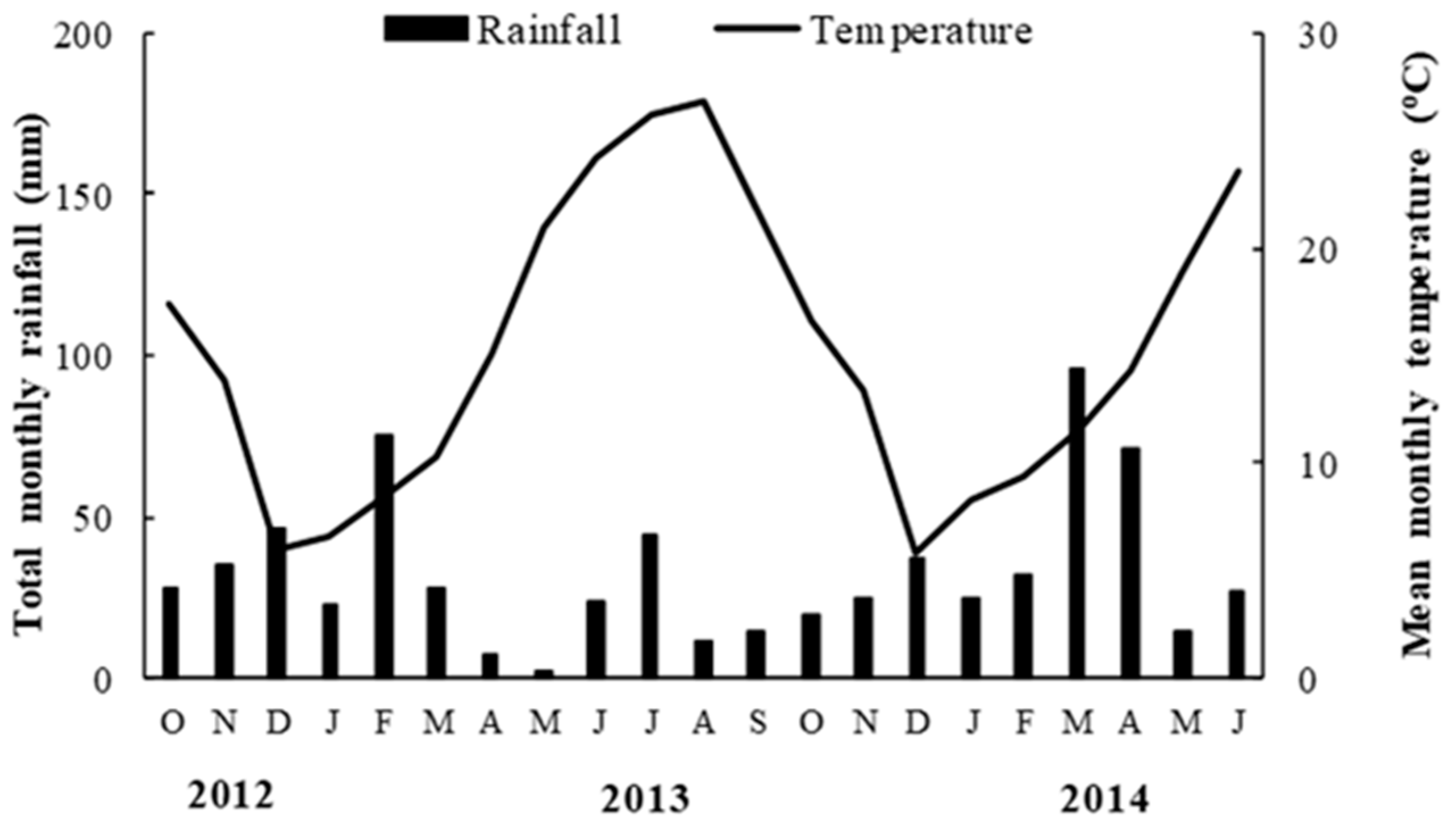
4.1. Experimental Site
4.1.1. Genotypes
4.1.2. Experimental Procedure
4.1.3. Data Collection
4.2. Gas Exchange Measurements
4.3. Carbon Isotope Discrimination (Δ)
4.4. Determination of Proline
4.5. Lipid Peroxidation and Hydrogen Peroxide Assays
4.6. RNA Isolation and cDNA Synthesis
4.7. Analyses of Gene Expression
4.8. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GY | grain yield |
| KI | potassium iodide |
| MDA | malondialdehyde |
| PCA | principal component analysis |
| Pro | Proline |
| TCA | trichloroacetic acid |
| TDW | total dry weight |
| WAS | weeks after seeding |
References
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Kiani-Pouya, A.; Rasoulia, F.; Rabbib, B.; Falakbolanda, Z.; Yong, M.; Chen, Z.-H.; Zhoua, M.; Shabala, S. Stomatal traits as a determinant of superior salinity tolerance in wild barley. J. Plant Physiol. 2020, 24, 153108. [Google Scholar] [CrossRef]
- Ouzounidou, G.; Ilias, I.; Giannakoula, A.; Theoharidou, I. Effect of water stress and NaCl triggered changes on yield, physiology, biochemistry of broad bean (Vicia faba) plants and on quality of harvested pods. Biologia 2014, 69, 1010–1017. [Google Scholar] [CrossRef]
- Sofy, M.R.; Elhindi, K.M.; Farouk, S.; Alotaibi, M.A. Zinc and Paclobutrazol Mediated Regulation of Growth, Upregulat-ing Antioxidant Aptitude and Plant Productivity of Pea Plants under Salinity. Plants 2020, 9, 1197. [Google Scholar] [CrossRef]
- Bor, M.; Özdemir, F.; Türkan, I. The effect of salt stress on lipid peroxidation and antioxidants in leaves of sugar beet Be-ta vulgaris L. and wild beet Beta maritima L. Plant Sci. 2003, 164, 77–84. [Google Scholar] [CrossRef]
- Moradi, F.; Ismail, A.M. Responses of Photosynthesis, Chlorophyll Fluorescence and ROS-Scavenging Systems to Salt Stress During Seedling and Reproductive Stages in Rice. Ann. Bot. 2007, 99, 1161–1173. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Chen, H.; Huang, J.; Zhang, Y.; Qi, M.; Liu, Y.; Li, T. Photosynthetic Response Mechanism of Soil Salinity-Induced Cross-Tolerance to Subsequent Drought Stress in Tomato Plants. Plants 2020, 9, 363. [Google Scholar] [CrossRef]
- Wang, W.B.; Kim, Y.H.; Lee, H.S.; Kim, K.Y.; Deng, X.P.; Kwak, S.S. Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiol. Biochem. 2009, 47, 570–577. [Google Scholar] [CrossRef]
- Yu, D.; Boughton, B.A.; Hill, C.B.; Feussner, I.; Roessner, U.; Rupasinghe, T.W.T. Insights into Oxidized Lipid Modifica-tion in Barley Roots as an Adaptation Mechanism to Salinity Stress. Front. Plant Sci. 2020, 11, 1. [Google Scholar] [CrossRef]
- Khan, N.A. NaCl-inhibited chlorophyll synthesis and associated changes in ethylene evolution and antioxidative en-zyme activities in wheat. Biol. Plant. 2006, 47, 437–440. [Google Scholar]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New Insights on Plant Salt Tolerance Mechanisms and Their Potential Use for Breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef]
- Mayer, K.; Martis, M.; Hedley, P.E.; Šimková, H.; Liu, H.; Morris, J.A.; Steuernagel, B.; Taudien, S.; Roessner, S.; Gundlach, H.; et al. Unlocking the Barley Genome by Chromosomal and Comparative Genomics. Plant Cell 2011, 23, 1249–1263. [Google Scholar] [CrossRef]
- Katerji, N.; Van Hoorn, J.; Hamdy, A.; Mastrorilli, M.; Fares, C.; Ceccarelli, S.; Grando, S.; Oweis, T. Classification and salt tolerance analysis of barley varieties. Agric. Water Manag. 2006, 85, 184–192. [Google Scholar] [CrossRef]
- Kaushal, R.; Ghosh, P.; Geilmann, H. Fingerprinting environmental conditions and related stress using stable isotopic composition of rice (Oryza sativa L.) grain organic matter. Ecol. Indic. 2016, 61, 941–951. [Google Scholar] [CrossRef]
- Papaefthimiou, D.; Tsaftaris, A.S. Characterization of a drought inducible trithorax-like H3K4 methyltransferase from barley. Biol. Plant. 2012, 56, 683–692. [Google Scholar] [CrossRef]
- Hernández, J.A. Salinity Tolerance in Plants: Trends and Perspectives. Int. J. Mol. Sci. 2019, 20, 2408. [Google Scholar] [CrossRef]
- Liu, L.; Nakamura, Y.; Taliman, N.A.; El Sabagh, A.; Moghaieb, R.; Saneoka, H. Differences in the Growth and Physiological Responses of the Leaves of Peucedanum japonicum and Hordeum vulgare Exposed to Salinity. Agriculture 2020, 10, 317. [Google Scholar] [CrossRef]
- Hammami, Z.; Gauffreteau, A.; BelhajFraj, M.; Sahli, A.; Jeuffroy, M.-H.; Rezgui, S.; Bergaoui, K.; McDonnell, R.; Trifa, Y. Predicting yield reduction in improved barley (Hordeum vulgare L.) varieties and landraces under salinity using selected tolerance traits. Field Crops Res. 2017, 211, 10–18. [Google Scholar] [CrossRef]
- Tavakoli, F.; Vazan, S.; Moradi, F.; Shiran, B.; Sorkheh, K. Differential Response of Salt-Tolerant and Susceptible Barley Genotypes to Salinity Stress. J. Crop Improv. 2010, 24, 244–260. [Google Scholar] [CrossRef]
- Hasanuzzaman; Shabala, L.; Zhou, M.; Brodribb, T.J.; Corkrey, R.; Shabala, S.; Brodribb, T.J. Factors determining stomatal and non-stomatal (residual) transpiration and their contribution towards salinity tolerance in contrasting barley genotypes. Environ. Exp. Bot. 2018, 153, 10–20. [Google Scholar] [CrossRef]
- Zhu, M.; Zhou, M.; Shabala, L.; Shabala, S. Linking osmotic adjustment and stomatal characteristics with salinity stress tolerance in contrasting barley accessions. Funct. Plant Biol. 2015, 42, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Rahnama, A.; James, R.A.; Poustini, K.; Munns, R. Stomatal conductance as a screen for osmotic stress tolerance in durum wheat growing in saline soil. Funct. Plant Biol. 2010, 37, 255–263. [Google Scholar] [CrossRef]
- Taiz, L.; Zeiger, E.; Moller, I.M.; Murphy, A. Plant Physiology and Development, 6th ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2015. [Google Scholar]
- Ansari, R.; Naqvi, M.S.; Khanzada, N.A.; Hubick, K.T. Carbon isotope discrimination in wheat cultivars under saline conditions. Pak. J. Bot. 1998, 30, 87–93. [Google Scholar]
- Isla, R.; Aragüés, R.; Royo, A. Validity of various physiological traits as screening criteria for salt tolerance in barley. Field Crops Res. 1998, 58, 97–107. [Google Scholar] [CrossRef]
- Farquhar, G.D.; Richards, R.A. Isotopic composition of plant carbon correlates with water-use efficiency of wheat geno-types. Funct. Plant Biol. 1984, 11, 539–552. [Google Scholar] [CrossRef]
- Merah, O.; Deléens, E.; Nachit, M.M.; Monneveux, P. Carbon isotope discrimination, leaf characteristics and grain yield of interspecific wheat lines and their durum parents under Mediterranean conditions. Cereal Res. Commun. 2001, 29, 143–149. [Google Scholar] [CrossRef]
- Shena, Q.; Yua, J.; Fua, L.; Wua, L.; Daia, F.; Jianga, L.; Wua, D.; Zhang, G. Ionomic, metabolomic and proteomic analyses reveal molecular mechanisms of root adaption to salt stress in Tibetan wild barley. Plant Physiol. Biochem. 2018, 123, 319–330. [Google Scholar] [CrossRef]
- Binott, J.J.; Owuoche, J.O.; Bartels, D. Physiological and molecular characterization of Kenyan barley (Hordeum vulgare L.) seedlings for salinity and drought tolerance. Euphytica 2017, 213, 139. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Allel, D.; Ben-Amar, A.; Abdelly, C. Leaf photosynthesis, chlorophyll fluorescence and ion content of barley (Hordeum vulgare) in response to salinity. J. Plant Nutr. 2017, 41, 497–508. [Google Scholar] [CrossRef]
- Giannakoula, A.; Ilias, I. The effect of water stress and salinity on growth and physiology of tomato (Lycopersicon escu-lentum Mill). Arch. Biol. Sci. 2013, 65, 611–620. [Google Scholar] [CrossRef]
- Zeeshan, M.; Lu, M.; Sehar, S.; Holford, P.; Wu, F. Comparison of Biochemical, Anatomical, Morphological, and Physiological Responses to Salinity Stress in Wheat and Barley Genotypes Deferring in Salinity Tolerance. Agronomy 2020, 10, 127. [Google Scholar] [CrossRef]
- Walia, H.; Wilson, C.; Wahid, A.; Condamine, P.; Cui, X.; Close, T.J. Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct. Integr. Genom. 2006, 6, 143–156. [Google Scholar] [CrossRef]
- Zhu, J.; Fan, Y.; Shabala, S.; Li, C.; Lv, C.; Guo, B.; Xu, R.; Zhou, M. Understanding Mechanisms of Salinity Tolerance in Barley by Proteomic and Biochemical Analysis of Near-Isogenic Lines. Int. J. Mol. Sci. 2020, 21, 1516. [Google Scholar] [CrossRef]
- Suprunova, T.; Krugman, T.; Distelfeld, A.; Fahima, T.; Nevo, E.; Korol, A. Identification of a novel gene (Hsdr4) in-volved in water-stress tolerance in wild barley. Plant Mol. Biol. 2007, 64, 17–34. [Google Scholar] [CrossRef]
- Jewell, M.C.; Campbell, B.C.; Godwin, I.D. Transgenic plants for abiotic stress resistance. In Transgenic Crop Plants; Chittaranjan, K., Charles, H.M., Albert, G.A., Timothy, C.H., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2010; pp. 67–132. [Google Scholar]
- Xu, D.; Duan, X.; Wang, B.; Hong, B.; Ho, T.; Wu, R. Expression of a Late Embryogenesis Abundant Protein Gene, HVA1, from Barley Confers Tolerance to Water Deficit and Salt Stress in Transgenic Rice. Plant Physiol. 1996, 110, 249–257. [Google Scholar] [CrossRef]
- Sivamani, E.; Bahieldin, A.; Wraith, J.M.; Alniemi, T.S.; Dyer, W.E.; Ho, T.-H.D.; Qu, R. Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci. 2000, 155, 1–9. [Google Scholar] [CrossRef]
- Kade, A.A.; Almesleman, M.; Baghdady, A.; Alzubi, H.; Alasaad, N.; Basha, N.A.; Dameriha, A.; Jacobsen, H.; Hassan, F. Isolation, Characterization of the hva1 Gene from Syrian Barley Varieties and Cloning into a Binary Plasmid Vector. Int. J. Bot. 2012, 8, 117–126. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.-S.; Lo, S.-F.; Sun, P.-K.; Lu, C.-A.; Ho, T.D.; Yu, S.-M. A late embryogenesis abundant protein HVA1 regulated by an inducible promoter enhances root growth and abiotic stress tolerance in rice without yield penalty. Plant Biotechnol. J. 2014, 13, 105–116. [Google Scholar] [CrossRef]
- Marakli, S.; Gozukirmizi, N. Analyses of abiotic stress and brassinosteroid-related some genes in barley roots grown under salinity stress and HBR treatments: Expression profiles and phylogeny. Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2017, 152, 324–332. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Alaraidh, I.A.; Alsahli, A.A.; Ali, H.M.; Alayafi, A.A.; Witczak, J.; Ahmad, M. Genetic Variation and Alleviation of Salinity Stress in Barley (Hordeum vulgare L.). Molecules 2018, 23, 2488. [Google Scholar] [CrossRef]
- Flowers, T.J.; Hajibagheri, M.A. Salinity tolerance in Hordeum vulgare: Ion concentrations in root cells of cultivars differ-ing in salt tolerance. Plant Soil 2001, 231, 1–9. [Google Scholar] [CrossRef]
- Lancashire, P.D.; Bleiholder, H.; Langelüddecke, P.; Stauss, R.; van den Boom, T.; Weber, E.; Witzen-Berger, A. An uni-formdecimal code for growth stages of crops and weeds. Ann. Appl. Biol. 1991, 119, 561–601. [Google Scholar] [CrossRef]
- Von Caemmerer, S.; Farquhar, G.D. Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 1981, 153, 376–387. [Google Scholar] [CrossRef]
- Troll, W.; Lindsley, J. A photometric method for the determination of proline. J. Biol. Chem. 1955, 215, 655–660. [Google Scholar] [CrossRef]
- Heath, R.L.; Parcker, L. Photoperoxidation in isolated chloroplasts. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Alexieva, V.; Sergiev, I.; Mapelli, S.; Karanov, E. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Environ. 2001, 24, 1337–1344. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rapacz, M.; Stępień, A.; Skorupa, K. Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): The effects of developmental stage and leaf age. Acta Physiol. Plant. 2012, 34, 1723–1733. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time poly-merase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]

| Significance of F Ratio a | |||
|---|---|---|---|
| Source | df | TDW | GY |
| Year (Y) | 1 | NS | NS |
| Salinity (S) | 1 | *** | *** |
| YS | 1 | NS | NS |
| R(YS) | 12 | *** | ** |
| Variety (V) | 15 | *** | *** |
| YV | 15 | NS | NS |
| SV | 15 | *** | *** |
| YSV | 15 | NS | NS |
| Error | 180 | ||
| CV, % | 14.6 | 16.2 | |
| Correlation | TDW | GY | Δ | A | E | g |
|---|---|---|---|---|---|---|
| TDW | 1.000 | 0.695 | 0.789 | 0.757 | 0.792 | 0.535 |
| GY | 0.695 | 1.000 | 0.679 | 0.575 | 0.554 | 0.218 |
| Δ | 0.789 | 0.679 | 1.000 | 0.534 | 0.548 | 0.402 |
| A | 0.757 | 0.575 | 0.534 | 1.000 | 0.883 | 0.689 |
| E | 0.792 | 0.554 | 0.548 | 0.883 | 1.000 | 0.714 |
| g | 0.535 | 0.218 | 0.402 | 0.689 | 0.714 | 1.000 |
| TDW | 0.001 | 0.000 | 0.000 | 0.000 | 0.016 | |
| GY | 0.001 | 0.002 | 0.010 | 0.013 | 0.208 | |
| Δ | 0.000 | 0.002 | 0.017 | 0.014 | 0.061 | |
| A | 0.000 | 0.010 | 0.017 | 0.000 | 0.002 | |
| E | 0.000 | 0.013 | 0.014 | 0.000 | 0.001 | |
| g | 0.016 | 0.208 | 0.061 | 0.002 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasilakoglou, I.; Dhima, K.; Giannakoula, A.; Dordas, C.; Skiada, V.; Papadopoulou, K. Carbon Assimilation, Isotope Discrimination, Proline and Lipid Peroxidation Contribution to Barley (Hordeum vulgare) Salinity Tolerance. Plants 2021, 10, 299. https://doi.org/10.3390/plants10020299
Vasilakoglou I, Dhima K, Giannakoula A, Dordas C, Skiada V, Papadopoulou K. Carbon Assimilation, Isotope Discrimination, Proline and Lipid Peroxidation Contribution to Barley (Hordeum vulgare) Salinity Tolerance. Plants. 2021; 10(2):299. https://doi.org/10.3390/plants10020299
Chicago/Turabian StyleVasilakoglou, Ioannis, Kico Dhima, Anastasia Giannakoula, Christos Dordas, Vasiliki Skiada, and Kalliope Papadopoulou. 2021. "Carbon Assimilation, Isotope Discrimination, Proline and Lipid Peroxidation Contribution to Barley (Hordeum vulgare) Salinity Tolerance" Plants 10, no. 2: 299. https://doi.org/10.3390/plants10020299
APA StyleVasilakoglou, I., Dhima, K., Giannakoula, A., Dordas, C., Skiada, V., & Papadopoulou, K. (2021). Carbon Assimilation, Isotope Discrimination, Proline and Lipid Peroxidation Contribution to Barley (Hordeum vulgare) Salinity Tolerance. Plants, 10(2), 299. https://doi.org/10.3390/plants10020299







