Selection Gain of Maize Haploid Inducers for the Tropical Savanna Environments
Abstract
:1. Introduction
2. Materials and Methods
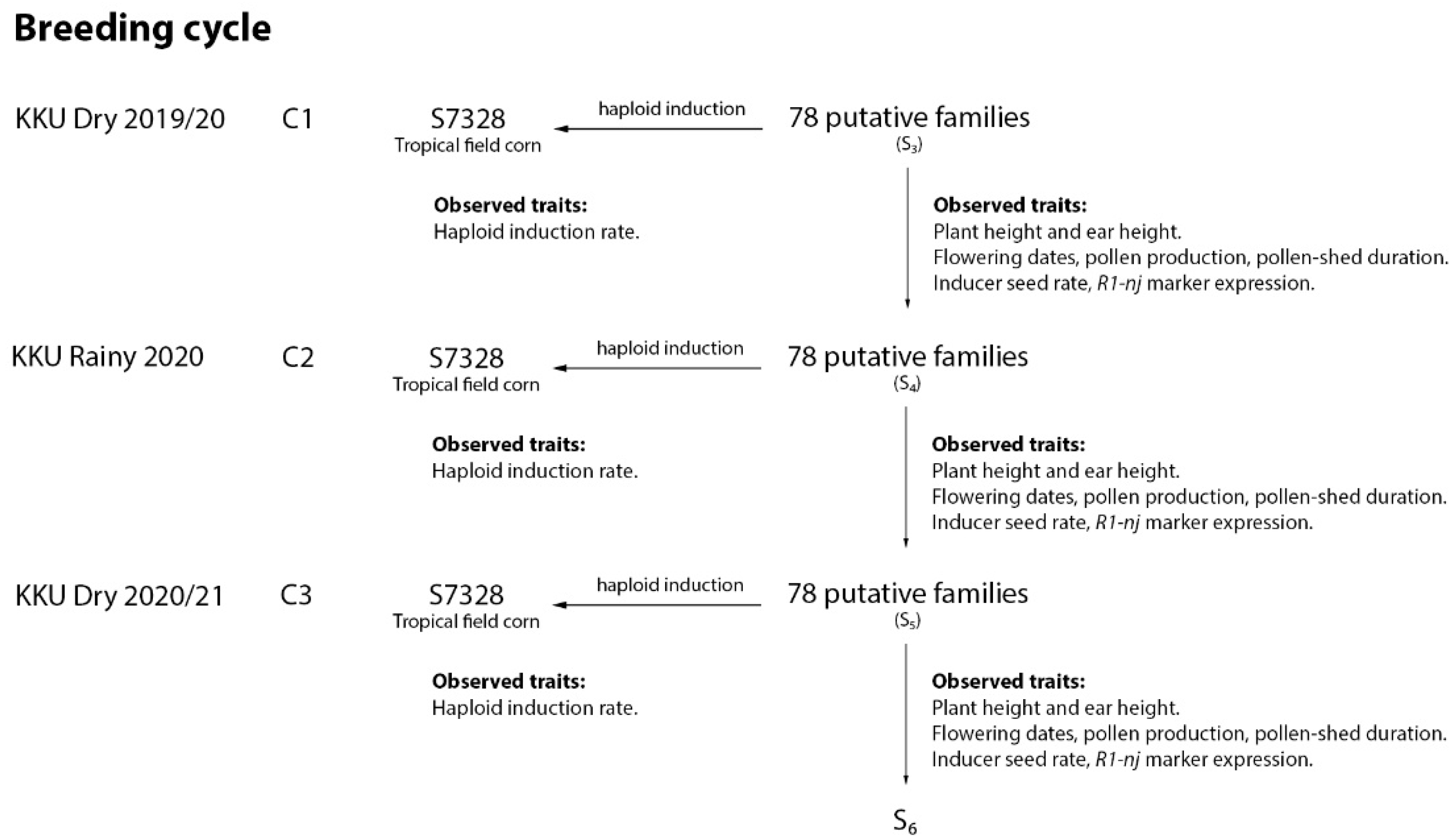
2.1. Plant Materials and Establishment of Base Populations
2.2. Population Improvement and In Vivo Haploid Induction
2.3. Field Data Collection
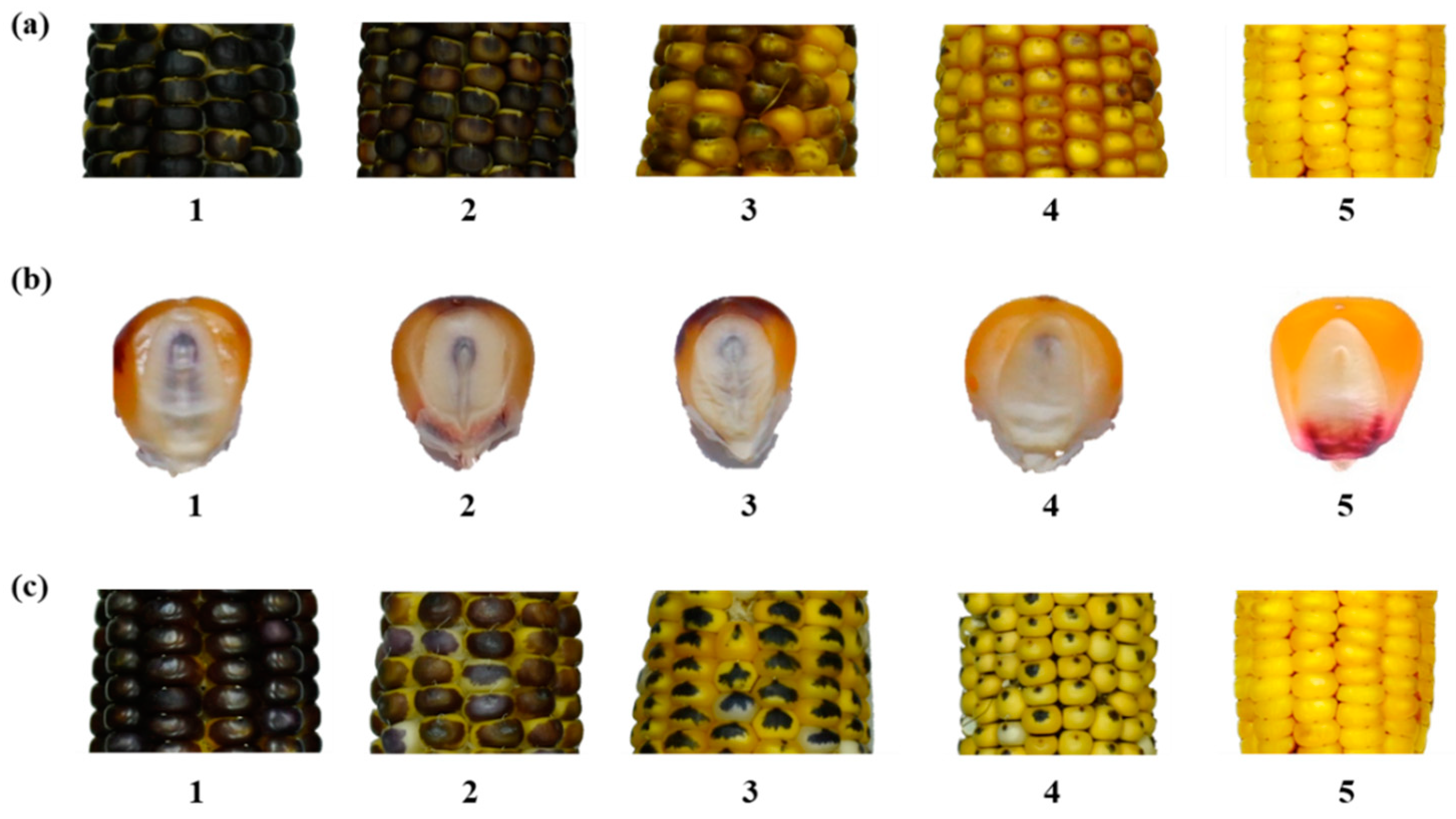
2.4. Visual Assessment of R1-nj Anthocyanin Expression on Ploidy Discrimination
2.5. Statistical Analysis
3. Results
3.1. Analysis of Variance
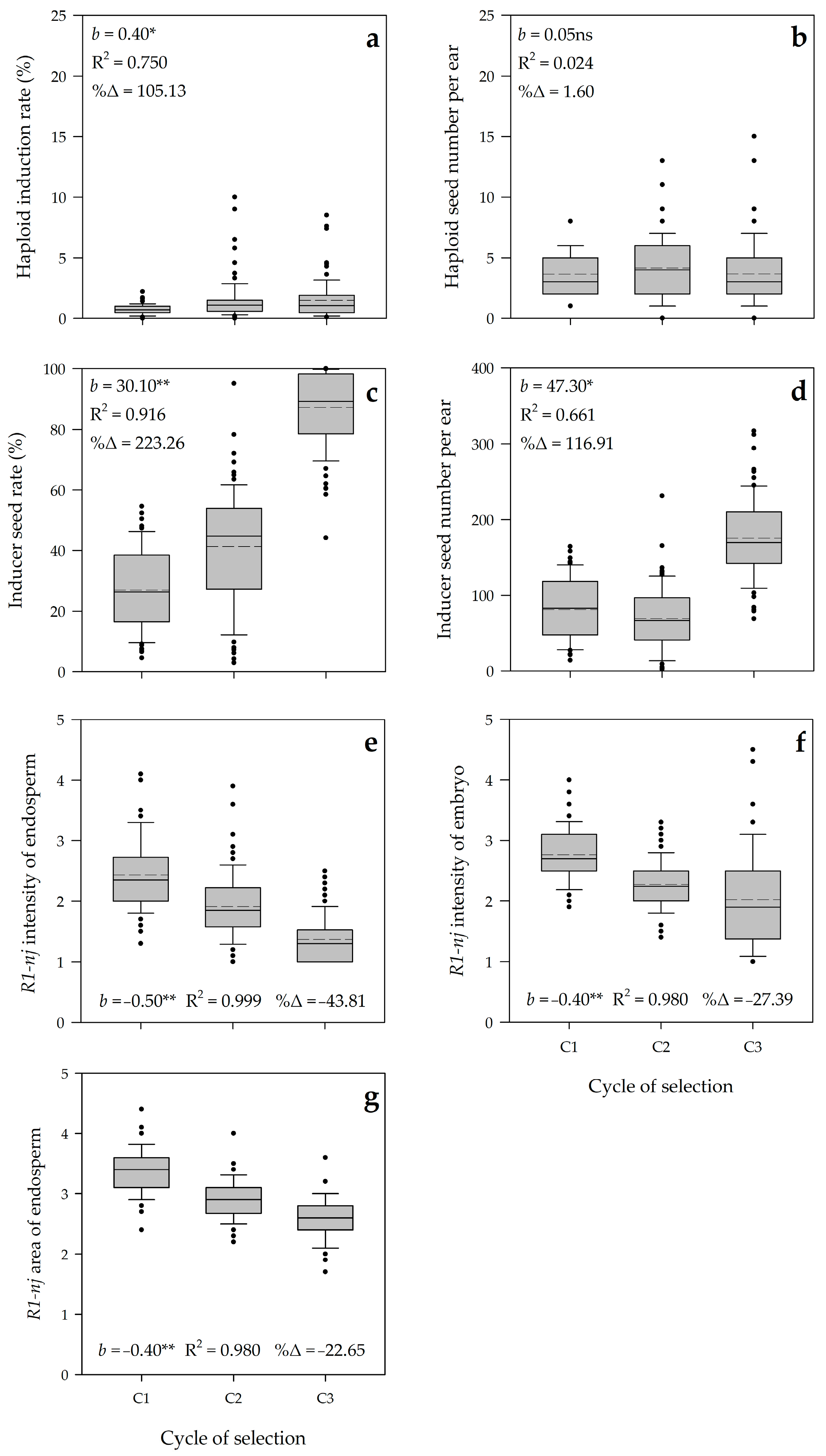
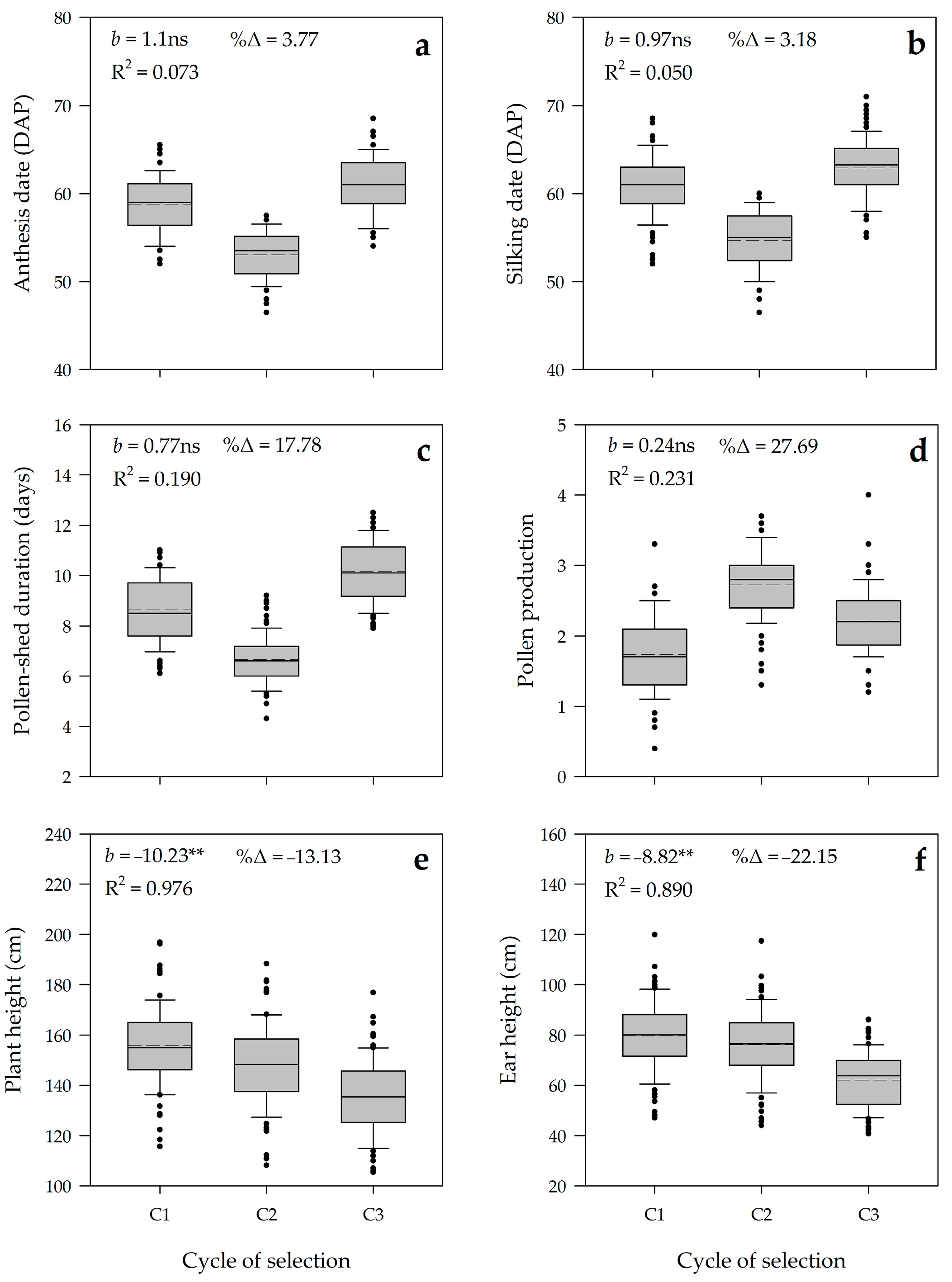
3.2. Realized Genetic Gain
3.3. Genetic Parameters and Predicted Genetic Gain
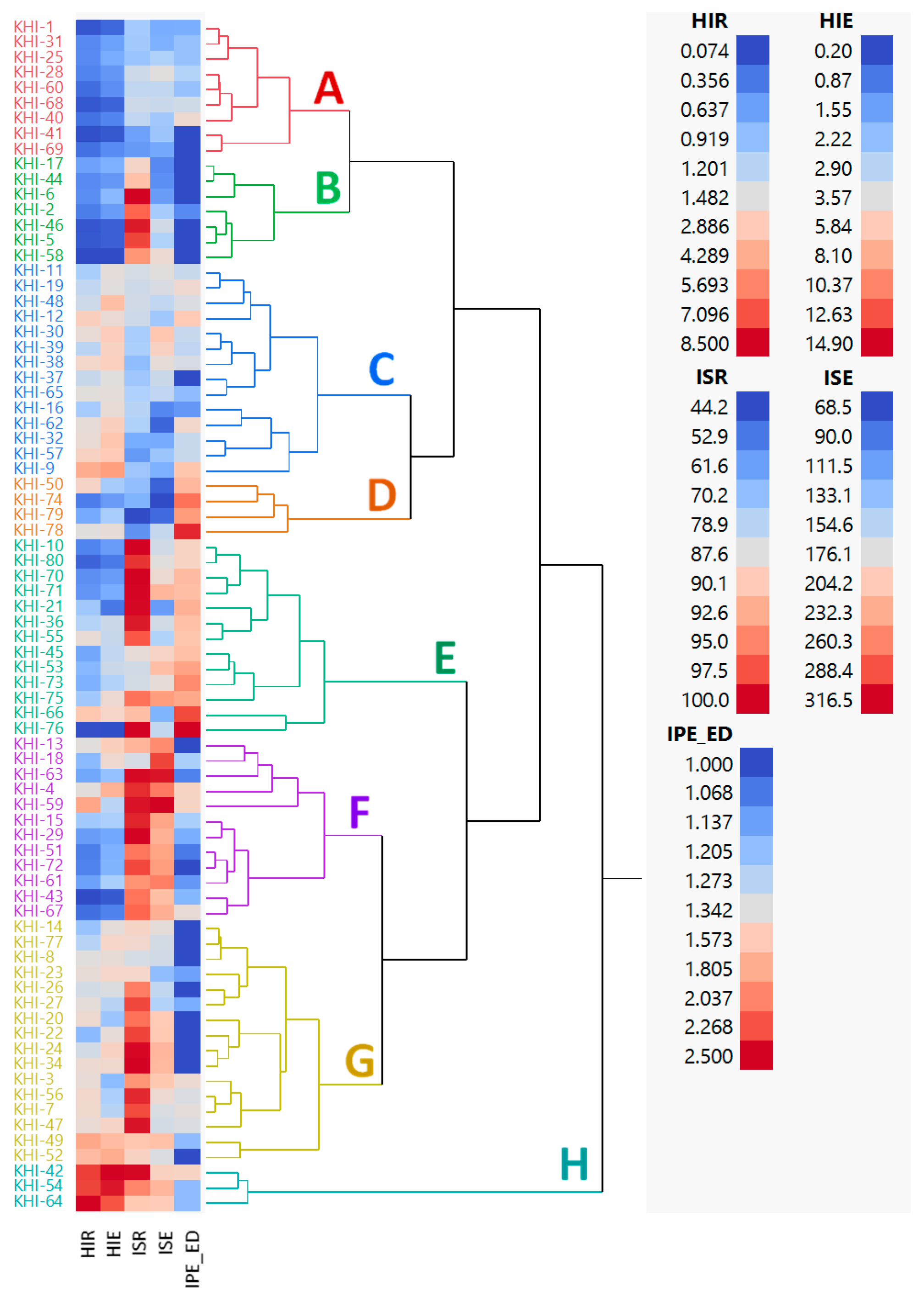
3.4. Phenotypic Variation among Families
4. Discussion
4.1. Analysis of Variance
4.2. Genetic Parameters and Genetic Gains Reveal the Effectiveness of Modified Ear-to-Row with Intra-Family Selection
4.3. The Impact of Breeding Strategy on Family Distribution and Phenotypic Coefficient Variation (PVC) among Families
4.4. Future Breeding Strategies for Improving Genetic Gain on HIR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hallauer, A.R.; Carena, M.J.; de Miranda Filho, J.B. Quantitative Genetics in Maize Breeding; Springer Science & Business Media: New York, NY, USA, 2010. [Google Scholar]
- Worku, M.; Makumbi, D.; Beyene, Y.; Das, B.; Mugo, S.; Pixley, K.; Bänziger, M.; Owino, F.; Olsen, M.; Asea, G.; et al. Grain yield performance and flowering synchrony of CIMMYT’s tropical maize (Zea mays L.) parental inbred lines and single crosses. Euphytica 2016, 211, 395–409. [Google Scholar] [CrossRef] [Green Version]
- Geiger, H.H.; Gordillo, G.A. Doubled haploids in hybrid maize breeding. Maydica 2009, 54, 485–499. [Google Scholar]
- Chase, S.S. Production of homozygous diploids of maize from monoploids. Agron. J. 1952, 44, 263–267. [Google Scholar] [CrossRef] [Green Version]
- Ao, G.M.; Zhao, S.X.; Li, G.H. In vitro induction of haploid plantlets from unpollinated ovaries of corn (Zea mays L.). Acta Genet. Sin. 1982, 9, 281–283. [Google Scholar]
- Chaikam, V.; Molenaar, W.; Melchinger, A.E.; Prasanna, M.B. Doubled haploid technology for line development in maize: Technical advances and prospects. Theor. Appl. Genet. 2019, 132, 3227–3243. [Google Scholar] [CrossRef] [Green Version]
- Coe, E.H. A line of maize with high haploid frequency. Am. Nat. 1959, 93, 381–382. [Google Scholar] [CrossRef]
- Tyrnov, V.S.; Zavalishina, A.N. High-frequency induction of matroclinal haploids in maize. Dokl. Biol. Sci.—Akad. Nauk. SSSR 1984, 276, 353–355. [Google Scholar]
- Zabirova, E.R.; Chumak, M.V.; Shatskaia, O.A.; Scherbak, V.S. Technology of the mass accelerated production of homozygous lines (in Rusian). Kukuruza Sorgo 1996, N4, 17–19. [Google Scholar]
- Chalyk, S.T. Creating new haploid-inducing lines of maize. Maize Genet. Coop. News Lett. 1999, 73, 53–54. [Google Scholar]
- Chen, S.; Song, T. Identification haploid with high oil xenia effect in maize. Acta Agron. Sin. 2003, 29, 587–590. [Google Scholar]
- Röber, F.K.; Gordillo, G.A.; Geiger, H.H. In vivo haploid induction in maize- Performance of new inducers and significance of doubled haploid lines in hybrid breeding. Maydica 2005, 50, 275–283. [Google Scholar]
- Barret, P.; Brinkmann, M.; Beckert, M. A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor. Appl. Genet. 2008, 117, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.T.; Coe, E.H. Doubled haploids. In Molecular Genetic Approaches to Maize Improvement. Biotechnology in Agriculture and Forestry; Kriz, A.L., Larkins, B.A., Eds.; Springer: Berlin, Germany, 2009; Volume 63. [Google Scholar]
- Rotarenco, V.A.; Dicu, G.; State, D.; Fuia, S. New inducers of maternal haploids in maize. Maize Genet. Coop. News Lett. 2010, 84, 15. [Google Scholar]
- De La Fuente, G.N. Improvement to the Maize (Zea mays L.) In Vivo Maternal Doubled Haploid System. Ph.D. Thesis, Iowa State University, Ames, Iowa, 2015. [Google Scholar]
- Prigge, V.; Schipprack, W.; Mahuku, G.; Atlin, G.N.; Melchinger, A.E. Development of in vivo haploid inducers for tropical maize breeding programs. Euphytica 2012, 185, 481–490. [Google Scholar] [CrossRef]
- Chaikam, V.; Nair, S.K.; Martinez, L.; Lopez, L.A.; Utz, H.F.; Melchinger, A.E.; Prasanna, B.M. Marker-assisted breeding of improved maternal haploid inducers in maize for the tropical/subtropical regions. Front. Plant Sci. 2018, 9, 1527. [Google Scholar] [CrossRef] [Green Version]
- Beck, H.E.; Zimmermann, N.E.; McVicar, T.R.; Vergopolan, N.; Berg, A.; Wood, E.F. Present and future Köppen-Geiger climate classification maps at 1-km resolution. Sci. Data 2018, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Lashermes, P.; Beckert, M. Genetic control of maternal haploidy in maize (Zea mays L.) and selection of haploid inducing lines. Theor. Appl. Genet. 1988, 76, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Prigge, V.; Xu, X.; Li, L.; Babu, R.; Chen, S.; Atlin, G.N.; Melchinger, A.E. New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 2012, 190, 781–793. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Hazel, L.N.; Lush, J.L. The efficiency of three methods of selection. J. Hered. 1942, 33, 393–399. [Google Scholar] [CrossRef]
- Hopkins, C.G. Improvement in the chemical composition of the corn kernel. Illinois Agric. Exp. Stn. Bull. 1899, 55, 1039–1057. [Google Scholar] [CrossRef] [Green Version]
- Compton, W.A.; Bahadur, K. Ten cycles of progress from modified ear-to-row selection in corn. Crop Sci. 1977, 17, 378–380. [Google Scholar] [CrossRef]
- Webel, O.D.; Lonnquist, J.H. An evaluation of modified ear-to-row selection in a population of corn (Zea mays L.). Crop Sci. 1967, 7, 651–655. [Google Scholar] [CrossRef]
- Paterniani, E. Selection among and within half-sib families in a Brazilian population of maize (Zea mays L.). Crop Sci 1967, 7, 212–216. [Google Scholar] [CrossRef]
- Khamphasan, P.; Lomthaisong, K.; Harakotr, B.; Scott, M.P.; Lertrat, K.; Suriharn, B. Effects of mass selection on husk and cob color in five purple corn populations segregating for purple husks. Agriculture 2020, 10, 311. [Google Scholar] [CrossRef]
- Thai Agricultural Practice, Department of Agriculture, Thailand. Available online: http://www.doa.go.th (accessed on 5 July 2021).
- Nanda, D.K.; Chase, S.S. An embryo marker for detecting monoploids of maize (Zea mays L.). Crop Sci. 1966, 6, 213–215. [Google Scholar] [CrossRef]
- Prigge, V.; Sánchez, C.; Dhillon, B.S.; Schipprank, W.; Araus, J.L.; Bänziger, M.; Melchinger, A.E. Doubled haploids in tropical maize: I. Effect of inducers and source germplasm on in vivo haploid induction rates. Crop Sci. 2011, 51, 1498–1506. [Google Scholar] [CrossRef]
- Chaikam, V.; Nair, S.K.; Babu, R.; Martinez, L.; Tejomurtula, J.; Prasanna, M.B. Analysis of effectiveness of R1-nj anthocyanin marker for in vivo haploid identification in maize and molecular markers for predicting the inhibition of R1-nj expression. Theor. Appl. Genet. 2015, 128, 159–171. [Google Scholar] [CrossRef]
- SAS Institute. SAS for Windows Version 9.0; SAS Institute: Cary, NC, USA, 2002. [Google Scholar]
- Gomez, K.A.; Gomez, A.A. Statistical Procedure for Agricultural Research; John Wiley and Sons: Singapore, 1984. [Google Scholar]
- Bernardo, R. Breeding for Quantitative Traits in Plants; Stemma Press: Woodbury, MN, USA, 2020. [Google Scholar]
- Hallauer, A.R.; Miranda, J.B. Quantitative Genetics in Maize Breeding, 2nd ed.; Iowa State University Press: Ames, IA, USA, 1988. [Google Scholar]
- Singh, R.K.; Chaudhary, B.D. Biometrical Methods in Quantitative Genetic Analysis; Kalyani Publishers: New Delhi, India, 2004. [Google Scholar]
- Souza, A.R.R.; Miranda, G.V.; Pereira, M.G.; de Souza, L.V. Predicting the genetic gain in the Brazilian white maize landrace. Ciência Rural 2009, 39, 19–24. [Google Scholar] [CrossRef] [Green Version]
- SAS Institute. JMP Trial 15.0.0 (403428); SAS Institute: Cary, NC, USA, 2021. [Google Scholar]
- Wright, S. Evolution in Mendelian populations. Genetics 1930, 16, 97–159. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2004, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Hussanun, S.; Suriharn, B.; Lertrat, K. Yield and early maturity response to four cycles of modified mass selection in purple waxy corn. Turk. J. Field Crops 2014, 19, 84–89. [Google Scholar] [CrossRef] [Green Version]
- Aman, M.A.; Sarkar, K.R. Selection for haploidy inducing potential in maize. Indian J. Genet. Plant Breed. 1978, 38, 452–457. [Google Scholar]
- Rutkoski, J.E. A practical guide to genetic gain. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 157, pp. 217–249. [Google Scholar]
- Maphumulo, S.G.; Derera, J.; Qwabe, F.; Fato, P.; Gasura, E.; Mafongoya, P. Heritability and genetic gain for grain yield and path coefficient analysis of some agronomic traits in early-maturing maize hybrids. Euphytica 2015, 206, 225–244. [Google Scholar] [CrossRef]
- Zinaw, D.; Seltene, A.; Addisu, G. Genetic variability and heritability of soybean (Glycine max (L.) Merrill) genotypes in Pawe district, Metekel zone, Benishangule Gumuz regional state, northwestern Ethiopia. J. Agric. Res. 2013, 2, 240–245. [Google Scholar]
- Eberhart, S.A. Least squares method for comparing progress among recurrent selection methods. Crop Sci. 1964, 4, 230–231. [Google Scholar] [CrossRef]
- Shimelis, H.; Shiringani, R. Variance components and heritabilities of yield and agronomic traits among cowpea genotypes. Euphytica 2010, 176, 383–389. [Google Scholar] [CrossRef]
- Gapare, W.J. Predicted and realized genetic gain in Eucalyptus grandis breeding seedling orchard in Zimbabwe. S. Afr. For. J. 2000, 189, 11–15. [Google Scholar]
- Bello, O.B.; Ige, S.A.; Azeez, M.A.; Afolabi, M.S.; Abdulmaliq, S.Y.; Mahamood, J. Heritability and genetic advance for grain yield and its component characters in maize (Zea mays L.). Int. J. Plant Res. 2012, 2, 138–145. [Google Scholar]
- Ribeiro, C.B.; Rezende, B.A.; Filho, J.S.S.B.; da Silva, E.D.B.; Dias, K.O.G.; Nunes, J.A.R.; Carvalho, P.L.S.; Mota, S.F.; Bustamante, F.O.; Diniz, R.P.; et al. Breeding strategies for tropical maize targeting in vivo haploid inducers. Crop Breed. Appl. Biotechnol. 2020, 20, 1–8. [Google Scholar] [CrossRef]
- Almeida, V.C.; Ursula Trentin, H.; Frei, U.K.; Lübberstedt, T. Genomic prediction of maternal haploid induction rate in maize. Plant Genome 2020, 13, e20014. [Google Scholar] [CrossRef] [Green Version]
- Shatskaya, O.A. Haploid inductors isolation in maize: Three cycles of selection on high frequency of induction of matroclinal haploids. Sel’skokhozyaistvennaya Biol. Agric. Biol. 2010, 5, 79–86. [Google Scholar]
- Kebede, A.Z.; Dhillon, B.S.; Schipprack, W.; Araus, J.L.; Bänziger, M.; Semagn, K.; Alvarado, G.; Melchinger, A.E. Effect of source germplasm and season on the in vivo haploid induction rate in tropical maize. Euphytica 2011, 180, 219–226. [Google Scholar] [CrossRef]
- De La Fuente, G.N.; Frei, U.K.; Trampe, B.; Nettleton, D.; Zhang, W.; Lübberstedt, T. A diallel analysis of a maize donor population response to in vivo maternal haploid induction: I. Inducibility. Crop Sci. 2018, 58, 1830–1837. [Google Scholar] [CrossRef] [Green Version]
- Eder, J.; Chalyk, S. In vivo haploid induction in maize. Theor. Appl. Genet. 2002, 104, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Rotarenco, V.; Dicu, G.; Sarmaniuc, M. Induction of maternal haploids in maize. Maize Genet. Coop. News Lett. 2009, 83, 15–17. [Google Scholar]
- Prasanna, B.M. Doubled haploid (DH) technology in maize breeding: An overview. In Doubled Haploid Technology in Maize Breeding: Theory and Practice; Prasanna, B.M., Chaikam, V., Mahuku, G., Eds.; CIMMYT: Mexico City, Mexico, 2012; pp. 1–8. [Google Scholar]
- Masuka, B.; Atlin, G.N.; Olsen, M.; Magorokosho, C.; Labuschagne, M.; Crossa, J.; Bänziger, M.; Pixley, K.V.; Vivek, B.S.; von Biljon, A.; et al. Gains in maize genetic improvement in eastern and southern Africa: I. CIMMYT hybrid breeding pipeline. Crop Sci. 2017, 57, 168–179. [Google Scholar] [CrossRef] [Green Version]
- Uliana Trentin, H.; Frei, U.K.; Lübberstedt, T. Breeding maize maternal haploid inducers. Plants 2020, 9, 614. [Google Scholar] [CrossRef]
- Ribaut, J.M.; Hoisington, D. Marker-assisted selection: New tools and strategies. Trends Plant Sci. 1998, 3, 236–239. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Xu, X.; Miao, J.; Li, L.; Zhang, D.; Mi, X.; Liu, C.; Tian, X.; Melchinger, E.A.; Chen, S. Fine mapping of qhir1 influencing in vivo haploid induction in maize. Theor. Appl. Genet. 2013, 126, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, W.; Zhong, Y.; Dong, X.; Hu, H.; Tian, X.; Wang, L.; Chen, B.; Chen, C.; Melchinger, A.E.; et al. Fine mapping of qhir8 affecting in vivo haploid induction in maize. Theor. Appl. Genet. 2015, 128, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Xu, X.; Li, L.; Liu, C.; Tian, X.; Li, W.; Chen, S. Marker-assisted selection and evaluation of high oil in vivo haploid inducers in maize. Mol. Breed. 2014, 34, 1147–1158. [Google Scholar] [CrossRef]



| Traits | Source of Variation | CV (%) | |||
|---|---|---|---|---|---|
| Cycle (C) | Family (F) | C × F | Pooled Error | ||
| df | 2 | 77 | 154 | 231 | |
| Agronomic traits | |||||
| Anthesis date | 2647.4 ** | 39.7 ** | 9.9 ** | 3.9 | 3.4 |
| Silking date | 2805.5 ** | 49.2 ** | 10.5 ** | 5.0 | 3.7 |
| Pollen-shed duration | 480.2 ** | 4.3 ** | 1.8 ns | 1.9 | 16.1 |
| Pollen production | 38.9 * | 0.9 ** | 0.3 ns | 0.4 | 22.6 |
| Plant height | 16,743.8 * | 1153.1 ** | 188.1 ns | 181.0 | 9.2 |
| Ear height | 13,651.3 * | 853.4 ** | 81.2 * | 63.2 | 10.9 |
| Haploid induction ability | |||||
| Haploid induction rate 1 | 29.2 * | 6.8 ** | 2.4 ** | 0.6 | 29.7 |
| Haploid seed number per ear 1 | 9.8 * | 19.7 ** | 7.3 ** | 2.8 | 24.4 |
| Inducer seed rate | 154,663.5 ** | 656.5 ** | 360.8 ** | 57.0 | 23.5 |
| Inducer seed number per ear | 528,600.6 ** | 7153.1 ** | 2593.8 ** | 709.2 | 24.5 |
| R1-nj intensity of endosperm | 44.1 * | 1.0 ** | 0.3ns | 0.3 | 18.1 |
| R1-nj intensity of embryo | 22.9 ** | 0.9 ** | 0.6ns | 0.5 | 19.3 |
| R1-nj area of endosperm | 22.8 ** | 0.4 ** | 0.2ns | 0.2 | 15.7 |
| Traits | GCV (%) | PCV (%) | GA | %GA | |||
|---|---|---|---|---|---|---|---|
| Haploid induction ability | |||||||
| Haploid induction rate | 0.74 | 1.37 | 0.54 | 70.05 | 95.55 | 1.30 | 105.81 |
| Haploid seed number per ear | 2.05 | 4.85 | 0.42 | 38.00 | 58.44 | 1.92 | 50.90 |
| Inducer seed rate | 299.73 | 356.73 | 0.84 | 33.40 | 36.43 | 32.69 | 63.06 |
| Inducer seed number per ear | 3221.95 | 3931.15 | 0.82 | 52.19 | 57.65 | 105.86 | 97.34 |
| R1-nj intensity of endosperm | 0.33 | 0.62 | 0.54 | 30.24 | 41.28 | 0.87 | 45.63 |
| R1-nj intensity of embryo | 0.21 | 0.69 | 0.31 | 19.52 | 35.22 | 0.52 | 22.28 |
| R1-nj area of endosperm | 0.10 | 0.32 | 0.33 | 10.93 | 19.12 | 0.38 | 12.87 |
| Agronomic traits | |||||||
| Anthesis date | 17.88 | 21.77 | 0.82 | 7.34 | 8.10 | 7.89 | 13.70 |
| Silking date | 21.12 | 26.13 | 0.81 | 7.70 | 8.57 | 8.51 | 14.27 |
| Pollen-shed duration | 1.19 | 3.04 | 0.39 | 12.84 | 20.55 | 1.40 | 16.53 |
| Pollen production | 0.26 | 0.64 | 0.41 | 23.00 | 35.91 | 0.68 | 30.36 |
| Plant height | 486.04 | 667.05 | 0.73 | 15.04 | 17.62 | 38.77 | 26.45 |
| Ear height | 395.07 | 458.28 | 0.86 | 27.36 | 29.47 | 38.02 | 52.33 |
| Cycle | ISR | ISE | HIR | HIE | IED | IEM | AED |
|---|---|---|---|---|---|---|---|
| C1 | 49.08 a | 49.80 b | 57.63 b | 44.95 b | 23.59 a | 16.05 b | 10.91 a |
| C2 | 45.51 a | 60.99 a | 118.09 a | 61.18 b | 29.31 a | 17.14 b | 12.03 a |
| C3 | 14.53 b | 29.88 c | 109.43 a | 80.74 a | 27.47 a | 40.22 a | 13.12 a |
| Cycle | AD | SD | PSD | PPD | PH | EH |
|---|---|---|---|---|---|---|
| C1 | 5.73 a | 5.81 a | 14.80 a | 30.70 a | 10.38 a | 17.24 a |
| C2 | 5.22 a | 5.93 a | 14.55 a | 17.10 b | 11.11 a | 18.08 a |
| C3 | 5.35 a | 5.45 a | 11.47 b | 22.01 b | 11.22 a | 18.32 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dermail, A.; Chankaew, S.; Lertrat, K.; Lübberstedt, T.; Suriharn, K. Selection Gain of Maize Haploid Inducers for the Tropical Savanna Environments. Plants 2021, 10, 2812. https://doi.org/10.3390/plants10122812
Dermail A, Chankaew S, Lertrat K, Lübberstedt T, Suriharn K. Selection Gain of Maize Haploid Inducers for the Tropical Savanna Environments. Plants. 2021; 10(12):2812. https://doi.org/10.3390/plants10122812
Chicago/Turabian StyleDermail, Abil, Sompong Chankaew, Kamol Lertrat, Thomas Lübberstedt, and Khundej Suriharn. 2021. "Selection Gain of Maize Haploid Inducers for the Tropical Savanna Environments" Plants 10, no. 12: 2812. https://doi.org/10.3390/plants10122812
APA StyleDermail, A., Chankaew, S., Lertrat, K., Lübberstedt, T., & Suriharn, K. (2021). Selection Gain of Maize Haploid Inducers for the Tropical Savanna Environments. Plants, 10(12), 2812. https://doi.org/10.3390/plants10122812







