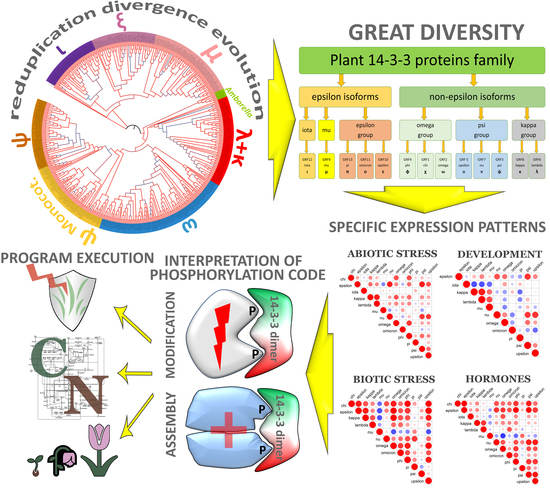
Evolution of 14-3-3 Proteins in Angiosperm Plants: Recurring Gene Duplication and Loss
Abstract
1. Introduction
2. Results
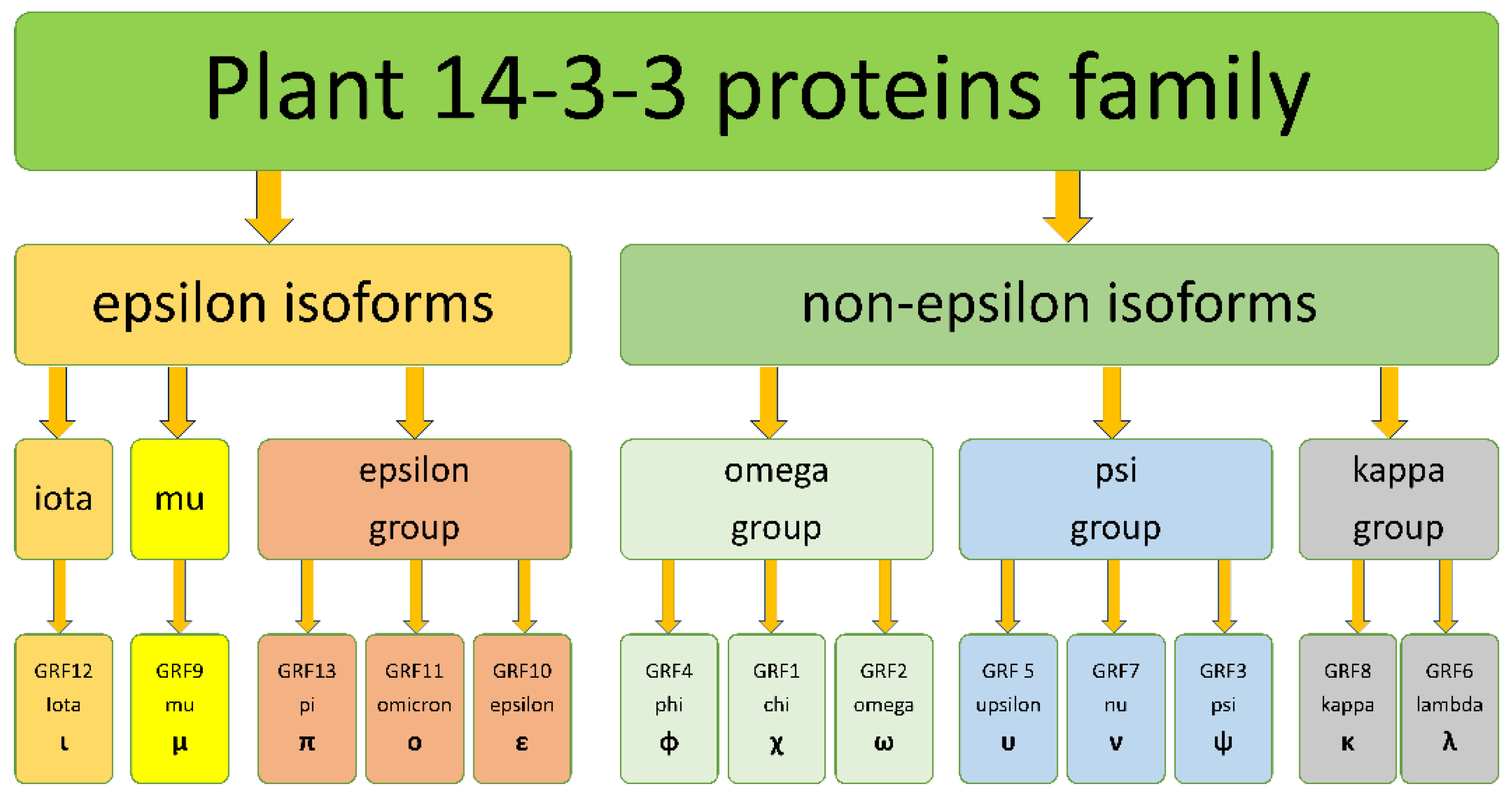
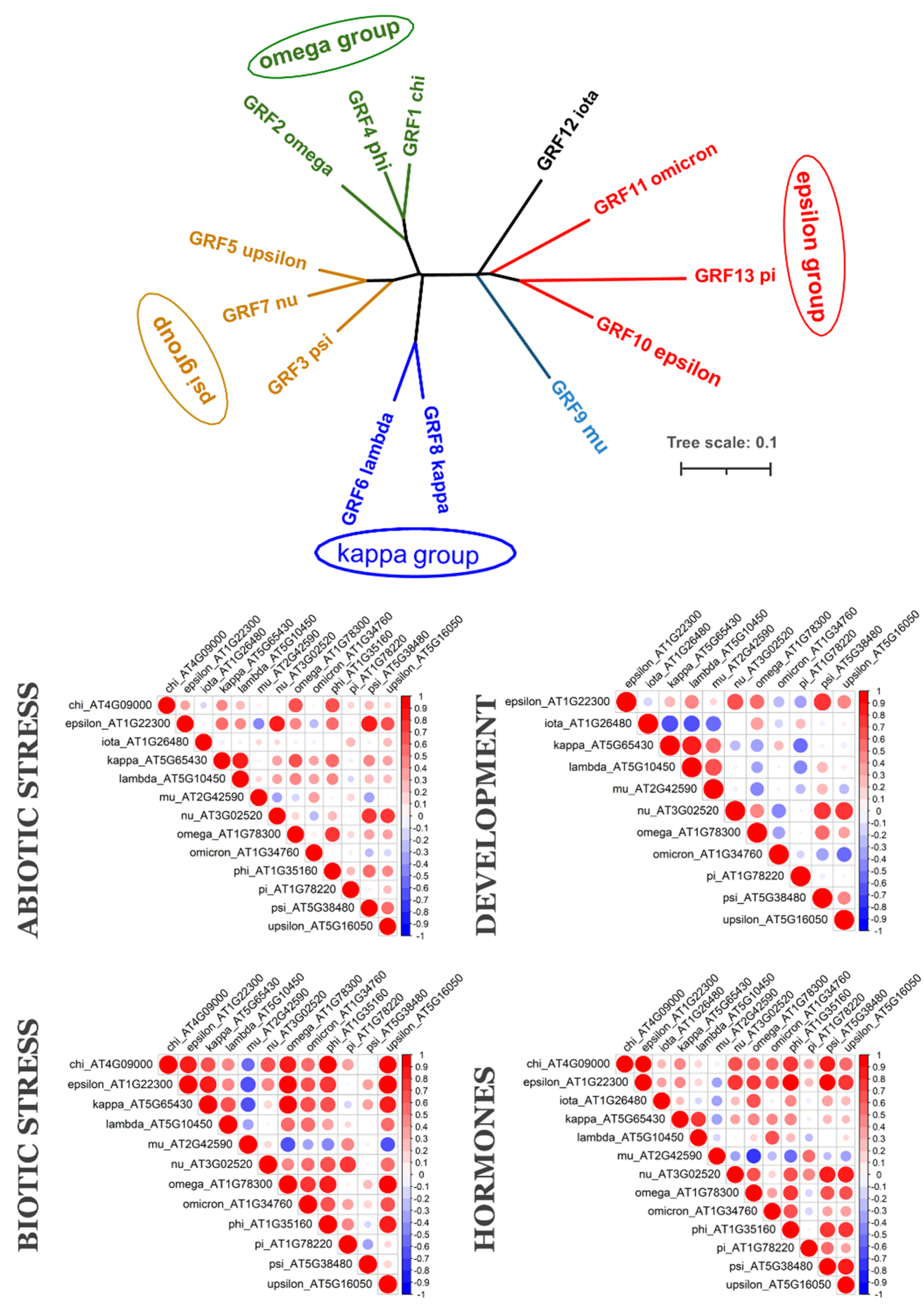
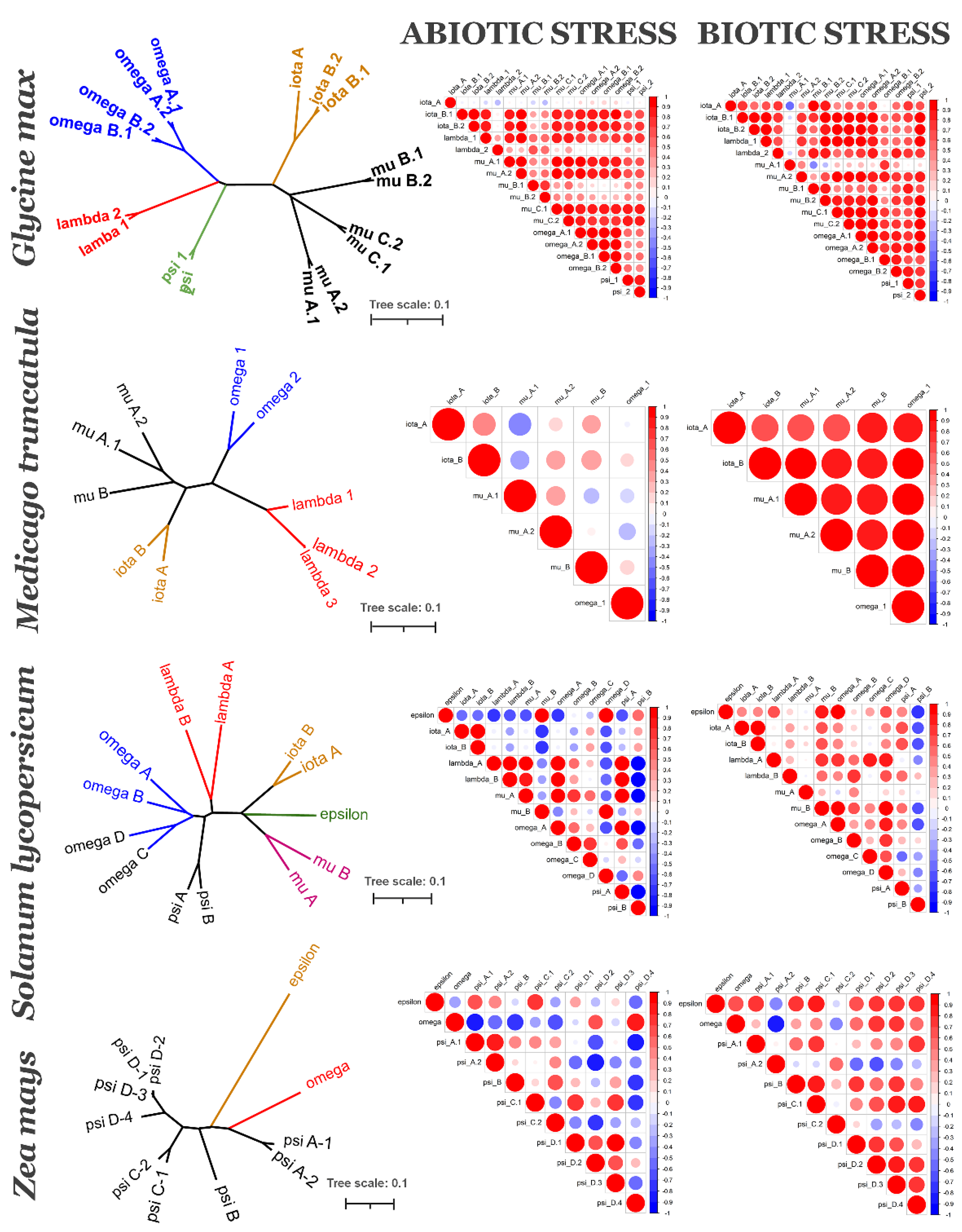
2.1. Phylogenetic Analysis of 14-3-3 Proteins Family
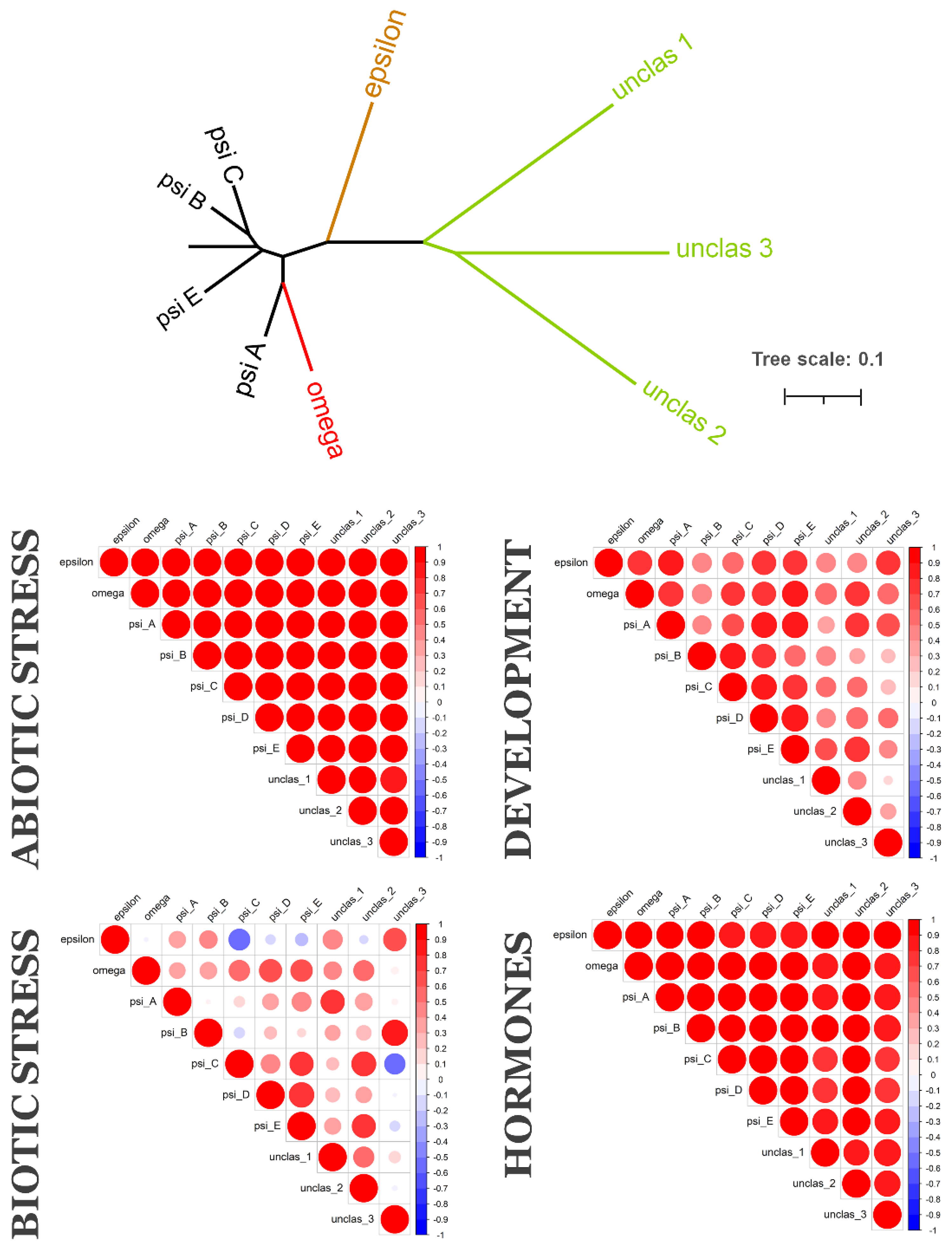
2.2. Coexpression of 14-3-3 Protein Isoforms in Plants
3. Discussion
4. Materials and Methods
4.1. 14-3-3 Sequences Search
4.2. Phylogenetic Analysis
4.3. Coexpression and Spatiotemporal Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dougherty, M.K.; Morrison, D.K. Unlocking the Code of 14-3-3. J. Cell Sci. 2004, 117, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.; Perez, V. Specific acidic proteins of the nervous system. In Physiological and Biochemical Aspects of Nervous Integration; Prentice-Hall: Englewood Cliffs, NJ, USA, 1968; pp. 343–359. [Google Scholar]
- Lu, G.; DeLisle, A.J.; De Vetten, N.C.; Ferl, R.J. Brain Proteins in Plants: An Arabidopsis Homolog to Neurotransmitter Pathway Activators Is Part of a DNA Binding Complex. Proc. Natl. Acad. Sci. USA 1992, 89, 11490–11494. [Google Scholar] [CrossRef]
- De Vetten, N.C.; Lu, G.; Ferl, R.J. A Maize Protein Associated with the G-Box Binding Complex Has Homology to Brain Regulatory Proteins. Plant Cell 1992, 4, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Aitken, A.; Bertsch, U.; Soll, J. A Plant Homologue to Mammalian Brain 14-3-3 Protein and Protein Kinase C Inhibitor. FEBS Lett. 1992, 296, 222–224. [Google Scholar] [CrossRef]
- Denison, F.C.; Paul, A.L.; Zupanska, A.K.; Ferl, R.J. 14-3-3 Proteins in Plant Physiology. Semin Cell Dev. Biol. 2011, 22, 720–727. [Google Scholar] [CrossRef]
- Cotelle, V.; Leonhardt, N. 14-3-3 Proteins in Guard Cell Signaling. Front. Plant Sci. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- de Boer, A.H.; van Kleeff, P.J.M.; Gao, J. Plant 14-3-3 Proteins as Spiders in a Web of Phosphorylation. Protoplasma 2013, 250, 425–440. [Google Scholar] [CrossRef]
- Zhao, X.; Li, F.; Li, K. The 14-3-3 Proteins: Regulators of Plant Metabolism and Stress Responses. Plant Biol. 2021, 23, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, S.; Ding, G.; Wang, B.; Li, Y.; Zhao, Y.; Shao, Q.; Feng, J.; Liu, S.; Qin, L.; et al. The Role of 14-3-3 Proteins in Cell Signalling Pathways and Virus Infection. J. Cell. Mol. Med. 2021, 25, 4173–4182. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.L.; Denison, F.C.; Schultz, E.R.; Zupanska, A.K.; Ferl, R.J. 14-3-3 Phosphoprotein Interaction Networks—Does Isoform Diversity Present Functional Interaction Specification? Front. Plant Sci. 2012, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Aitken, A.; Collinge, D.B.; van Heusden, B.P.; Isobe, T.; Roseboom, P.H.; Rosenfeld, G.; Soll, J. 14-3-3 Proteins: A Highly Conserved, Widespread Family of Eukaryotic Proteins. Trends Biochem. Sci. 1992, 17, 498–501. [Google Scholar] [CrossRef]
- Sribar, J.; Sherman, N.E.; Prijatelj, P.; Faure, G.; Gubensek, F.; Fox, J.W.; Aitken, A.; Pungercar, J.; Krizaj, I. The Neurotoxic Phospholipase A2 Associates, through a Non-Phosphorylated Binding Motif, with 14-3-3 Protein Gamma and Epsilon Isoforms. Biochem. Biophys. Res. Commun. 2003, 302, 691–696. [Google Scholar] [CrossRef]
- Rosenquist, M.; Sehnke, P.; Ferl, R.J.; Sommarin, M.; Larsson, C. Evolution of the 14-3-3 Protein Family: Does the Large Number of Isoforms in Multicellular Organisms Reflect Functional Specificity? J. Mol. Evol. 2000, 51, 446–458. [Google Scholar] [CrossRef]
- DeLille, J.M.; Sehnke, P.C.; Ferl, R.J. The Arabidopsis 14-3-3 Family of Signaling Regulators. Plant Physiol. 2001, 126, 35–38. [Google Scholar] [CrossRef]
- Konagaya, K.I.; Matsushita, Y.; Kasahara, M.; Nyunoya, H. Members of 14-3-3 Protein Isoforms Interacting with the Resistance Gene Product N and the Elicitor of Tobacco Mosaic Virus. J. Gen. Plant Pathol. 2004, 70, 221–231. [Google Scholar] [CrossRef]
- Li, M.; Ren, L.; Xu, B.; Yang, X.; Xia, Q.; He, P.; Xiao, S.; Guo, A.; Hu, W.; Jin, Z. Genome-Wide Identification, Phylogeny, and Expression Analyses of the 14-3-3 Family Reveal Their Involvement in the Development, Ripening, and Abiotic Stress Response in Banana. Front. Plant Sci. 2016, 7, 1442. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, Y.; Yuan, L.; Bian, Y.; Wang, L.; Zhen, S.; Hu, Y.; Yan, Y. Molecular Characterization of the 14-3-3 Gene Family in Brachypodium distachyon L. Reveals High Evolutionary Conservation and Diverse Responses to Abiotic Stresses. Front. Plant Sci. 2016, 7, 1099. [Google Scholar] [CrossRef]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. 14-3-3 Proteins in Plant Hormone Signaling: Doing Several Things at Once. Front. Plant Sci. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski, M.; Oecking, C. Five New 14-3-3 Isoforms from Nicotiana tabacum L.: Implications for the Phylogeny of Plant 14-3-3 Proteins. Planta 1997, 204, 127–130. [Google Scholar] [CrossRef]
- Sehnke, P.C.; Rosenquist, M.; Alsterfjord, M.; DeLille, J.; Sommarin, M.; Larsson, C.; Ferl, R.J. Evolution and Isoform Specificity of Plant 14-3-3 Proteins. Plant Mol. Biol. 2002, 50, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Wang, T.; Xie, Y.; Zhang, J.; Hu, J. Genome-Wide Identification, Classification, and Expression Analysis of 14-3-3 Gene Family in Populus. PLoS ONE 2015, 10, e0123225. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Wang, S.; Xiang, W.; Yang, H.; Tahir, M.M.; Zheng, S.; An, N.; Han, M.; Zhao, C.; Zhang, D. Genome-Wide Identification of the 14–3-3 Gene Family and Its Participation in Floral Transition by Interacting with TFL1/FT in Apple. BMC Genom. 2021, 22, 41. [Google Scholar] [CrossRef]
- Kumar, R. Differential Abundance and Transcription of 14-3-3 Proteins during Vegetative Growth and Sexual Reproduction in Budding Yeast. Sci. Rep. 2018, 8, 2145. [Google Scholar] [CrossRef]
- The Angiosperm Phylogeny Group. An Update of the Angiosperm Phylogeny Group Classification for the Orders and Families of Flowering Plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Yim, W.C.; Yu, Y.; Song, K.; Jang, C.S.; Lee, B.M. PLANEX: The Plant Co-Expression Database. BMC Plant Biol. 2013, 13, 1–9. [Google Scholar] [CrossRef]
- Tseng, K.; Li, G.; Hung, Y.; Chow, C.; Wu, N. EXPath 2.0: An Updated Database for Integrating High-Throughput Gene Expression Data with Biological Pathways. Plant Cell Physiol. 2020, 61, 1818–1827. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Kudo, T.; Terashima, S.; Saito, M.; Nambara, E.; Yano, K. CATchUP: A Web Database for Spatiotemporally Regulated Genes Special Online Collection—Database Paper. Plant Cell Physiol. 2017, 58, 1–18. [Google Scholar] [CrossRef][Green Version]
- Wu, K.; Rooney, M.F.; Ferl, R.J. The Arabidopsis 14-3-3 Multigene Family. Plant Physiol. 1997, 114, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, M.; Alsterfjord, M.; Larsson, C.; Sommarin, M. Data Mining the Arabidopsis Genome Reveals Fifteen 14-3-3 Genes. Expression Is Demonstrated for Two out of Five Novel Genes. Plant Physiol. 2001, 127, 142–149. [Google Scholar] [CrossRef]
- DePamphilis, C.W.; Palmer, J.D.; Rounsley, S.; Sankoff, D.; Schuster, S.C.; Ammiraju, J.S.S.; Barbazuk, W.B.; Chamala, S.; Chanderbali, A.S.; Determann, R.; et al. The Amborella Genome and the Evolution of Flowering Plants. Science 2013, 342, 1467–1477. [Google Scholar] [CrossRef]
- Landis, J.B.; Soltis, D.E.; Li, Z.; Marx, H.E.; Barker, M.S.; Tank, D.C.; Soltis, P.S. Impact of Whole-genome Duplication Events on Diversification Rates in Angiosperms. Am. J. Bot. 2018, 105, 348–363. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Fares, M.A. Evolutionary Dynamics and Functional Specialization of Plant Paralogs Formed by Whole and Small-Scale Genome Duplications. Mol. Biol. Evol. 2012, 29, 3541–3551. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene Duplication and Evolution in Recurring Polyploidization-Diploidization Cycles in Plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral Polyploidy in Seed Plants and Angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- McKain, M.R.; Tang, H.; McNeal, J.R.; Ayyampalayam, S.; Davis, J.I.; dePamphilis, C.W.; Givnish, T.J.; Pires, J.C.; Stevenson, D.W.; Leebens-Mack, J.H. A Phylogenomic Assessment of Ancient Polyploidy and Genome Evolution across the Poales. Genome Biol. Evol. 2016, 8, 1150–1164. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The Banana (Musa acuminata) Genome and the Evolution of Monocotyledonous Plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Droc, G.; Larivière, D.; Guignon, V.; Yahiaoui, N.; This, D.; Garsmeur, O.; Dereeper, A.; Hamelin, C.; Argout, X.; Dufayard, J.-F.; et al. The Banana Genome Hub. Database J. Biol. Databases Curation 2013, 2013, 1–14. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Amosova, A.V.; Belyakov, E.A.; Zhurbenko, P.M.; Mikhailova, Y.V.; Punina, E.O.; Shneyer, V.S.; Loskutov, I.G.; Muravenko, O.V. Genetic Consequences of Interspecific Hybridization, Its Role in Speciation and Phenotypic Diversity of Plants. Russ. J. Genet. 2019, 55, 278–294. [Google Scholar] [CrossRef]
- Feng, G.; Burleigh, J.G.; Braun, E.L.; Mei, W.; Barbazuk, W.B. Evolution of the 3R-MYB Gene Family in Plants. Genome Biol. Evol. 2017, 9, 1013. [Google Scholar] [CrossRef]
- Swatek, K.N.; Graham, K.; Agrawal, G.K.; Thelen, J.J. The 14-3-3 Isoforms Chi and Epsilon Differentially Bind Client Proteins from Developing Arabidopsis Seed. J. Proteome Res. 2011, 10, 4076–4087. [Google Scholar] [CrossRef]
- Pallucca, R.; Visconti, S.; Camoni, L.; Cesareni, G.; Melino, S.; Panni, S.; Torreri, P.; Aducci, P. Specificity of ε and Non-ε Isoforms of Arabidopsis 14-3-3 Proteins towards the H+-ATPase and Other Targets. PLoS ONE 2014, 9, e90764. [Google Scholar] [CrossRef] [PubMed]
- CCSB Interactome Database. Available online: http://interactome.dfci.harvard.edu/A_thaliana/index.php?page=download (accessed on 31 October 2021).
- Consortium, A.I.M.; Dreze, M.; Carvunis, A.-R.; Charloteaux, B.; Galli, M.; Pevzner, S.J.; Tasan, M.; Ahn, Y.-Y.; Balumuri, P.; Barabási, A.-L.; et al. Evidence for Network Evolution in an Arabidopsis Interactome Map. Science 2011, 333, 601–607. [Google Scholar] [CrossRef]
- Yang, X.; Yang, S.; Qi, H.; Wang, T.; Li, H.; Zhang, Z. PlaPPISite: A Comprehensive Resource for Plant Protein-Protein Interaction Sites. BMC Plant Biol. 2020, 20, 61. [Google Scholar] [CrossRef]
- PlaPPISite. Available online: http://zzdlab.com/plappisite/index.php (accessed on 31 October 2021).
- Li, H.; Yang, S.; Wang, C.; Zhou, Y.; Zhang, Z. AraPPISite: A Database of Fine-Grained Protein–Protein Interaction Site Annotations for Arabidopsis thaliana. Plant Mol. Biol. 2016, 92, 105–116. [Google Scholar] [CrossRef]
- Liu, Z.; Jia, Y.; Ding, Y.; Shi, Y.; Li, Z.; Guo, Y.; Gong, Z.; Yang, S. Plasma Membrane CRPK1-Mediated Phosphorylation of 14-3-3 Proteins Induces Their Nuclear Import to Fine-Tune CBF Signaling during Cold Response. Mol. Cell 2017, 66, 117–128.e5. [Google Scholar] [CrossRef]
- Visconti, S.; D’Ambrosio, C.; Fiorillo, A.; Arena, S.; Muzi, C.; Zottini, M.; Aducci, P.; Marra, M.; Scaloni, A.; Camoni, L. Overexpression of 14-3-3 Proteins Enhances Cold Tolerance and Increases Levels of Stress-Responsive Proteins of Arabidopsis Plants. Plant Sci. 2019, 289, 110215. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Du, Y.; Jiang, L.; Liu, J.Y. Molecular Analysis and Expression Patterns of the 14-3-3 Gene Family from Oryza sativa. J. Biochem. Mol. Biol. 2007, 40, 349–357. [Google Scholar] [CrossRef]
- Beringer, J.E.; Brewin, N.; Johnston, A.W.B.; Schulman, H.M.; Hopwood, D.A. The Rhizobium-legume symbiosis. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 1979, 204, 219–233. [Google Scholar] [CrossRef]
- Moore, R.; Peregganan, M. The Evolutionary Dynamics of Plant Duplicate Genes. Curr. Opin. Plant Biol. 2005, 8, 122–128. [Google Scholar] [CrossRef]
- Charon, C.; Bruggeman, Q.; Thareau, V.; Henry, Y. Gene Duplication within the Green Lineage: The Case of TEL Genes. J. Exp. Bot. 2012, 63, 5061–5077. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Software for Systematics and Evolution MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.89). Available online: https://github.com/taiyun/corrplot (accessed on 10 December 2021).
- R Core Team. R: A Language and Environment for Statistical Computing 2021; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]

| Subfamily of 14-3-3 Protein | Species | Gene ID | Classification | Specific Expression Location and Conditions |
|---|---|---|---|---|
| Omega group | A. thaliana | AT4G09000 | Chi (GRF (1) | Nonhair root epidermal cell |
| G. max | Glyma.14G176900 | Omega B.2 | Shoot system; drought environment | |
| Psi group | S. bicolor | Sobic.005G145200 | Psi | Anther |
| Kappa/lambda group | A.thaliana | AT5G10450 | Lambda (GRF 6) | Whole plant |
| G. max | Glyma.04G092600 | Lambda 1 | Mature leaves in full sunlight | |
| M. truncatula | Medtr3g100620.1 | Lambda 3 | Flower, lamina | |
| Epsilon group | S. bicolor | Sobic.005G183200 | Epsilon | Root; abscisic acid treatment |
| Iota Group | S. lycopersicon | Solyc05g012420 | Iota B | Flower bud |
| G. max | Glyma.20G025900 | Iota A | Flower | |
| G. max | Glyma.02G115900 | Iota B.2 | Mature leaves in full sunlight | |
| Mu group | G. max | Glyma.05G158100 | Mu A.1 | Root noduleplant embryo axis |
| G. max | Glyma.12G229200 | Mu B.1 | Inner integument | |
| G. max | Glyma.13G270600 | Mu B.2 | Seed coat |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikhaylova, Y.V.; Puzanskiy, R.K.; Shishova, M.F. Evolution of 14-3-3 Proteins in Angiosperm Plants: Recurring Gene Duplication and Loss. Plants 2021, 10, 2724. https://doi.org/10.3390/plants10122724
Mikhaylova YV, Puzanskiy RK, Shishova MF. Evolution of 14-3-3 Proteins in Angiosperm Plants: Recurring Gene Duplication and Loss. Plants. 2021; 10(12):2724. https://doi.org/10.3390/plants10122724
Chicago/Turabian StyleMikhaylova, Yulia V., Roman K. Puzanskiy, and Maria F. Shishova. 2021. "Evolution of 14-3-3 Proteins in Angiosperm Plants: Recurring Gene Duplication and Loss" Plants 10, no. 12: 2724. https://doi.org/10.3390/plants10122724
APA StyleMikhaylova, Y. V., Puzanskiy, R. K., & Shishova, M. F. (2021). Evolution of 14-3-3 Proteins in Angiosperm Plants: Recurring Gene Duplication and Loss. Plants, 10(12), 2724. https://doi.org/10.3390/plants10122724