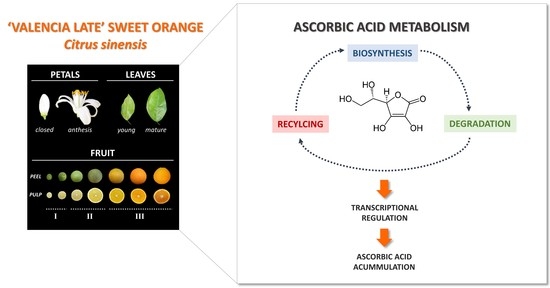
Ascorbic Acid Content and Transcriptional Profiling of Genes Involved in Its Metabolism during Development of Petals, Leaves, and Fruits of Orange (Citrus sinensis cv. Valencia Late)
Abstract
1. Introduction
2. Results
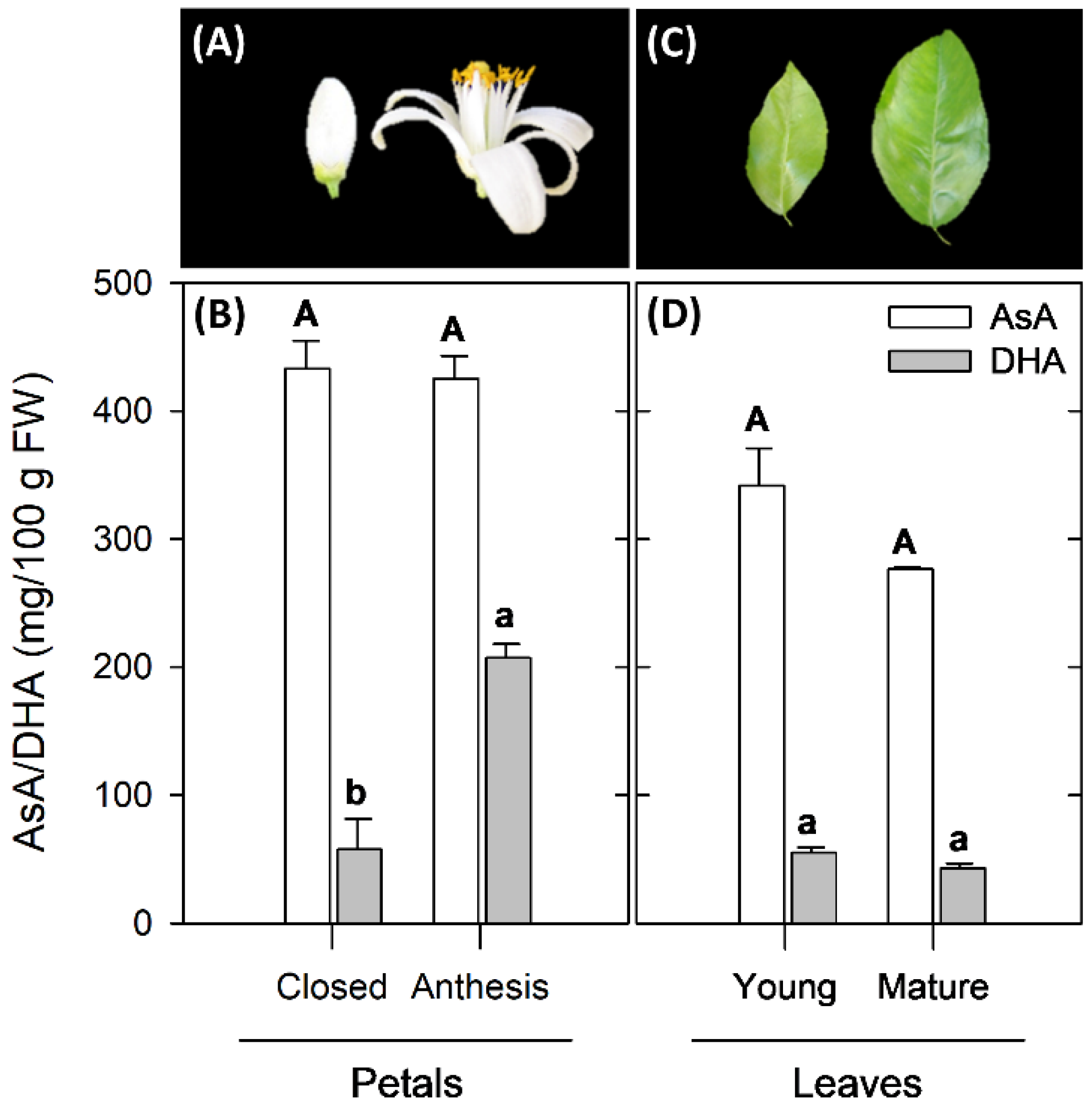
2.1. Changes in Ascorbic Acid Content during Development of Petals and Leaves of Valencia Late Oranges
2.2. Changes in Fruit Growth, Peel Coloration, and Ascorbic Acid Content in the Flavedo and Pulp during Fruit Development of Valencia Late Orange
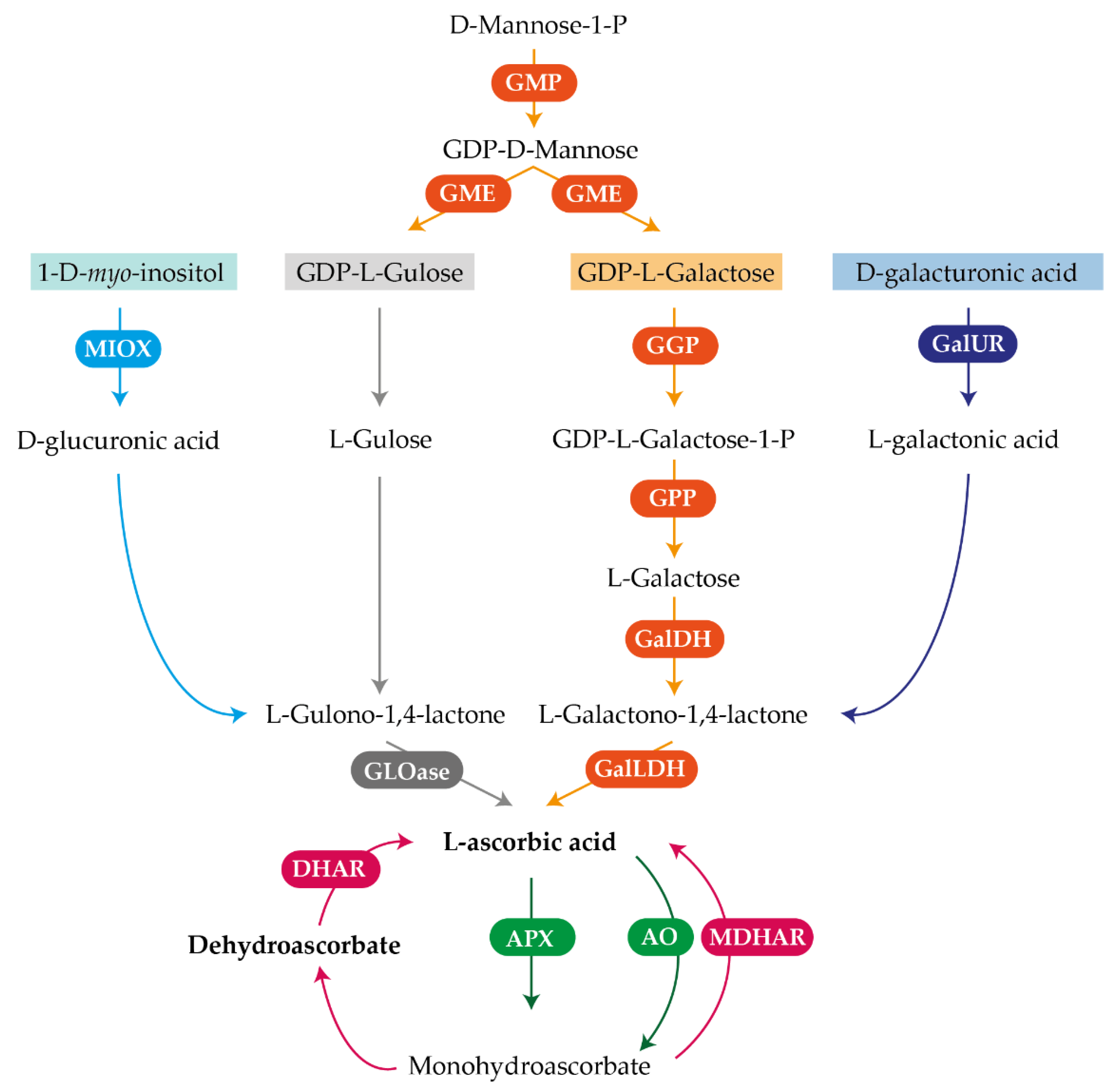
2.3. Transcriptional Analysis of Genes Involved in the AsA Metabolic Pathways
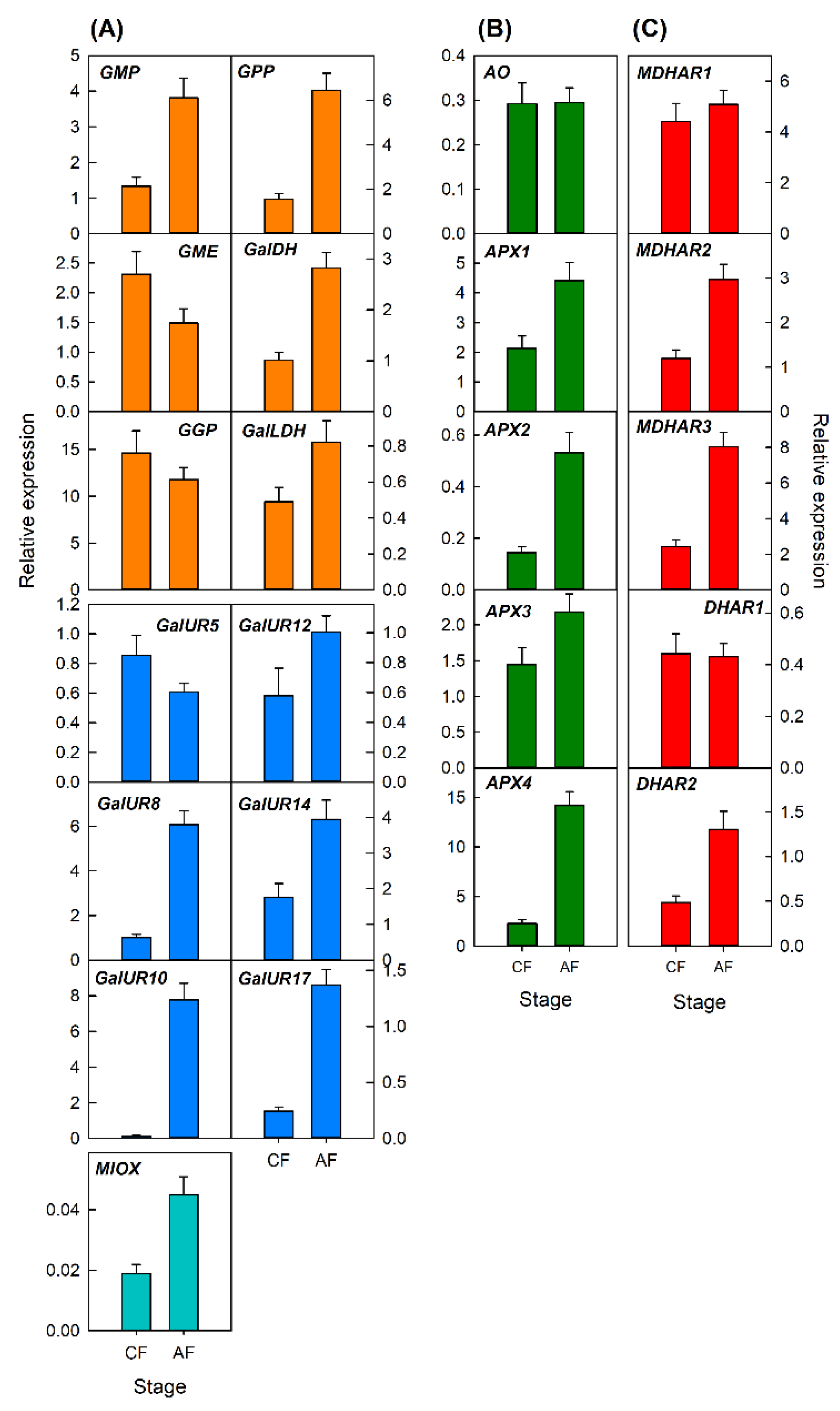
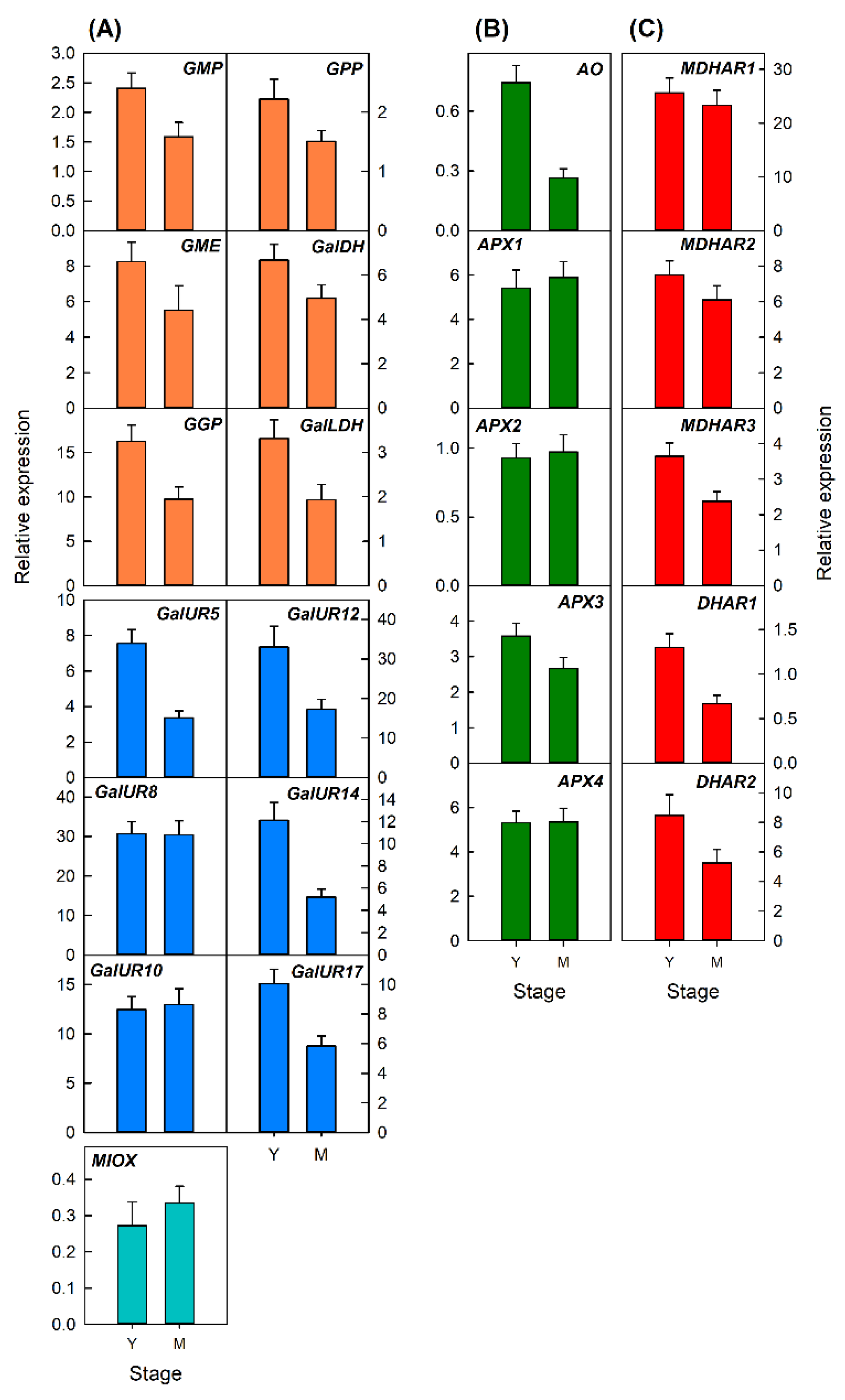
2.3.1. Transcriptional Analysis of Genes Involved in AsA Metabolism during Development of Petals and Leaves of Valencia Late Oranges
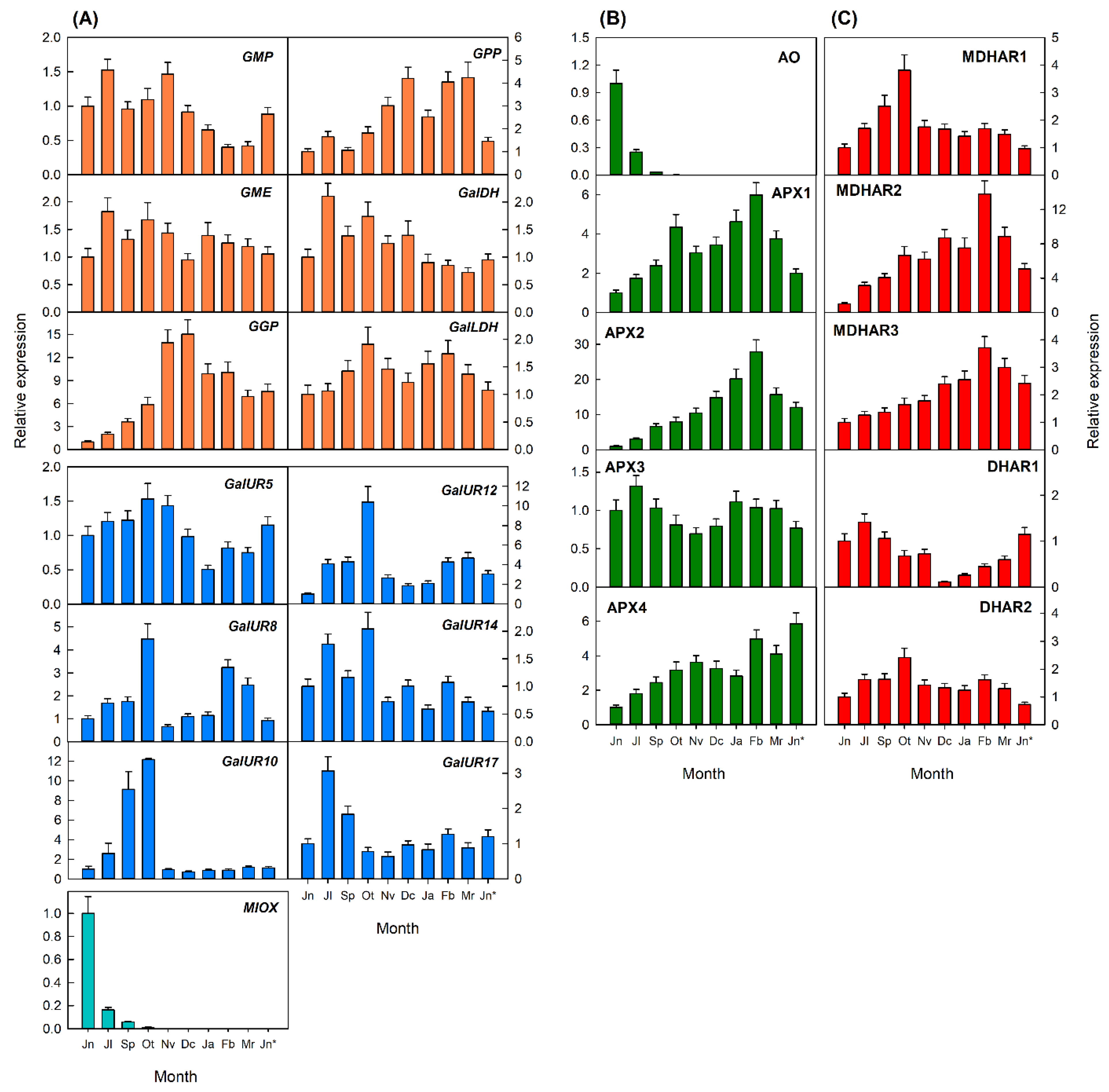
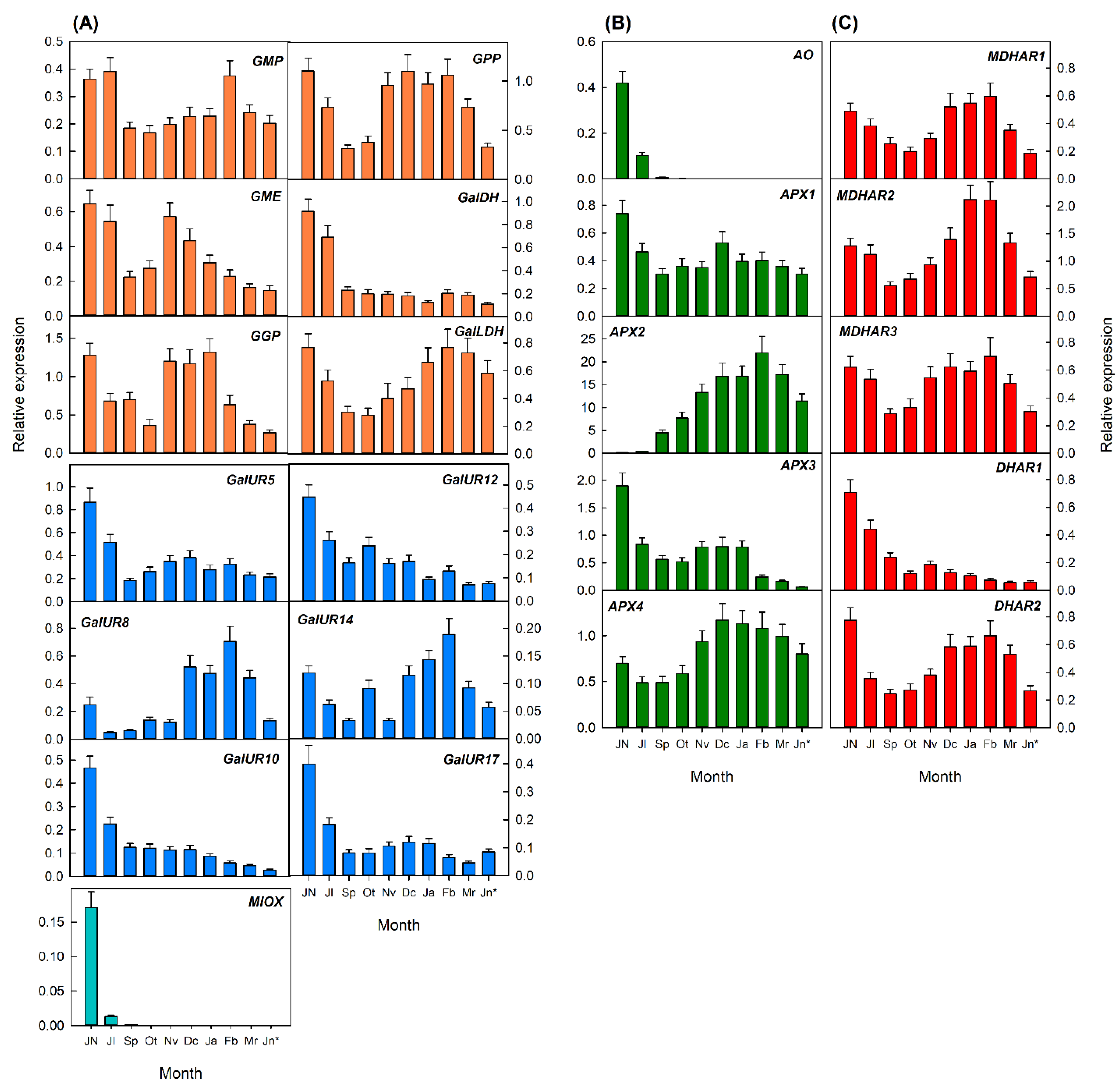
2.3.2. Transcriptional Analysis of Genes Involved in AsA Metabolism in Flavedo and Pulp during Fruit Development of Valencia Late Oranges
2.4. Co-Expression Networks of Genes Involved in AsA Metabolism in Flavedo and Pulp of Fruits
2.5. Cis-Acting Regulatory Elements Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Ascorbic Acid Extraction and Analysis by HPLC
4.3. RNA Extraction and cDNA Synthesis
4.4. Gene Expression Analysis by Real Time PCR
4.5. Search for Putative Transcription Factor Binding Sites in the Promoter Region of Genes Involved in AsA Metabolism
4.6. Co-Expression Gene Networks
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davey, M.W.; Van Montagu, M.; Inzé, D.; Sanmartin, M.; Kanellis, A.; Smirnoff, N.; Benzie, I.J.J.; Strain, J.J.; Favell, D.; Fletcher, J. Plant L-ascorbic acid: Chemistry, function, metabolism, bioavailability and effects of processing. J. Sci. Food Agric. 2000, 80, 825–860. [Google Scholar] [CrossRef]
- Grosso, G.; Bei, R.; Mistretta, A.; Marventano, S.; Calabrese, G.; Masuelli, L.; Giganti, M.G.; Modesti, A.; Galvano, F.; Gazzolo, D. Effects of Vitamin C on health: A review of evidence. Front. Biosci. 2013, 18, 1017–1029. [Google Scholar] [CrossRef]
- Mellidou, I.; Kanellis, A.K. Genetic Control of Ascorbic Acid Biosynthesis and Recycling in Horticultural Crops. Front. Chem. 2017, 5, 50. [Google Scholar] [CrossRef]
- Fenech, M.; Amaya, I.; Valpuesta, V.; Botella, M.A. Vitamin C Content in Fruits: Biosynthesis and Regulation. Front. Plant Sci. 2019, 9, 2006. [Google Scholar] [CrossRef]
- Mellidou, I.; Koukounaras, A.; Chatzopoulou, F.; Kostas, S.; Kanellis, A.K. Plant Vitamin C: One Single Molecule with a Plethora of Roles. In Fruit and Vegetable Phytochemicals: Chemistry and Human Health; Yahia, E.M., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2017; Volume 1, pp. 463–498. ISBN 9781119158042. [Google Scholar]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic Acid-A Potential Oxidant Scavenger and Its Role in Plant Development and Abiotic Stress Tolerance. Front. Plant Sci. 2017, 8, 613. [Google Scholar] [CrossRef] [PubMed]
- Mellidou, I.; Koukounaras, A.; Kostas, S.; Patelou, E.; Kanellis, A.K. Regulation of Vitamin C Accumulation for Improved Tomato Fruit Quality and Alleviation of Abiotic Stress. Genes 2021, 12, 694. [Google Scholar] [CrossRef]
- Hodges, D.M.; Lester, G.E.; Munro, K.D.; Toivonen, P.M.A. Oxidative stress: Importance for postharvest quality. HortScience 2004, 39, 924–929. [Google Scholar] [CrossRef]
- Mellidou, I.; Buts, K.; Hatoum, D.; Ho, Q.T.; Johnston, J.W.; Watkins, C.B.; Schaffer, R.J.; Gapper, N.E.; Giovannoni, J.J.; Rudell, D.R.; et al. Transcriptomic events associated with internal browning of apple during postharvest storage. BMC Plant Biol. 2014, 14, 328. [Google Scholar] [CrossRef] [PubMed]
- Cascia, G.; Bulley, S.M.; Punter, M.; Bowen, J.; Rassam, M.; Schotsmans, W.C.; Larrigaudière, C.; Johnston, J.W. Investigation of ascorbate metabolism during inducement of storage disorders in pear. Physiol. Plant. 2013, 147, 121–134. [Google Scholar] [CrossRef]
- Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radic. Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Ioannidi, E.; Kalamaki, M.S.; Engineer, C.; Pateraki, I.; Alexandrou, D.; Mellidou, I.; Giovannonni, J.; Kanellis, A.K. Expression profiling of ascorbic acid-related genes during tomato fruit development and ripening and in response to stress conditions. J. Exp. Bot. 2009, 60, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ma, F.; Zhang, M.; Pu, F. Distribution and metabolism of ascorbic acid in apple fruits (Malus domestica Borkh cv. Gala). Plant Sci. 2008, 174, 606–612. [Google Scholar] [CrossRef]
- Pignocchi, C.; Foyer, C.H. Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr. Opin. Plant Biol. 2003, 6, 379–389. [Google Scholar] [CrossRef]
- Gallie, D.R. The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Padayatty, S.J.; Levine, M. Vitamin C: The known and the unknown and Goldilocks. Oral Dis. 2016, 22, 463–493. [Google Scholar] [CrossRef] [PubMed]
- Martí, N.; Mena, P.; Cánovas, J.A.; Micol, V.; Saura, D. Vitamin C and the Role of Citrus Juices as Functional Food. Nat. Prod. Commun. 2009, 4, 677–700. [Google Scholar] [CrossRef]
- Bermejo, A.; Cano, A. Analysis of Nutritional Constituents in Twenty Citrus Cultivars from the Mediterranean Area at Different Stages of Ripening. Food Nutr. Sci. 2012, 03, 639–650. [Google Scholar] [CrossRef]
- Alós, E.; Rodrigo, M.J.; Zacarías, L. Differential transcriptional regulation of l-ascorbic acid content in peel and pulp of citrus fruits during development and maturation. Planta 2014, 239, 1113–1128. [Google Scholar] [CrossRef]
- Yang, X.-Y.; Xie, J.-X.; Wang, F.-F.; Zhong, J.; Liu, Y.-Z.; Li, G.-H.; Peng, S.-A. Comparison of ascorbate metabolism in fruits of two citrus species with obvious difference in ascorbate content in pulp. J. Plant Physiol. 2011, 168, 2196–2205. [Google Scholar] [CrossRef]
- Caruso, P.; Russo, M.P.; Caruso, M.; Di Guardo, M.; Russo, G.; Fabroni, S.; Timpanaro, N.; Licciardello, C. A Transcriptional Analysis of the Genes Involved in the Ascorbic Acid Pathways Based on a Comparison of the Juice and Leaves of Navel and Anthocyanin-Rich Sweet Orange Varieties. Plants 2021, 10, 1291. [Google Scholar] [CrossRef]
- Magwaza, L.S.; Mditshwa, A.; Tesfay, S.Z.; Opara, U.L. An overview of preharvest factors affecting vitamin C content of citrus fruit. Sci. Hortic. 2017, 216, 12–21. [Google Scholar] [CrossRef]
- Lado, J.; Alós, E.; Rodrigo, M.J.; Zacarías, L. Light avoidance reduces ascorbic acid accumulation in the peel of Citrus fruit. Plant Sci. 2015, 231, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Arbona, V.; Flors, V.; Jacas, J.; García-Agustín, P.; Gómez-Cadenas, A. Enzymatic and Non-enzymatic Antioxidant Responses of Carrizo citrange, a Salt-Sensitive Citrus Rootstock, to Different Levels of Salinity. Plant Cell Physiol. 2003, 44, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Rivas, F.; Gravina, A.; Agustí, M. Girdling effects on fruit set and quantum yield efficiency of PSII in two Citrus cultivars. Tree Physiol. 2007, 27, 527–535. [Google Scholar] [CrossRef] [PubMed]
- De Campos, M.K.F.; de Carvalho, K.; de Souza, F.S.; Marur, C.J.; Pereira, L.F.P.; Filho, J.C.B.; Vieira, L.G.E. Drought tolerance and antioxidant enzymatic activity in transgenic ‘Swingle’ citrumelo plants over-accumulating proline. Environ. Exp. Bot. 2011, 72, 242–250. [Google Scholar] [CrossRef]
- Seday, U.; Gulsen, O.; Uzun, A.; Toprak, G. Response of citrus rootstocks to different salinity levels for morphological and antioxidative enzyme activites. J. Anim. Plant Sci. 2014, 24, 512–520. [Google Scholar]
- Kostopoulou, Z.; Therios, I.; Roumeliotis, E.; Kanellis, A.K.; Molassiotis, A. Melatonin combined with ascorbic acid provides salt adaptation in Citrus aurantium L. seedlings. Plant Physiol. Biochem. 2015, 86, 155–165. [Google Scholar] [CrossRef]
- Strobbe, S.; De Lepeleire, J.; Van Der Straeten, D. From in planta Function to Vitamin-Rich Food Crops: The ACE of Biofortification. Front. Plant Sci. 2018, 9, 1862. [Google Scholar] [CrossRef]
- Broun, P. Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 2004, 7, 202–209. [Google Scholar] [CrossRef]
- Capell, T.; Christou, P. Progress in plant metabolic engineering. Curr. Opin. Biotechnol. 2004, 15, 148–154. [Google Scholar] [CrossRef]
- Hu, T.; Ye, J.; Tao, P.; Li, H.; Zhang, J.; Zhang, Y.; Ye, Z. The tomato HD-Zip I transcription factor SlHZ24 modulates ascorbate accumulation through positive regulation of the D-mannose/L-galactose pathway. Plant J. 2016, 85, 16–29. [Google Scholar] [CrossRef]
- Cano, A.; Medina, A.; Bermejo, A. Bioactive compounds in different citrus varieties. Discrimination among cultivars. J. Food Compos. Anal. 2008, 21, 377–381. [Google Scholar] [CrossRef]
- Bermejo, A.; Pardo, J.L.; Morales, J.; Cano, A. Comparative Study of Bioactive Components and Quality from Juices of Different Mandarins: Discriminant Multivariate Analysis of Their Primary and Secondary Metabolites. Agric. Sci. 2016, 07, 341–351. [Google Scholar] [CrossRef][Green Version]
- Dhuique-Mayer, C.; Caris-Veyrat, C.; Ollitrault, P.; Curk, F.; Amiot, M.-J. Varietal and Interspecific Influence on Micronutrient Contents in Citrus from the Mediterranean Area. J. Agric. Food Chem. 2005, 53, 2140–2145. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Chen, L.-L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.-B.; Hao, B.-H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Arrom, L.; Munné-Bosch, S. Tocopherol composition in flower organs of Lilium and its variations during natural and artificial senescence. Plant Sci. 2010, 179, 289–295. [Google Scholar] [CrossRef]
- Rogers, H.J. Is there an important role for reactive oxygen species and redox regulation during floral senescence? Plant Cell Environ. 2012, 35, 217–233. [Google Scholar] [CrossRef]
- Li, M.; Ma, F.; Guo, C.; Liu, J. Ascorbic acid formation and profiling of genes expressed in its synthesis and recycling in apple leaves of different ages. Plant Physiol. Biochem. 2010, 48, 216–224. [Google Scholar] [CrossRef]
- Li, H.; Huang, W.; Wang, G.-L.; Wang, W.-L.; Cui, X.; Zhuang, J. Transcriptomic analysis of the biosynthesis, recycling, and distribution of ascorbic acid during leaf development in tea plant (Camellia sinensis (L.) O. Kuntze). Sci. Rep. 2017, 7, 46212. [Google Scholar] [CrossRef]
- Alquézar, B.; Rodrigo, M.J.; Lado, J.; Zacarías, L. A comparative physiological and transcriptional study of carotenoid biosynthesis in white and red grapefruit (Citrus paradisi Macf.). Tree Genet. Genomes 2013, 9, 1257–1269. [Google Scholar] [CrossRef]
- Assefa, A.D.; Saini, R.K.; Keum, Y.-S. Fatty acids, tocopherols, phenolic and antioxidant properties of six citrus fruit species: A comparative study. J. Food Meas. Charact. 2017, 11, 1665–1675. [Google Scholar] [CrossRef]
- Rey, F.; Zacarías, L.; Rodrigo, M.J. Regulation of Tocopherol Biosynthesis During Fruit Maturation of Different Citrus Species. Front. Plant Sci. 2021, 12, 743993. [Google Scholar] [CrossRef]
- Tadeo, F.R.; Terol, J.; Rodrigo, M.J.; Licciardello, C.; Sadka, A. Fruit growth and development. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Eds.; Woodhead Publishing-Elsevier: Sawston, UK, 2020; pp. 245–269. ISBN 9780128121634. [Google Scholar]
- Alós, E.; Rodrigo, M.J.; Zacarías, L. Transcriptomic analysis of genes involved in the biosynthesis, recycling and degradation of L-ascorbic acid in pepper fruits (Capsicum annuum L.). Plant Sci. 2013, 207, 2–11. [Google Scholar] [CrossRef]
- Imai, T.; Ban, Y.; Terakami, S.; Yamamoto, T.; Moriguchi, T. l-Ascorbate biosynthesis in peach: Cloning of six l-galactose pathway-related genes and their expression during peach fruit development. Physiol. Plant. 2009, 136, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, X.; Wang, P.; Ma, F. Ascorbic Acid Accumulation and Expression of Genes Involved in Its Biosynthesis and Recycling in Developing Apple Fruit. J. Am. Soc. Hortic. Sci. 2011, 136, 231–238. [Google Scholar] [CrossRef]
- Liang, D.; Zhu, T.; Ni, Z.; Lin, L.; Tang, Y.; Wang, Z.; Wang, X.; Wang, J.; Lv, X.; Xia, H. Ascorbic acid metabolism during sweet cherry (Prunus avium) fruit development. PLoS ONE 2017, 12, e0172818. [Google Scholar] [CrossRef]
- Wang, L.; Meng, X.; Yang, D.; Ma, N.; Wang, G.; Meng, Q. Overexpression of tomato GDP-l-galactose phosphorylase gene in tobacco improves tolerance to chilling stress. Plant Cell Rep. 2014, 33, 1441–1451. [Google Scholar] [CrossRef]
- Mellidou, I.; Chagné, D.; Laing, W.A.; Keulemans, J.; Davey, M.W. Allelic Variation in Paralogs of GDP-l-Galactose Phosphorylase Is a Major Determinant of Vitamin C Concentrations in Apple Fruit. Plant Physiol. 2012, 160, 1613–1629. [Google Scholar] [CrossRef]
- Laing, W.A.; Martínez-Sánchez, M.; Wright, M.A.; Bulley, S.M.; Brewster, D.; Dare, A.P.; Rassam, M.; Wang, D.; Storey, R.; Macknight, R.C.; et al. An Upstream Open Reading Frame Is Essential for Feedback Regulation of Ascorbate Biosynthesis in Arabidopsis. Plant Cell 2015, 27, 772–786. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef]
- Ayadi, M.; Delaporte, V.; Li, Y.-F.; Zhou, D.-X. Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis. FEBS Lett. 2004, 562, 147–154. [Google Scholar] [CrossRef]
- Foyer, C.H.; Kerchev, P.I.; Hancock, R.D. The ABA-INSENSITIVE-4 (ABI4) transcription factor links redox, hormone and sugar signaling pathways. Plant Signal. Behav. 2012, 7, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Heiber, I.; Cai, W.; Baier, M. Linking Chloroplast Antioxidant Defense to Carbohydrate Availability: The Transcript Abundance of Stromal Ascorbate Peroxidase Is Sugar-Controlled via Ascorbate Biosynthesis. Mol. Plant 2014, 7, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, J.; Li, S.; Kakan, X.; Zhou, Y.; Miao, Y.; Wang, F.; Qin, H.; Huang, R. Ascorbic Acid Integrates the Antagonistic Modulation of Ethylene and Abscisic Acid in the Accumulation of Reactive Oxygen Species. Plant Physiol. 2019, 179, 1861–1875. [Google Scholar] [CrossRef]
- Guo, W.L.; Chen, R.G.; Gong, Z.H.; Yin, Y.X.; Ahmed, S.S.; He, Y.M. Exogenous abscisic acid increases antioxidant enzymes and related gene expression in pepper (Capsicum annuum) leaves subjected to chilling stress. Genet. Mol. Res. 2012, 11, 4063–4080. [Google Scholar] [CrossRef]
- Rossel, J.B.; Walter, P.B.; Hendrickson, L.; Chow, W.S.; Poole, A.; Mullineaux, P.M.; Pogson, B.J. A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 2006, 29, 269–281. [Google Scholar] [CrossRef]
- Miret, J.A.; Munné-Bosch, S. Abscisic acid and pyrabactin improve vitamin C contents in raspberries. Food Chem. 2016, 203, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, L.; Luo, Z.; Mou, W.; Mao, L.; Ying, T. Comparative Transcriptome Analysis Reveals the Influence of Abscisic Acid on the Metabolism of Pigments, Ascorbic Acid and Folic Acid during Strawberry Fruit Ripening. PLoS ONE 2015, 10, e0130037. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Marcos, J.F.; Alférez, F.; Mallent, M.D.; Zacarías, L. Characterization of Pinalate, a novel Citrus sinensis mutant with a fruit-specific alteration that results in yellow pigmentation and decreased ABA content. J. Exp. Bot. 2003, 54, 727–738. [Google Scholar] [CrossRef]
- Pastori, G.M.; Kiddle, G.; Antoniw, J.; Bernard, S.; Veljovic-Jovanovic, S.; Verrier, P.J.; Noctor, G.; Foyer, C.H. Leaf Vitamin C Contents Modulate Plant Defense Transcripts and Regulate Genes That Control Development through Hormone Signaling. Plant Cell 2003, 15, 939–951. [Google Scholar] [CrossRef]
- Attolico, A.D.; De Tullio, M.C. Increased ascorbate content delays flowering in long-day grown Arabidopsis thaliana (L.) Heynh. Plant Physiol. Biochem. 2006, 44, 462–466. [Google Scholar] [CrossRef]
- Ye, J.; Hu, T.; Yang, C.; Li, H.; Yang, M.; Ijaz, R.; Ye, Z.; Zhang, Y. Transcriptome Profiling of Tomato Fruit Development Reveals Transcription Factors Associated with Ascorbic Acid, Carotenoid and Flavonoid Biosynthesis. PLoS ONE 2015, 10, e0130885. [Google Scholar] [CrossRef]
- Stewart, I.; Wheaton, T.A. Carotenoids in citrus. Their accumulation induced by ethylene. J. Agric. Food Chem. 1972, 20, 448–449. [Google Scholar] [CrossRef]
- Rodrigo, M.-J.; Marcos, J.F.; Zacarías, L. Biochemical and Molecular Analysis of Carotenoid Biosynthesis in Flavedo of Orange (Citrus sinensis L.) during Fruit Development and Maturation. J. Agric. Food Chem. 2004, 52, 6724–6731. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Quandt, K.; Frech, K.; Karas, H.; Wingender, E.; Werner, T. Matlnd and Matlnspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995, 23, 4878–4884. [Google Scholar] [CrossRef]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alós, E.; Rey, F.; Gil, J.V.; Rodrigo, M.J.; Zacarias, L. Ascorbic Acid Content and Transcriptional Profiling of Genes Involved in Its Metabolism during Development of Petals, Leaves, and Fruits of Orange (Citrus sinensis cv. Valencia Late). Plants 2021, 10, 2590. https://doi.org/10.3390/plants10122590
Alós E, Rey F, Gil JV, Rodrigo MJ, Zacarias L. Ascorbic Acid Content and Transcriptional Profiling of Genes Involved in Its Metabolism during Development of Petals, Leaves, and Fruits of Orange (Citrus sinensis cv. Valencia Late). Plants. 2021; 10(12):2590. https://doi.org/10.3390/plants10122590
Chicago/Turabian StyleAlós, Enriqueta, Florencia Rey, José Vicente Gil, María Jesús Rodrigo, and Lorenzo Zacarias. 2021. "Ascorbic Acid Content and Transcriptional Profiling of Genes Involved in Its Metabolism during Development of Petals, Leaves, and Fruits of Orange (Citrus sinensis cv. Valencia Late)" Plants 10, no. 12: 2590. https://doi.org/10.3390/plants10122590
APA StyleAlós, E., Rey, F., Gil, J. V., Rodrigo, M. J., & Zacarias, L. (2021). Ascorbic Acid Content and Transcriptional Profiling of Genes Involved in Its Metabolism during Development of Petals, Leaves, and Fruits of Orange (Citrus sinensis cv. Valencia Late). Plants, 10(12), 2590. https://doi.org/10.3390/plants10122590