Exogenous Myo-Inositol Alleviates Salt Stress by Enhancing Antioxidants and Membrane Stability via the Upregulation of Stress Responsive Genes in Chenopodium quinoa L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup and Treatment
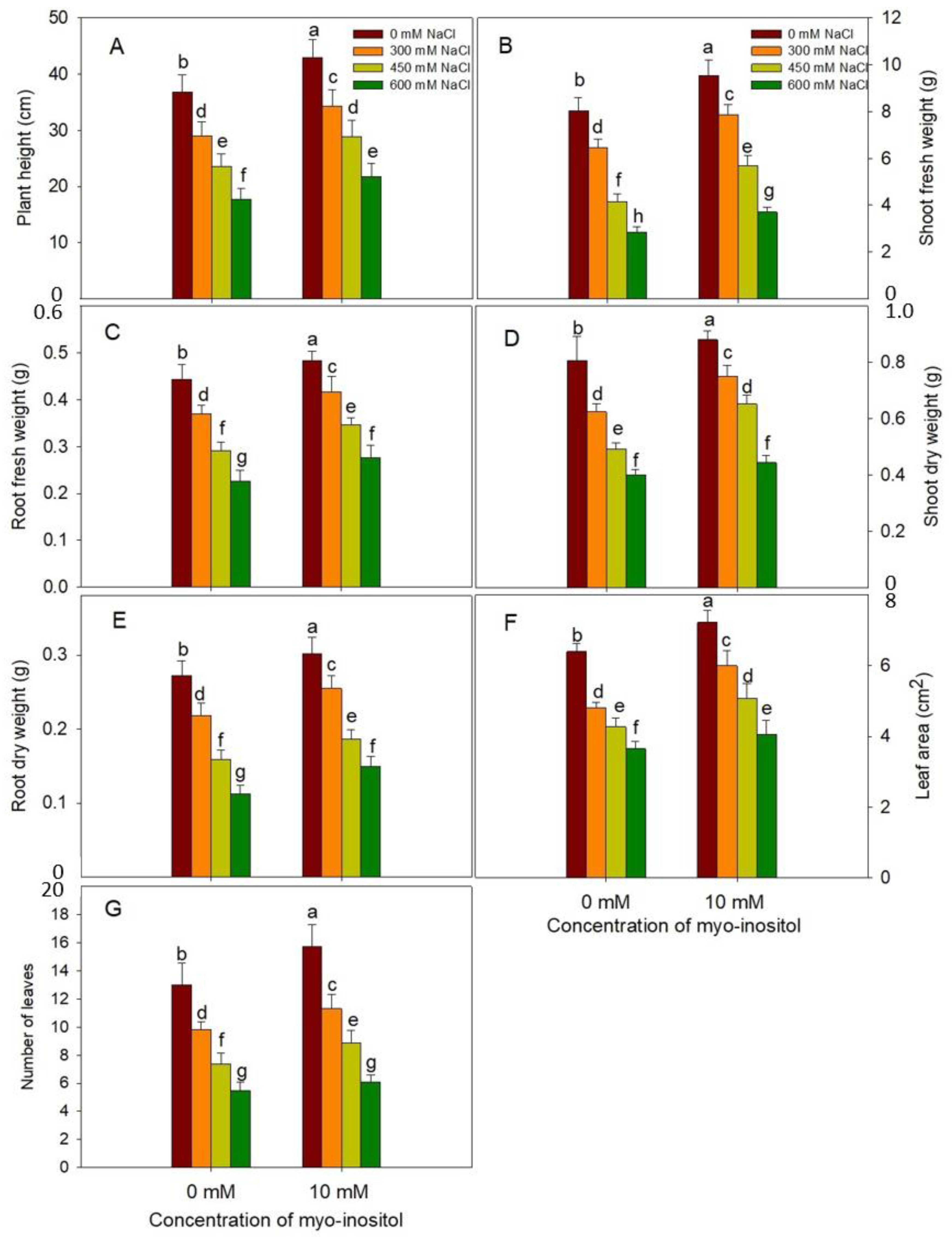
2.2. Growth Measurements
2.3. Measurement of Photosynthetic Pigments, Gas Exchange Parameters, and PSII Activity
2.4. Estimation of Stress Biomarkers
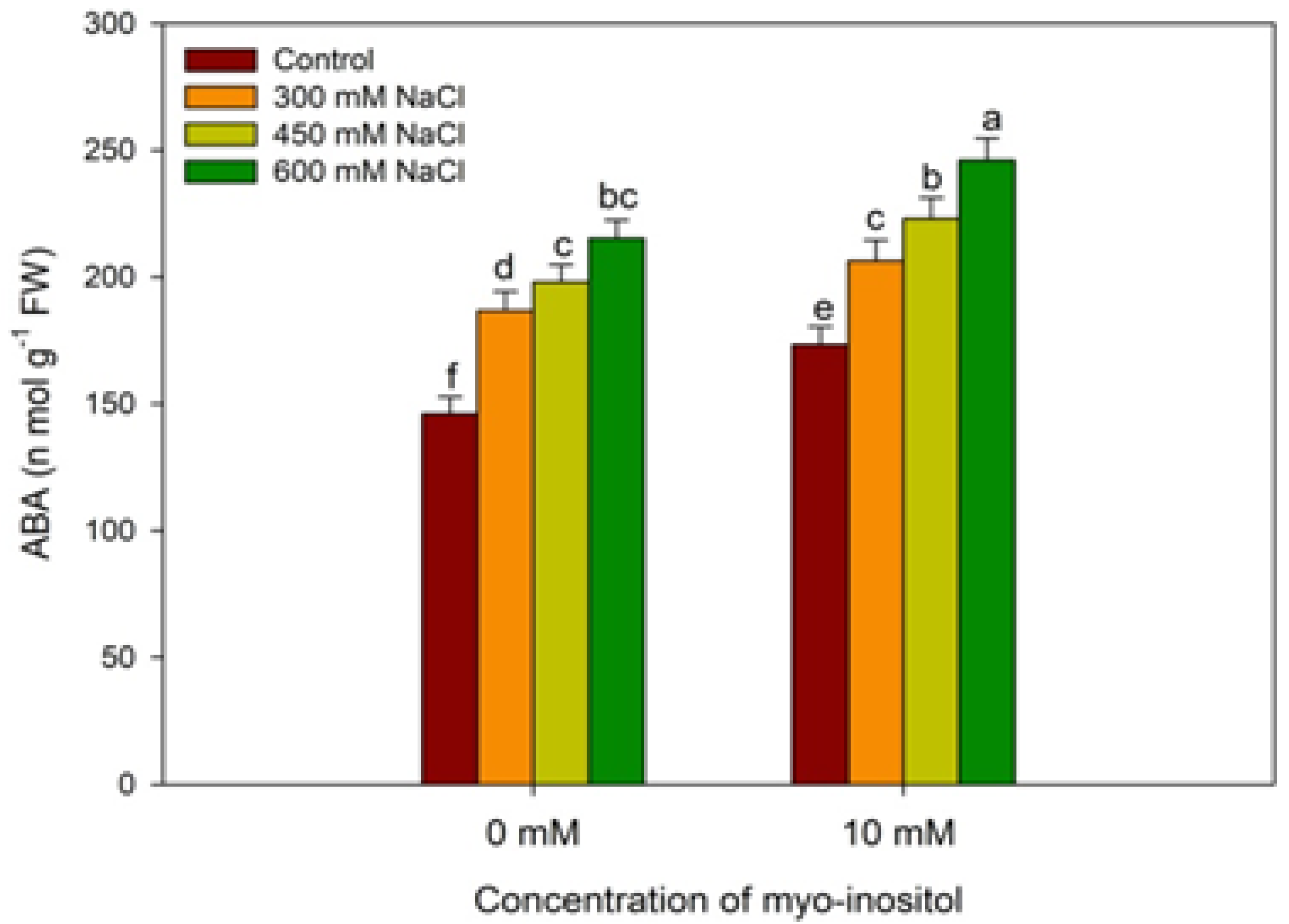
2.5. Estimation of Abscisic Acid
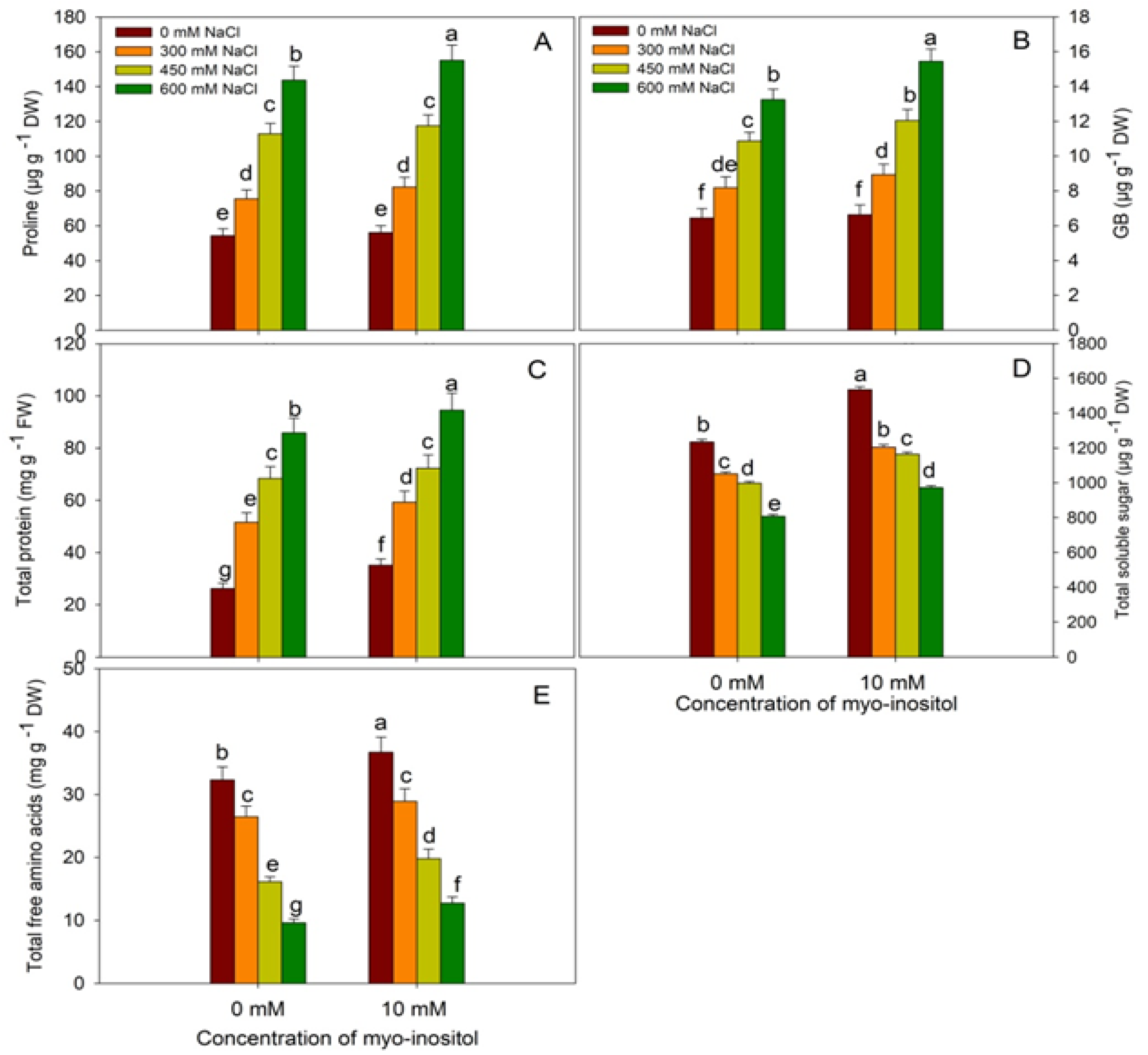
2.6. Estimation of Osmolytes
2.7. Measurements of RWC and LWP
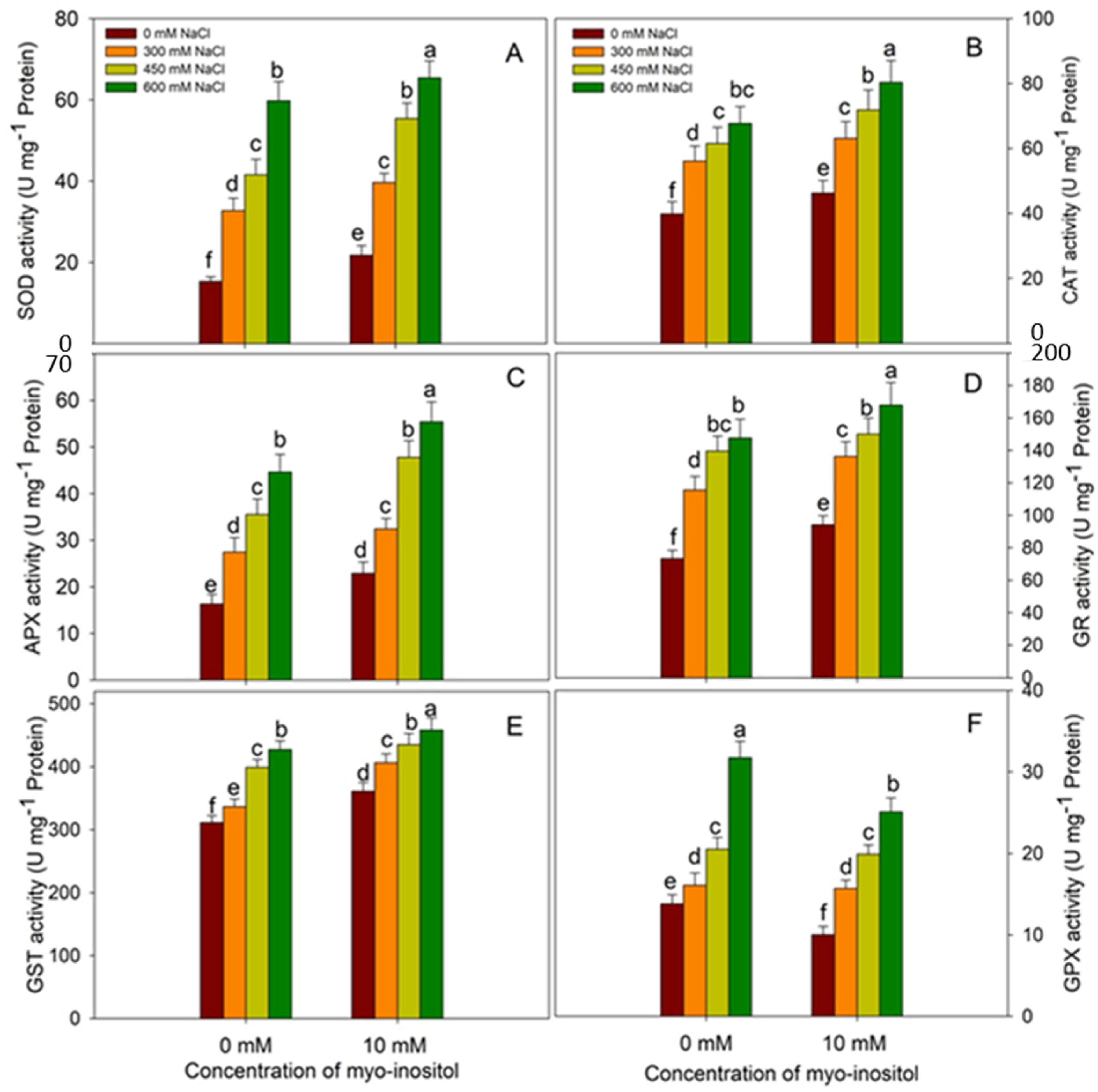
2.8. Assay of Antioxidant Enzymes
2.9. Estimation of Non-Enzymatic Antioxidants
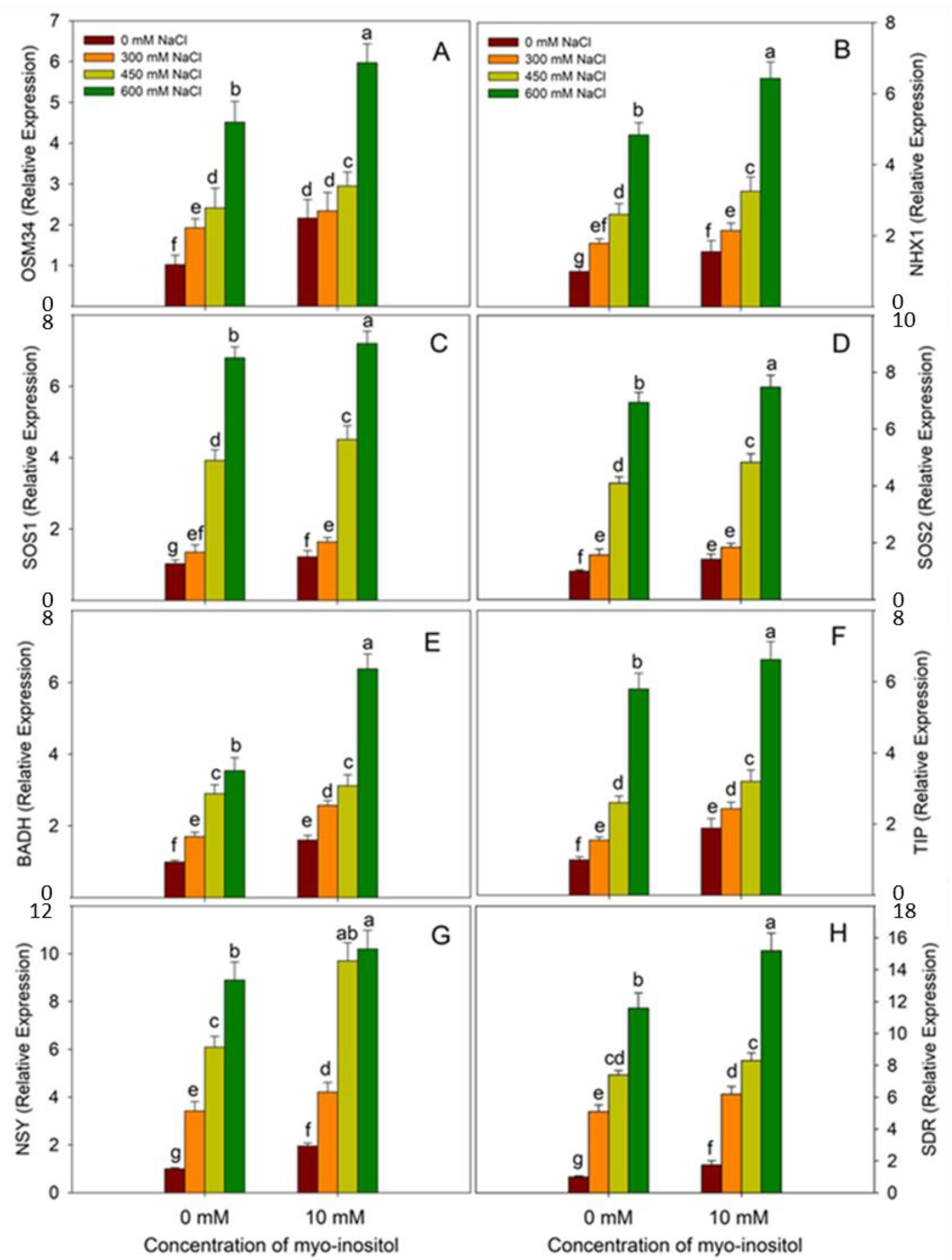
2.10. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
2.11. Statistical Analysis
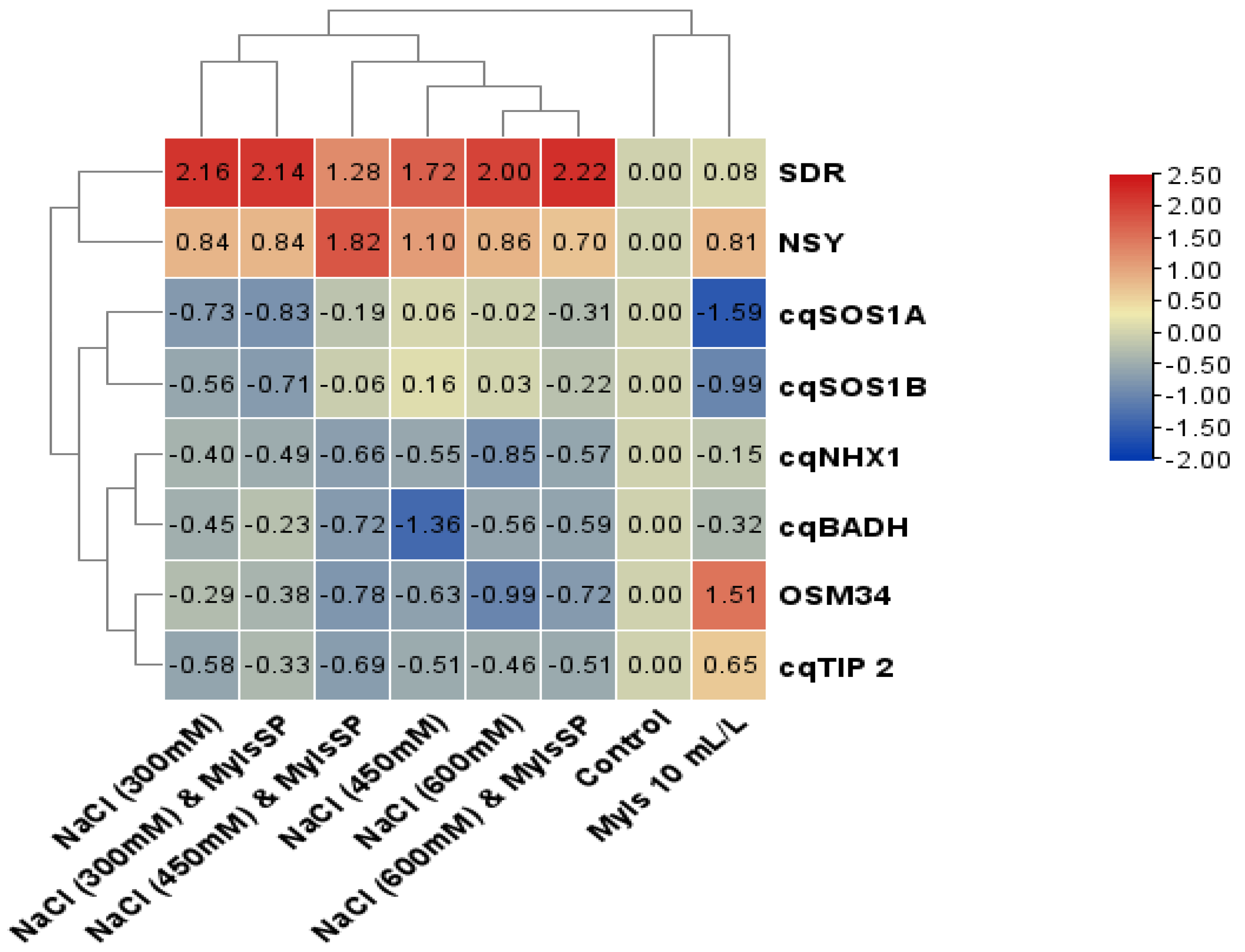
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zahra, N.; Raza, Z.A.; Mahmood, S. Effect of Salinity Stress on Various Growth and Physiological Attributes of Two Contrasting Maize Genotypes. Braz. Arc. Biol. Technol. 2020, 63, 2020. [Google Scholar] [CrossRef]
- Ali, M.M.; Jeddi, K.; Attia, M.S.; Elsayed, S.M.; Yusuf, M.; Osman, M.S.; Soliman, M.H.; Hessini, K. Wuxal amino (Bio stimulant) improved growth and physiological performance of tomato plants under salinity stress through adaptive mechanisms and antioxidant potential. Saudi J. Biol. Sci. 2021, 28, 3204–3213. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075. [Google Scholar] [CrossRef]
- Fatma, M.; Masood, A.; Per, T.S.; Khan, N.A. Nitric oxide alleviates salt stress inhibited photosynthetic performance by interacting with sulfur assimilation in mustard. Front. Plant Sci. 2016, 7, 521. [Google Scholar] [CrossRef] [Green Version]
- Fariduddin, Q.; Zaid, A.; Mohammad, F. Plant Growth Regulators and Salt Stress: Mechanism of Tolerance Trade-Off. In Salt Stress, Microbes, and Plant Interactions: Causes and Solution; Akhtar, M., Ed.; Springer: Singapore, 2019. [Google Scholar] [CrossRef]
- Elkelish, A.A.; Soliman, M.H.; Alhaithloul, H.A.; El-Esawi, M.A. Selenium protects wheat seedlings against salt stress-mediated oxidative damage by up-regulating antioxidants and osmolytes metabolism. Plant Physiol. Biochem. 2019, 137, 144–153. [Google Scholar] [CrossRef] [PubMed]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef] [Green Version]
- Omidbakhshfard, M.A.; Omranian, N.; Ahmadi, F.S.; Nikoloski, Z.; Mueller-Roeber, B. Effect of salt stress on genes encoding translation-associated proteins in Arabidopsis thaliana. Plant Signal. Behav. 2012, 7, 1095–1102. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Liu, S.; Lu, Z.; Pang, S.; Wang, L.; Wang, L.; Li, W. Gene expression profiles and flavonoid accumulation during salt stress in Ginkgo biloba seedlings. Plants 2020, 9, 1162. [Google Scholar] [CrossRef]
- Jogawat, A. Osmolytes and their role in abiotic stress tolerance in plants. Mol. Plant Abiotic Stress Biol. Biotechnol. 2019, 91–104. [Google Scholar] [CrossRef]
- Zaid, A.; Wani, S.H. Reactive Oxygen Species Generation, Scavenging and Signaling in Plant Defense Responses. In Bioactive Molecules in Plant Defense; Jogaiah, S., Abdelrahman, M., Eds.; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Islam, S.; Zaid, A.; Mohammad, F. Role of Triacontanol in Counteracting the Ill Effects of Salinity in Plants: A Review. J. Plant Growth Regul. 2021, 40, 1–10. [Google Scholar] [CrossRef]
- Volkov, V.; Beilby, M.J. Editorial: Salinity tolerance in plants: Mechanisms and regulation of ion transport. Front. Plant Sci. 2017, 8, 1795. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Ali, M.A.; Soliman, M.H.; Alqarawi, A.A.; Abd_Allah, E.F.; Fang, X.-W. Insights into 28-homobrassinolide (HBR)-mediated redox homeostasis, AsA–GSH cycle, and methylglyoxal detoxification in soybean under drought-induced oxidative stress. J. Plant Interact. 2020, 15, 371–385. [Google Scholar] [CrossRef]
- Patel, M.K.; Kumar, M.; Li, W.; Luo, Y.; Burritt, D.J.; Alkan, N.; Tran, L.S.P. Enhancing salt tolerance of plants: From metabolic reprogramming to exogenous chemical treatments and molecular approaches. Cells 2020, 9, 2492. [Google Scholar] [CrossRef] [PubMed]
- Abdel Razik, E.S.; Alharbi, B.M.; Pirzadah, T.B.; Alnusairi, G.S.H.; Soliman, M.H.; Hakeem, K.R. γ-Aminobutyric acid (GABA) mitigates drought and heat stress in sunflower (Helianthusannuus L.) by regulating its physiological, biochemical and molecular pathways. Physiol. Plant. 2020, 172, 505–527. [Google Scholar] [CrossRef]
- Michell, R.H. Inositol derivatives: Evolution and functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Case, K.C.; Salsaa, M.; Yu, W.; Greenberg, M.L. Regulation of inositol biosynthesis: Balancing health and pathophysiology. In Lipid Signalling in Human Diseases; Gomez-Cambronero, J., Frohman, M.A., Eds.; Springer: Cham, Switzerland, 2018; Volume 259, pp. 221–260. [Google Scholar]
- Eckardt, N.A. Myo-inositol biosynthesis genes in Arabidopsis: Differential patterns of gene expression and role in cell death. Plant Cell 2010, 22, 537. [Google Scholar] [CrossRef] [Green Version]
- Valluru, R.; Van den Ende, W. Myo-inositol and beyond—Emerging networks under stress. Plant Sci. 2011, 181, 387–400. [Google Scholar] [CrossRef]
- Jia, Q.; Kong, D.; Li, Q.; Sun, S.; Song, J.; Zhu, Y.; Liang, K.; Ke, Q.; Lin, W.; Huang, J. The function of inositol phosphatases in plant tolerance to abiotic stress. Int. J. Mol. Sci. 2019, 20, 3999. [Google Scholar] [CrossRef] [Green Version]
- Murphy, A.M.; Otto, B.; Brearley, C.A.; Carr, J.P.; Hanke, D.E. A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 2008, 56, 638–652. [Google Scholar] [CrossRef]
- Meng, P.H.; Raynaud, C.; Tcherkez, G.; Blanchet, S.; Massoud, K.; Domenichini, S.; Henry, Y.; Soubigou-Taconnat, L.; Lelarge-Trouverie, C.; Saindrenan, P.; et al. Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 2009, 4, e7364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Zhou, K.; Ren, G.; Yang, S.; Liu, Y.; Zhang, Z.; Li, Y.; Gong, X.; Ma, F. Myo-inositol mediates reactive oxygen species-induced programmed cell death via salicylic acid-dependent and ethylene-dependent pathways in apple. Hortic. Res. 2020, 7, 138. [Google Scholar] [CrossRef] [PubMed]
- Graf, B.L.; Rojas-Silva, P.; Rojo, L.E.; Delatorre-Herrera, J.; Baldeón, M.E.; Raskin, I. Innovations in Health Value and Functional Food Development of Quinoa (Chenopodium quinoa Willd.): Quinoa health value and functional food. Comp. Rev. Food Sci. Food Saf. 2015, 14, 431–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobsen, S.E.; Mujica, A.; Jensen, C.R. The resistance of quinoa (Chenopodium quinoa Willd.) to adverse abiotic factors. Food Rev. Int. 2003, 19, 99–109. [Google Scholar] [CrossRef]
- Koyro, H.W.; Lieth, H.; Eisa, S.S. Salt tolerance of Chenopodium quinoa wild, grains of the andes: Influence of salinity on biomass production, yield, composition of reserves in the seeds, water and solute relations. In Mangroves and Halophytes: Restoration and Utilisation. Tasks for Vegetation Sciences; Lieth, H., Sucre, M.G., Herzog, B., Eds.; Springer: Dordrecht, The Netherlands, 2008; Volume 43. [Google Scholar] [CrossRef]
- Maleki, P.; Bahrami, H.A.; Saadat, S.; Sharifi, F.; Dehghany, F.; Salehi, M. Salinity threshold value of quinoa (Chenopodium quinoa Willd.) at various growth stages and the appropriate irrigation method by saline water. Commun. Soil Sci. Plant Anal. 2018, 49, 1815–1825. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef] [Green Version]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Elstner, E.F.; Heupel, A. Inhibition of nitrite formation from hydroxyl ammonium-chloride: A simple assay for superoxide dismutase. Anal. Biochem. 1976, 70, 616–620. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Blum, A.; Ebercon, A. Cell membrane stability as a measure of drought and heat tolerance in wheat. Crop. Sci. 1981, 21, 43–47. [Google Scholar] [CrossRef]
- Sairam, R.K. Effect of moisture stress on physiological activities of two contrasting wheat genotypes. Indian J. Exp. Biol. 1994, 32, 584–593. [Google Scholar]
- Siciliano, I.; Amaral Carneiro, G.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Jasmonic acid, abscisic acid, and salicylic acid are involved in the phytoalexin responses of rice to Fusariumfujikuroi, a high gibberellin producer pathogen. J. Agric. Food Chem. 2015, 63, 8134–8142. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Grieve, C.M.; Grattan, S.R. Rapid assay for determination of water soluble quaternary ammonium compounds. Plant. Soil 1983, 70, 303. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Irigoyen, J.J.; Einerich, D.W.; Sanchez-Diaz, M. Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicagosativa) plants. Physiol. Plant. 1992, 84, 55–60. [Google Scholar] [CrossRef]
- Moore, S.; Stein, W.H. Photometric ninhydrin method for use in the chromatography of amino acids. J. Biol. Chem. 1948, 176, 367–388. [Google Scholar] [CrossRef]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Misra, H.; Fridovich, I. The Purification and Properties of Superoxide Dismutase from Neurosporacrassa. J. Bacteriol. 1972, 247, 3410. [Google Scholar]
- Aebi, H. Catalase in vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar]
- Nakano, Y.; Asada, K. Purification of ascorbate peroxidase in spinach chloroplast; its inactivation in ascorbate depleted medium and reactivation by monodehydroascorbate radical. Plant Cell Physiol. 1987, 28, 131–140. [Google Scholar]
- Smith, I.K.; Vierheller, T.L.; Thorne, C.A. Assay of glutathione reductase in crude tissue homogenates using 5, 50-dithiobis (2-nitrobenzoic acid). Anal. Biochem. 1998, 175, 408–413. [Google Scholar] [CrossRef]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione-S-transferase, the first enzymatic step in mercapturic acidformation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [CrossRef]
- Hossain, M.A.; Hasanuzzaman, M.; Fujita, M. Up-regulation of antioxidant and glyoxalase systems by exogenous glycine betaine and proline in mung bean confer tolerance to cadmium stress. Physiol. Mol. Biol. Plants 2010, 16, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Jagota, S.K.; Dani, H.M. A new colorimetric technique for the estimation of vitamin C using Folinphenolreagent. Anal. Biochem. 1982, 127, 178–182. [Google Scholar] [CrossRef]
- Anderson, M.E. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 1985, 113, 548–555. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCt Method. Methods 2001, 25, 4. [Google Scholar] [CrossRef]
- Wolny, E.; Skalska, A.; Braszewska, A.; Mur, L.A.J.; Hasterok, R. Defining the Cell Wall, Cell Cycle and Chromatin Landmarks in the Responses of Brachypodiumdistachyon to Salinity. Int. J. Mol. Sci. 2021, 22, 949. [Google Scholar] [CrossRef]
- Singh, D.; Roy, B.K. Salt stress affects mitotic activity and modulates antioxidant systems in onion roots. Braz. J. Bot. 2016, 39, 67–76. [Google Scholar] [CrossRef]
- Dell’Aversana, E.; Hessini, K.; Ferchichi, S.; Fusco, G.M.; Woodrow, P.; Ciarmiello, L.F.; Abdelly, C.; Carillo, P. Salinity Duration Differently Modulates Physiological Parameters and Metabolites Profile in Roots of Two Contrasting Barley Genotypes. Plants 2021, 10, 307. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Zhou, K.; Li, Y.; Chen, X.; Liu, B.; Li, C.; Gong, X.; Ma, F. Exogenous myo-inositol alleviates salinity-induced stress in Malushupehensis Rehd. Plant Physiol. Biochem. 2018, 133, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rosales, M.P.; Galvez, F.J.; Huertas, R.; Aranda, M.N.; Baghour, M.; Cagnac, O.; Venema, K. Plant NHX cation/proton antiporters. Plant Signal. Behav. 2009, 4, 265–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Ma, N.; Wang, C.; Fan, H.; Wang, M.; Zhang, J.; Cao, J.; Wang, D. A Golgi-Localized Sodium/Hydrogen Exchanger Positively Regulates Salt Tolerance by Maintaining Higher K+/Na+ Ratio in Soybean. Front. Plant Sci. 2021, 12, 638340. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Amin, I.; Rasool, S.; Mir, M.A.; Wani, W.; Masoodi, K.Z.; Ahmad, P. Ion homeostasis for salinity tolerance in plants: A molecular approach. Physiol. Plant. 2021, 171, 578–594. [Google Scholar] [CrossRef]
- Yang, Z.; Li, J.L.; Liu, L.N.; Xie, Q.; Sui, N. Photosynthetic regulation under salt stress and salt-tolerance mechanism of sweet sorghum. Front. Plant Sci. 2020, 10, 1722. [Google Scholar] [CrossRef]
- Sathee, L.; Sairam, R.K.; Chinnusamy, V.; Jha, S.K. Differential transcript abundance of salt overly sensitive (SOS) pathway genes is a determinant of salinity stress tolerance of wheat. Acta Physiol. Plant. 2015, 37, 169. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, L.; Wang, J.; Zhong, Y.; Cheng, Z.M. Isolation and expression analysis of Salt Overly Sensitive gene family in grapevine (Vitisvinifera) in response to salt and PEG stress. PLoS ONE 2019, 14, e0212666. [Google Scholar] [CrossRef] [Green Version]
- Bashir, M.A.; Silvestri, C.; Ahmad, T.; Hafiz, I.A.; Abbasi, N.A.; Manzoor, A.; Cristofori, V.; Rugini, E. Osmotin: A cationic protein leads to improve biotic and abiotic stress tolerance in plants. Plants 2020, 9, 992. [Google Scholar] [CrossRef]
- Goel, D.; Singh, A.K.; Yadav, V.; Babbar, S.B.; Bansal, K.C. Overexpression of osmotin gene confers tolerance to salt and drought stresses in transgenic tomato (Solanumlycopersicum L.). Protoplasma 2010, 245, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Maeshima, M. Tonoplast Transporters: Organization and function. Ann. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 469–497. [Google Scholar] [CrossRef]
- Liu, L.H.; Ludewig, U.; Gassert, B.; Frommer, W.B.; von Wirén, N. Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol. 2003, 133, 1220–1228. [Google Scholar] [CrossRef] [Green Version]
- Boursiac, Y.; Chen, S.; Luu, D.T.; Sorieul, M.; van den Dries, N.; Maurel, C. Early effects of salinity on water transport in Arabidopsis roots. molecular and cellular features of aquaporin expression. Plant Physiol. 2005, 139, 790–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamchoy, K.; Pumirat, P.; Reamtong, O.; Pakotiprapha, D.; Leartsakulpanich, U.; Boonyuen, U. Functional analysis of BPSS2242 reveals its detoxification role in Burkholderi apseudomallei under salt stress. Sci. Rep. 2020, 10, 10453. [Google Scholar] [CrossRef]
- Cheng, W.H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.C.; Arroyo, A.; Leon, P.; Nambara, A.; Asami, T.; Seo, M.; et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signalling and abscisic acid biosynthesis and functions. Plant Cell. 2002, 14, 2723–2743. [Google Scholar] [CrossRef]
- Bouvier, F.; D’harlingue, A.; Backhaus, R.A.; Kumagai, M.H.; Camara, B. Identification of neoxanthin synthase as a carotenoid cyclaseparalog. Eur. J. Biochem. 2000, 267, 6346–6352. [Google Scholar] [CrossRef] [Green Version]
- Neuman, H.; Galpaz, N., Jr.; Cunningham, F.X.; Zamir, D.; Hirschberg, J. The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis. Plant J. 2014, 78, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Giossi, C.; Cartaxana, P.; Cruz, S. Photoprotective Role of Neoxanthin in Plants and Algae. Molecules 2020, 25, 4617. [Google Scholar] [CrossRef]
- Kwon, O.K.; Mekapogu, M.; Kim, K.S. Effect of salinity stress on photosynthesis and related physiological responses in carnation (Dianthus caryophyllus). Hortic. Environ. Biotechnol. 2019, 60, 831–839. [Google Scholar] [CrossRef]
- Taibi, K.; Taibi, F.; Abderrahim, L.A.; Ennajah, A.; Belkhodja, M.; Mulet, J.M. Effect of salt stress on growth, chlorophyll content, lipid peroxidation and antioxidant defence systems in Phaseolus vulgaris L. S. Afr. J. Bot. 2016, 105, 306–312. [Google Scholar] [CrossRef]
- Najar, R.; Aydi, S.; Sassi-Aydi, S.; Zarai, A.; Abdelly, C. Effect of salt stress on photosynthesis and chlorophyll fluorescence in Medicagotruncatula. Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2018, 153, 88–97. [Google Scholar]
- Qin, C.; Ahanger, M.A.; Zhou, J.; Ahmed, N.; Wei, C.; Yuan, S.; Ashraf, M.; Zhang, L. Beneficial role of acetylcholine in chlorophyll metabolism and photosynthetic gas exchange in Nicotiana benthamiana seedlings under salinity stress. Plant Biol. 2020, 22, 357–365. [Google Scholar] [CrossRef]
- Sudhir, P.; Murthy, S. Effects of salt stress on basic processes of photosynthesis. Photosynthetica 2004, 42, 481–486. [Google Scholar] [CrossRef]
- Qin, C.; Ahanger, M.A.; Lin, B.; Huang, Z.; Zhou, J.; Ahmed, N.; Ai, S.; Mustafa, N.S.A.; Ashraf, M.; Zhang, L. Comparative transcriptomic analysis reveals the regulatory effects of acetylcholine on salt tolerance of Nicotiana benthamiana. Phytochemistry 2021, 181, 112582. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.N.; Ribeiro, R.V.; Ferreira-Silva, S.L.; Viegas, R.A.; Silveira, J.A.G. Comparative effects of salinity and water stress on photosynthesis, water relations and growth of Jatrophacurcas plants. J. Arid. Environ. 2010, 74, 1130–1137. [Google Scholar] [CrossRef]
- Turan, S.; Tripathy, B.C. Salt-stress induced modulation of chlorophyll biosynthesis during de-etiolation of rice seedlings. Physiol. Plant 2015, 153, 477–491. [Google Scholar] [CrossRef]
- Sarropoulou, V.; Dimassi-Theriou, K.; Therios, I. Effects of exogenous indole-3-butyric acid and myo-inositol on in vitro rooting, vegetative growth and biochemical changes in leaves and roots in the sweet cherry rootstock MxM 14 using shoot tip explants. Exp. Plant Physiol. 2015, 27, 191–201. [Google Scholar] [CrossRef]
- Pandit, P.R.; Fulekar, M.H.; Karuna, M.S.L. Effect of salinity stress on growth, lipid productivity, fatty acid composition, and biodiesel properties in Acutodesmusobliquus and Chlorella vulgaris. Environ. Sci. Poll. Res. 2017, 24, 13437–13451. [Google Scholar] [CrossRef]
- Begum, N.; Akhtar, K.; Ahanger, M.A.; Iqbal, M.; Wang, P.; Mustafa, N.S.; Zhang, L. Arbuscularmycorrhizal fungi improve growth, essential oil, secondary metabolism, and yield of tobacco (Nicotianatabacum L.) under drought stress conditions. Environ. Sci. Pollut. Res. 2021, 28, 45276–45295. [Google Scholar] [CrossRef]
- Munawar, W.; Hameed, A.; Khan, M.K.R. Differential Morphophysiological and Biochemical Responses of Cotton Genotypes Under Various Salinity Stress Levels During Early Growth Stage. Front. Plant Sci. 2021, 12, 622309. [Google Scholar] [CrossRef] [PubMed]
- Ahanger, M.A.; Mir, R.A.; Alyemeni, M.N.; Ahmad, P. Combined effects of brassinosteroid and kinetin mitigates salinity stress in tomato through the modulation of antioxidant and osmolyte metabolism. Plant Physiol. Biochem. 2020, 147, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Sarker, U.; Oba, S. The response of salinity stress-induced A. tricolor to growth, anatomy, physiology, non-enzymatic and enzymatic antioxidants. Front. Plant Sci. 2020, 11, 559876. [Google Scholar] [CrossRef] [PubMed]
- Dumanovic, J.; Nepovimova, E.; Natic, M.; Kuca, K.; Jacevic, V. The Significance of Reactive Oxygen Species and Antioxidant Defense System in Plants: A Concise Overview. Front. Plant Sci. 2021, 11, 552969. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fu, J.; Shi, D.; Peng, Y. Myo-inositol enhances drought tolerance in creeping bentgrass through alteration of osmotic adjustment, photosynthesis, and antioxidant defense. Crop. Sci. 2020, 60, 2149–2158. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Al-Mahmud, J.; Fujita, M. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [Green Version]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic acid-a potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant Sci. 2017, 8, 613. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Lisko, K.A.; Torres, R.; Harris, R.S.; Belisle, M.; Vaughan, M.M.; Jullian, B.; Chevone, B.I.; Mendes, P.; Nessler, C.L.; Lorence, A. Elevating vitamin C content via overexpression of myo-inositol oxygenase and l-gulono-1, 4-lactone oxidase in Arabidopsis leads to enhanced biomass and tolerance to abiotic stresses. Vitr. Cell Dev. Biol. Plant 2013, 49, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Tian, F.; Wang, W.; Liang, C.; Wang, X.; Wang, G.; Wang, W. Over accumulation of glycine betaine makes the function of the thylakoid membrane better in wheat under salt stress. Crop J. 2017, 5, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Hmidi, D.; Abdelly, C.; Athar, H.R.; Ashraf, M.; Messedi, D. Effect of salinity on osmotic adjustment, proline accumulation and possible role of ornithine-δ-aminotransferase in proline biosynthesis in Cakile maritime. Physiol. Mol. Biol. Plants 2018, 24, 1017–1033. [Google Scholar] [CrossRef] [PubMed]
- Giri, J. Glycine betaine and abiotic stress tolerance in plants. Plant Signal. Behav. 2011, 6, 1746–1751. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.; Divyanshu, K.; Kumar, S.; Swapnil, P.; Zehra, A.; Shukla, V.; Yadav, M.; Upadhyay, R.S. Regulation of L-proline biosynthesis, signal transduction, transport, accumulation and its vital role in plants during variable environmental conditions. Heliyon 2019, 5, e02952. [Google Scholar] [CrossRef] [Green Version]
- Rosa, M.; Prado, C.; Podazza, G.; Interdonato, R.; Gonzalez, J.A.; Hilal, M.; Prado, F.E. Soluble sugars—Metabolism, sensing and abiotic stress. Plant Signal. Behav. 2009, 4, 388–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Sun, Y.; Chen, S.; Jiang, J.; Chen, F.; Fang, W.; Liu, Z. The effect of exogenously applied nitric oxide on photosynthesis and antioxidant activity in heat stressed chrysanthemum. Biol. Plant 2011, 55, 737. [Google Scholar] [CrossRef]
- Khan, A.A.; Wang, T.; Hussain, T.; Amna Ali, F.; Shi, F.; Latef, A.A.H.A.; Ali, O.M.; Hayat, K.; Mehmood, S.; Zainab, N.; et al. Halotolerant-Koccuria rhizophila (14asp)-Induced Amendment of Salt Stress in Pea Plants by Limiting Na+ Uptake and Elevating Production of Antioxidants. Agronomy 2021, 11, 1907. [Google Scholar] [CrossRef]




| Gene Name | Primer Sequence (5′–3′) | |
|---|---|---|
| Osmotin-like protein (Osmotin-34) | F | GAACGGAGGGTGTCACAAAATC |
| R | CGTAGTGGGTCCACAAGTTCCT | |
| Tonoplast-localized Na+/H+ exchanger 1 (cqNHX1) | F | GCACTTCTGTTGCTGTGAGTTCCA |
| R | TGTGCCCTGACCTCGTAAACTGAT | |
| Salt overlay sensitive 1 (SOS1A) | F | CCTCATGATGCTTCCGACAA |
| R | CCGAGTCAAGTGCTTCATCA | |
| Salt overlay sensitive 1 (SOS1B) | F | ACCCTCATGATGCTTCTGATAC |
| R | TGCTTCATCAACTGATTGCAT | |
| Betaine aldehyde dehydrogenase (cqBADH) | F | GGTTACAGTCATTCAGACACCATCA |
| R | AACAAAGGGAGCCAAGCAGTT | |
| Tonoplast intrinsic protein 2 (TIP2) | F | AGTCCACCACCGATAAGAGGACCA |
| R | CCACATCCATGCAAATATGGAAAGAGGA | |
| Short-chain alcohol dehydrogenases/reductases (SDR) | F | CAATCTTGGCCAGCATCTCT |
| R | CCAGCTAACCCAGCATTGTT | |
| Neoxanthin synthase (NSY) | F | TTGTCTTGGACACCTGACACA |
| R | CTCCAGTCCGTCATGGAAAA | |
| β-Actin | F | GTGCCCATTTACGAAGGATA |
| R | GAAGACTCCATGCCGATCAT | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Mushhin, A.A.M.; Qari, S.H.; Fakhr, M.A.; Alnusairi, G.S.H.; Alnusaire, T.S.; ALrashidi, A.A.; Latef, A.A.H.A.; Ali, O.M.; Khan, A.A.; Soliman, M.H. Exogenous Myo-Inositol Alleviates Salt Stress by Enhancing Antioxidants and Membrane Stability via the Upregulation of Stress Responsive Genes in Chenopodium quinoa L. Plants 2021, 10, 2416. https://doi.org/10.3390/plants10112416
Al-Mushhin AAM, Qari SH, Fakhr MA, Alnusairi GSH, Alnusaire TS, ALrashidi AA, Latef AAHA, Ali OM, Khan AA, Soliman MH. Exogenous Myo-Inositol Alleviates Salt Stress by Enhancing Antioxidants and Membrane Stability via the Upregulation of Stress Responsive Genes in Chenopodium quinoa L. Plants. 2021; 10(11):2416. https://doi.org/10.3390/plants10112416
Chicago/Turabian StyleAl-Mushhin, Amina A. M., Sameer H. Qari, Marwa A. Fakhr, Ghalia S. H. Alnusairi, Taghreed S. Alnusaire, Ayshah Aysh ALrashidi, Arafat Abdel Hamed Abdel Latef, Omar M. Ali, Amir Abdullah Khan, and Mona H. Soliman. 2021. "Exogenous Myo-Inositol Alleviates Salt Stress by Enhancing Antioxidants and Membrane Stability via the Upregulation of Stress Responsive Genes in Chenopodium quinoa L." Plants 10, no. 11: 2416. https://doi.org/10.3390/plants10112416
APA StyleAl-Mushhin, A. A. M., Qari, S. H., Fakhr, M. A., Alnusairi, G. S. H., Alnusaire, T. S., ALrashidi, A. A., Latef, A. A. H. A., Ali, O. M., Khan, A. A., & Soliman, M. H. (2021). Exogenous Myo-Inositol Alleviates Salt Stress by Enhancing Antioxidants and Membrane Stability via the Upregulation of Stress Responsive Genes in Chenopodium quinoa L. Plants, 10(11), 2416. https://doi.org/10.3390/plants10112416









