Unique N-Terminal Interactions Connect F-BOX STRESS INDUCED (FBS) Proteins to a WD40 Repeat-like Protein Pathway in Arabidopsis
Abstract
:1. Introduction
2. Results
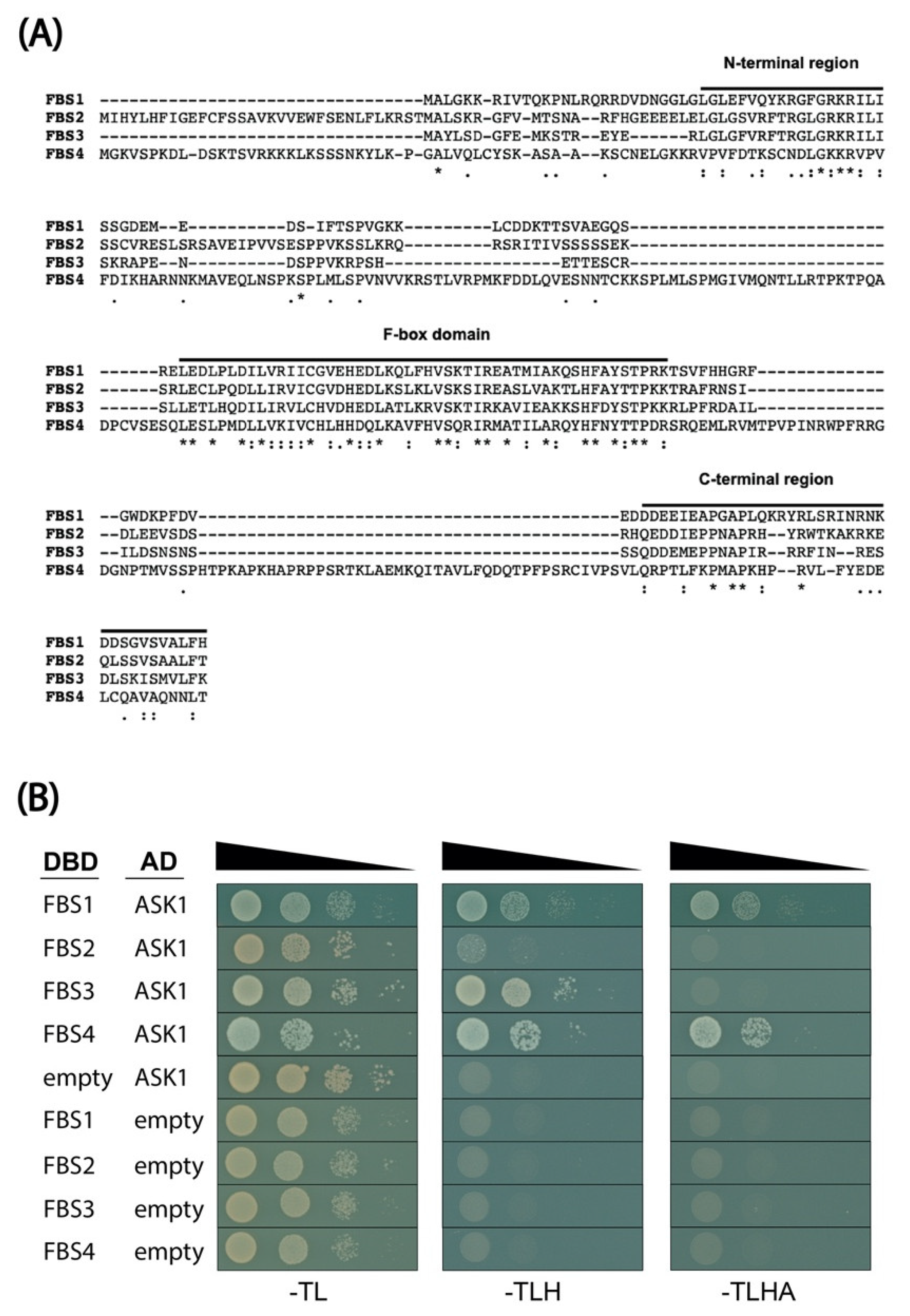
2.1. FBS Protein Interaction with ASK1
2.2. Identification of a New FBS1 Interactor
2.3. FBS Interactions with FBIPs
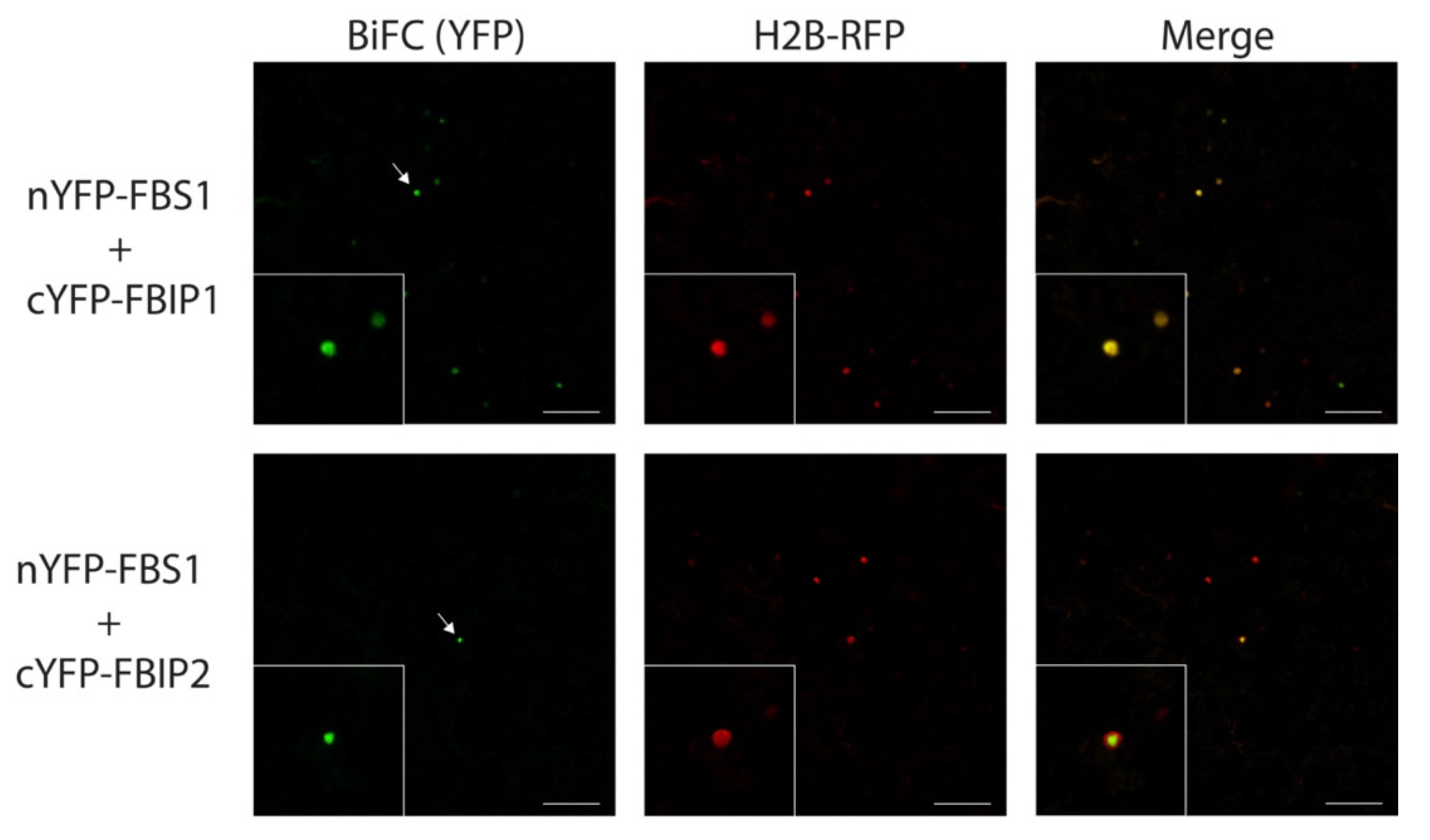
2.4. FBS Interactions with FBIP Occur in the Nucleus
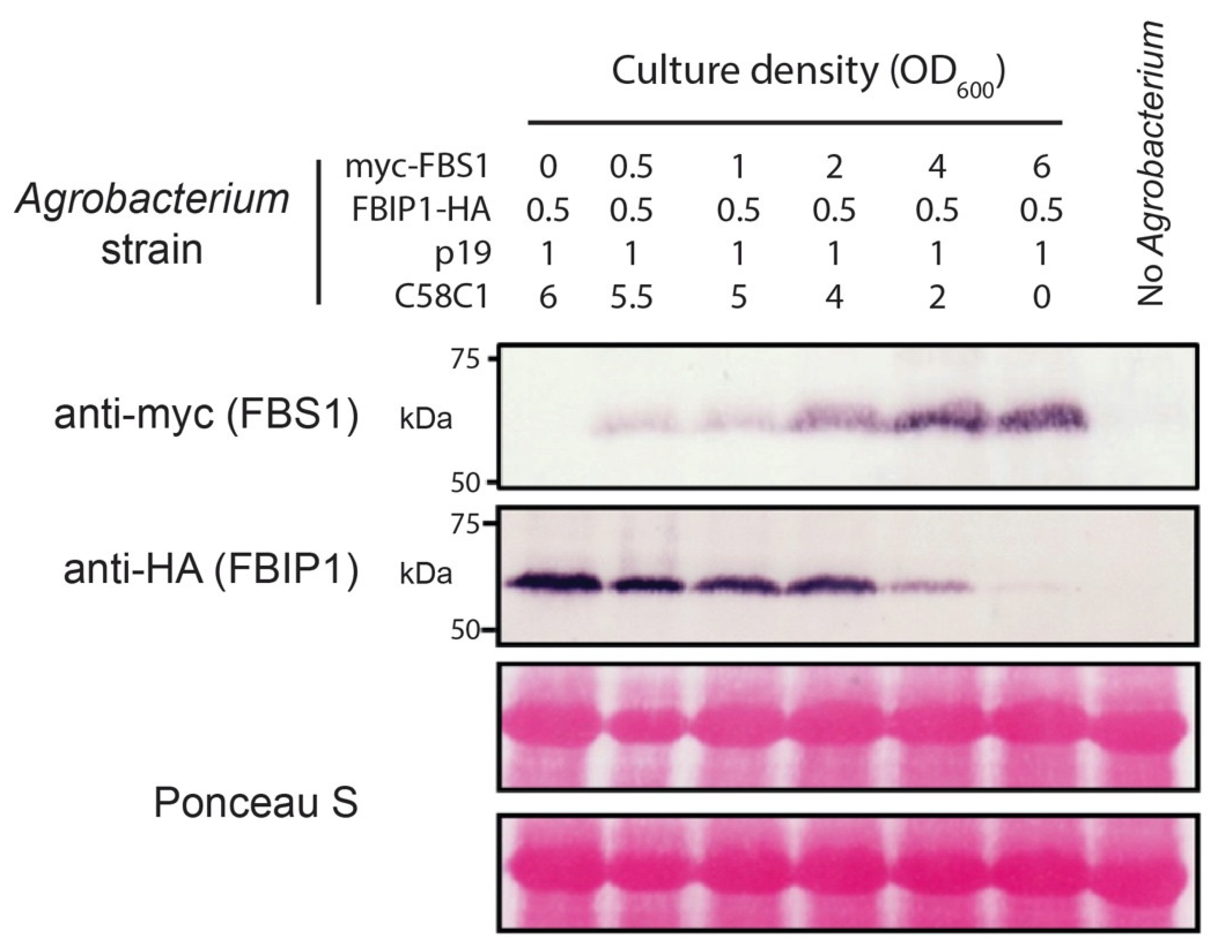
2.5. FBS1 Destabilizes FBIP1
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hua, Z.; Vierstra, R.D. The Cullin-RING Ubiquitin-Protein Ligases. Annu. Rev. Plant Biol. 2011, 62, 299–334. [Google Scholar] [CrossRef] [Green Version]
- Gagne, J.M.; Downes, B.P.; Shiu, S.H.; Durski, A.M.; Vierstra, R.D. The F-Box Subunit of the SCF E3 Complex Is Encoded by a Diverse Superfamily of Genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef] [Green Version]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate Perception by Inositol-Phosphate-Potentiated COI1-JAZ Co-Receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Fang, Q.; Zhou, F.; Zhang, Y.; Singh, S.; Huang, C. Degradation of STOP1 Mediated by the F-box Proteins RAH1 and RAE1 Balances Aluminum Resistance and Plant Growth in Arabidopsis Thaliana. Plant J. 2021, 106, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nolan, T.M.; Clark, N.M.; Jiang, H.; Montes-Serey, C.; Guo, H.; Bassham, D.C.; Walley, J.W.; Yin, Y. The F-Box E3 Ubiquitin Ligase BAF1 Mediates the Degradation of the Brassinosteroid-Activated Transcription Factor BES1 through Selective Autophagy in Arabidopsis. Plant Cell 2021, koab210. [Google Scholar] [CrossRef]
- Jin, J. Systematic Analysis and Nomenclature of Mammalian F-Box Proteins. Genes Dev. 2004, 18, 2573–2580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Liu, P.; Inuzuka, H.; Wei, W. Roles of F-Box Proteins in Cancer. Nat. Rev. Cancer 2014, 14, 233–247. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-M.; Feke, A.; Li, M.-W.; Adamchek, C.; Webb, K.; Pruneda-Paz, J.; Bennett, E.J.; Kay, S.A.; Gendron, J.M. Decoys Untangle Complicated Redundancy and Reveal Targets of Circadian Clock F-Box Proteins. Plant Physiol. 2018, 177, 1170–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, A.; Tateishi, Y.; Onoyama, I.; Okita, Y.; Nakayama, K.; Nakayama, K.I. Fbxw7β Resides in the Endoplasmic Reticulum Membrane and Protects Cells from Oxidative Stress. Cancer Sci. 2011, 102, 749–755. [Google Scholar] [CrossRef]
- Spruck, C.; Strohmaier, H.; Watson, M.; Smith, A.P.L.; Ryan, A.; Krek, W.; Reed, S.I. A CDK-Independent Function of Mammalian Cks1: Targeting of SCFSkp2 to the CDK Inhibitor P27Kip. Mol. Cell 2001, 7, 12. [Google Scholar] [CrossRef]
- Kirk, R.; Laman, H.; Knowles, P.P.; Murray-Rust, J.; Lomonosov, M.; Meziane, E.K.; McDonald, N.Q. Structure of a Conserved Dimerization Domain within the F-Box Protein Fbxo7 and the PI31 Proteasome Inhibitor. J. Biol. Chem. 2008, 283, 22325–22335. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.E.; Randle, S.J.; Laman, H. Beyond Ubiquitination: The Atypical Functions of Fbxo7 and Other F-Box Proteins. Open Biol. 2013, 3, 130131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE Is a Circadian Photoreceptor Stabilized by GIGANTEA in Blue Light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA Complex Formation Is Required for Day-Length Measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Yasuhara, M. Identification of ASK and Clock-Associated Proteins as Molecular Partners of LKP2 (LOV Kelch Protein 2) in Arabidopsis. J. Exp. Bot. 2004, 55, 2015–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoltowski, B.D.; Imaizumi, T. Structure and Function of the ZTL/FKF1/LKP2 Group Proteins in Arabidopsis. In The Enzymes; Elsevier: Amsterdam, The Netherlands, 2014; Volume 35, pp. 213–239. [Google Scholar]
- Más, P.; Kim, W.-Y.; Somers, D.E.; Kay, S.A. Targeted Degradation of TOC1 by ZTL Modulates Circadian Function in Arabidopsis Thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S.; Kim, S.K.; Lee, S.Y.; MacCoss, M.J.; Imaizumi, T. Distinct Roles of FKF1, GIGANTEA, and ZEITLUPE Proteins in the Regulation of CONSTANS Stability in Arabidopsis Photoperiodic Flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Calderon, M.T.; Sepulveda-Garcia, E.; Rocha-Sosa, M. Characterization of Novel F-Box Proteins in Plants Induced by Biotic and Abiotic Stress. Plant Sci. Int. J. Exp. Plant Biol. 2012, 185, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda-Garcia, E.; Rocha-Sosa, M. The Arabidopsis F-Box Protein AtFBS1 Interacts with 14-3-3 Proteins. Plant Sci. Int. J. Exp. Plant Biol. 2012, 195, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.E.; Keller, K.; Chan, K.X.; Gessel, M.M.; Thines, B.C. Transcriptome Analysis Uncovers Arabidopsis F-BOX STRESS INDUCED 1 as a Regulator of Jasmonic Acid and Abscisic Acid Stress Gene Expression. BMC Genom. 2017, 18, 533. [Google Scholar] [CrossRef]
- Catala, R.; Lopez-Cobollo, R.; Mar Castellano, M.; Angosto, T.; Alonso, J.M.; Ecker, J.R.; Salinas, J. The Arabidopsis 14-3-3 Protein RARE COLD INDUCIBLE 1A Links Low-Temperature Response and Ethylene Biosynthesis to Regulate Freezing Tolerance and Cold Acclimation. Plant Cell 2014, 26, 3326–3342. [Google Scholar] [CrossRef] [Green Version]
- Van Kleeff, P.J.; Jaspert, N.; Li, K.W.; Rauch, S.; Oecking, C.; de Boer, A.H. Higher Order Arabidopsis 14-3-3 Mutants Show 14-3-3 Involvement in Primary Root Growth Both under Control and Abiotic Stress Conditions. J. Exp. Bot. 2014, 65, 5877–5888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Lin, H.; Chen, S.; Becker, K.; Yang, Y.; Zhao, J.; Kudla, J.; Schumaker, K.S.; Guo, Y. Inhibition of the Arabidopsis Salt Overly Sensitive Pathway by 14-3-3 Proteins. Plant Cell 2014, 26, 1166–1182. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Jia, Y.; Ding, Y.; Shi, Y.; Li, Z.; Guo, Y.; Gong, Z.; Yang, S. Plasma Membrane CRPK1-Mediated Phosphorylation of 14-3-3 Proteins Induces Their Nuclear Import to Fine-Tune CBF Signaling during Cold Response. Mol. Cell 2017, 66, 117–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-Activated 14-3-3 Proteins as a Molecular Switch in Salt Stress Tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuroda, H.; Yanagawa, Y.; Takahashi, N.; Horii, Y.; Matsui, M. A Comprehensive Analysis of Interaction and Localization of Arabidopsis SKP1-like (ASK) and F-Box (FBX) Proteins. PLoS ONE 2012, 7, e50009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arabidopsis Interactome Mapping Consortium; Dreze, M.; Carvunis, A.-R.; Charloteaux, B.; Galli, M.; Pevzner, S.J.; Tasan, M.; Ahn, Y.-Y.; Balumuri, P.; Barabasi, A.-L.; et al. Evidence for Network Evolution in an Arabidopsis Interactome Map. Science 2011, 333, 601–607. [Google Scholar] [CrossRef] [Green Version]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The Interactive Data Analysis Centre for Arabidopsis Subcellular Protein Locations. Nucleic Acids Res. 2017, 45, D1064–D1074. [Google Scholar] [CrossRef] [Green Version]
- Jain, B.P.; Pandey, S. WD40 Repeat Proteins: Signalling Scaffold with Diverse Functions. Protein J 2018, 37, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; An, K.; Zhou, J.-B.; Wu, N.-S.; Wang, Y.; Ye, Z.-Q.; Wu, Y.-D. WDSPdb: An Updated Resource for WD40 Proteins. Bioinformatics 2019, 35, 4824–4826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Scholkopf, B.; Weigel, D.; Lohmann, J.U. A Gene Expression Map of Arabidopsis Thaliana Development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress Global Stress Expression Data Set: Protocols, Evaluation and Model Data Analysis of UV-B Light, Drought and Cold Stress Responses. Plant J. Cell Mol. Biol. 2007, 50, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Goda, H.; Sasaki, E.; Akiyama, K.; Maruyama-Nakashita, A.; Nakabayashi, K.; Li, W.; Ogawa, M.; Yamauchi, Y.; Preston, J.; Aoki, K.; et al. The AtGenExpress Hormone and Chemical Treatment Data Set: Experimental Design, Data Evaluation, Model Data Analysis and Data Access. Plant J. 2008, 55, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Rozwadowski, K. EAR Motif-Mediated Transcriptional Repression in Plants: An Underlying Mechanism for Epigenetic Regulation of Gene Expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Shyu, C.; Figueroa, P.; DePew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.E.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 Lacks a Canonical Degron and Has an EAR Motif That Mediates Transcriptional Repression of Jasmonate Responses in Arabidopsis. Plant Cell 2012, 24, 536–550. [Google Scholar] [CrossRef] [Green Version]
- Long, J.A. TOPLESS Regulates Apical Embryonic Fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA Connects the Co-Repressor TOPLESS to Jasmonate Signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [Green Version]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS Interactome: A Framework for Gene Repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional Corepressor TOPLESS Complexes with Pseudoresponse Regulator Proteins and Histone Deacetylases to Regulate Circadian Transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos Maraschin, F.; Memelink, J.; Offringa, R. Auxin-Induced, SCF TIR1—Mediated Poly-Ubiquitination Marks AUX/IAA Proteins for Degradation. Plant J. 2009, 59, 100–109. [Google Scholar] [CrossRef]
- Ke, J.; Ma, H.; Gu, X.; Thelen, A.; Brunzelle, J.S.; Li, J.; Xu, H.E.; Melcher, K. Structural Basis for Recognition of Diverse Transcriptional Repressors by the TOPLESS Family of Corepressors. Sci. Adv. 2015, 1, e1500107. [Google Scholar] [CrossRef] [Green Version]
- Long, Y.; Schiefelbein, J. Novel TTG1 Mutants Modify Root-Hair Pattern Formation in Arabidopsis. Front. Plant Sci. 2020, 11, 383. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; He, Z.; Lu, G.; Lee, S.C.; Alonso, J.; Ecker, J.R.; Luan, S. A WD40 Domain Cyclophilin Interacts with Histone H3 and Functions in Gene Repression and Organogenesis in Arabidopsis. Plant Cell 2007, 19, 2403–2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Jeong, J.C.; Zhu, Y.; Sokolchik, I.; Miyazaki, S.; Zhu, J.-K.; Hasegawa, P.M.; Bohnert, H.J.; Shi, H.; Yun, D.-J.; et al. Involvement of Arabidopsis HOS15 in Histone Deacetylation and Cold Tolerance. Proc. Natl. Acad. Sci. USA 2008, 105, 4945–4950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehdi, S.; Derkacheva, M.; Ramström, M.; Kralemann, L.; Bergquist, J.; Hennig, L. The WD40 Domain Protein MSI1 Functions in a Histone Deacetylase Complex to Fine-Tune Abscisic Acid Signaling. Plant Cell 2016, 28, 42–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krogan, N.T.; Hogan, K.; Long, J.A. APETALA2 Negatively Regulates Multiple Floral Organ Identity Genes in Arabidopsis by Recruiting the Co-Repressor TOPLESS and the Histone Deacetylase HDA19. Development 2012, 139, 4180–4190. [Google Scholar] [CrossRef] [Green Version]
- Leydon, A.R.; Wang, W.; Gala, H.P.; Gilmour, S.; Juarez-Solis, S.; Zahler, M.L.; Zemke, J.E.; Zheng, N.; Nemhauser, J.L. Repression by the Arabidopsis TOPLESS Corepressor Requires Association with the Core Mediator Complex. eLife 2021, 10, e66739. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–BHLH–WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-Option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [Green Version]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. Mechanisms and Function of Substrate Recruitment by F-Box Proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 369–381. [Google Scholar] [CrossRef]
- Wang, L.; Xu, Q.; Yu, H.; Ma, H.; Li, X.; Yang, J.; Chu, J.; Xie, Q.; Wang, Y.; Smith, S.M.; et al. Strigolactone and Karrikin Signaling Pathways Elicit Ubiquitination and Proteolysis of SMXL2 to Regulate Hypocotyl Elongation in Arabidopsis Thaliana. Plant Cell 2020, 32, 2251–2270. [Google Scholar] [CrossRef]
- Zhao, X.; Li, F.; Li, K. The 14-3-3 Proteins: Regulators of Plant Metabolism and Stress Responses. Plant Biol. J. 2021, 23, 531–539. [Google Scholar] [CrossRef]
- Welcker, M.; Larimore, E.A.; Swanger, J.; Bengoechea-Alonso, M.T.; Grim, J.E.; Ericsson, J.; Zheng, N.; Clurman, B.E. Fbw7 Dimerization Determines the Specificity and Robustness of Substrate Degradation. Genes Dev. 2013, 27, 2531–2536. [Google Scholar] [CrossRef] [Green Version]
- Barbash, O.; Lee, E.; Diehl, J. Phosphorylation-Dependent Regulation of SCFFbx4 Dimerization and Activity Involves a Novel Component. Oncogene 2011, 30, 1995–2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, T.; Cai, J.; Zhan, E.; Yang, Y.; Zhao, J.; Guo, Y.; Zhou, H. Stability and Localization of 14-3-3 Proteins Are Involved in Salt Tolerance in Arabidopsis. Plant Mol. Biol. 2016, 92, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; mi Park, Y.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Maier, A.; Lee, J.-H.; Laubinger, S.; Saijo, Y.; Wang, H.; Qu, L.-J.; Hoecker, U.; Deng, X.W. Biochemical Characterization of Arabidopsis Complexes Containing constitutively photomorphogenic1 and suppressor of phya Proteins in Light Control of Plant Development. Plant Cell 2008, 20, 2307–2323. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of Series of Gateway Binary Vectors, PGWBs, for Realizing Efficient Construction of Fusion Genes for Plant Transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sepulveda-Garcia, E.; Fulton, E.C.; Parlan, E.V.; O’Connor, L.E.; Fleming, A.A.; Replogle, A.J.; Rocha-Sosa, M.; Gendron, J.M.; Thines, B. Unique N-Terminal Interactions Connect F-BOX STRESS INDUCED (FBS) Proteins to a WD40 Repeat-like Protein Pathway in Arabidopsis. Plants 2021, 10, 2228. https://doi.org/10.3390/plants10102228
Sepulveda-Garcia E, Fulton EC, Parlan EV, O’Connor LE, Fleming AA, Replogle AJ, Rocha-Sosa M, Gendron JM, Thines B. Unique N-Terminal Interactions Connect F-BOX STRESS INDUCED (FBS) Proteins to a WD40 Repeat-like Protein Pathway in Arabidopsis. Plants. 2021; 10(10):2228. https://doi.org/10.3390/plants10102228
Chicago/Turabian StyleSepulveda-Garcia, Edgar, Elena C. Fulton, Emily V. Parlan, Lily E. O’Connor, Anneke A. Fleming, Amy J. Replogle, Mario Rocha-Sosa, Joshua M. Gendron, and Bryan Thines. 2021. "Unique N-Terminal Interactions Connect F-BOX STRESS INDUCED (FBS) Proteins to a WD40 Repeat-like Protein Pathway in Arabidopsis" Plants 10, no. 10: 2228. https://doi.org/10.3390/plants10102228
APA StyleSepulveda-Garcia, E., Fulton, E. C., Parlan, E. V., O’Connor, L. E., Fleming, A. A., Replogle, A. J., Rocha-Sosa, M., Gendron, J. M., & Thines, B. (2021). Unique N-Terminal Interactions Connect F-BOX STRESS INDUCED (FBS) Proteins to a WD40 Repeat-like Protein Pathway in Arabidopsis. Plants, 10(10), 2228. https://doi.org/10.3390/plants10102228





