A Strategy for the Production and Molecular Validation of Agrobacterium-Mediated Intragenic Octoploid Strawberry
Abstract
:1. Introduction
2. Results
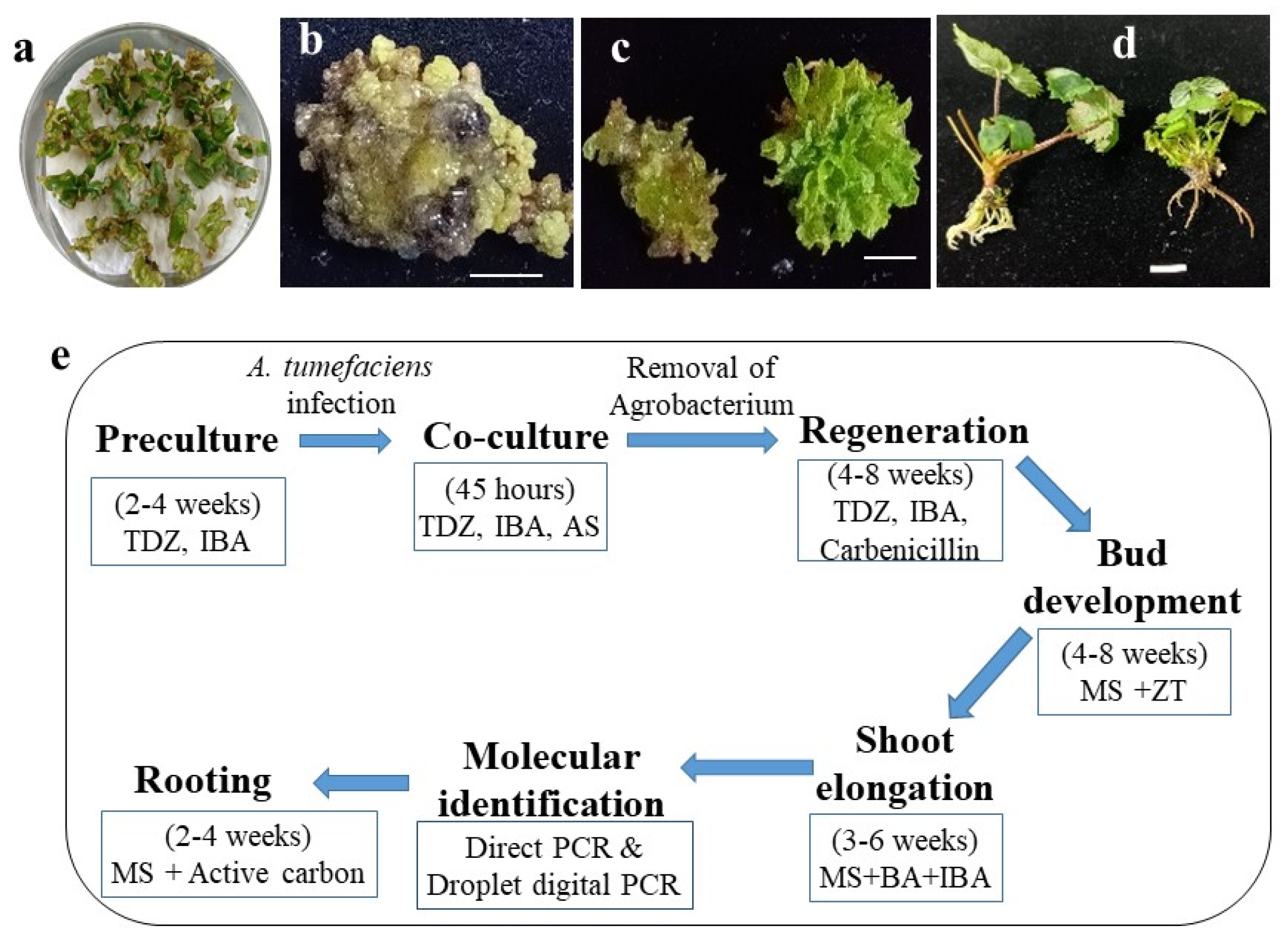
2.1. Optimization of Shoot Regeneration from Leaf Explants of the Strawberry cv. ‘Shanghai Angel’
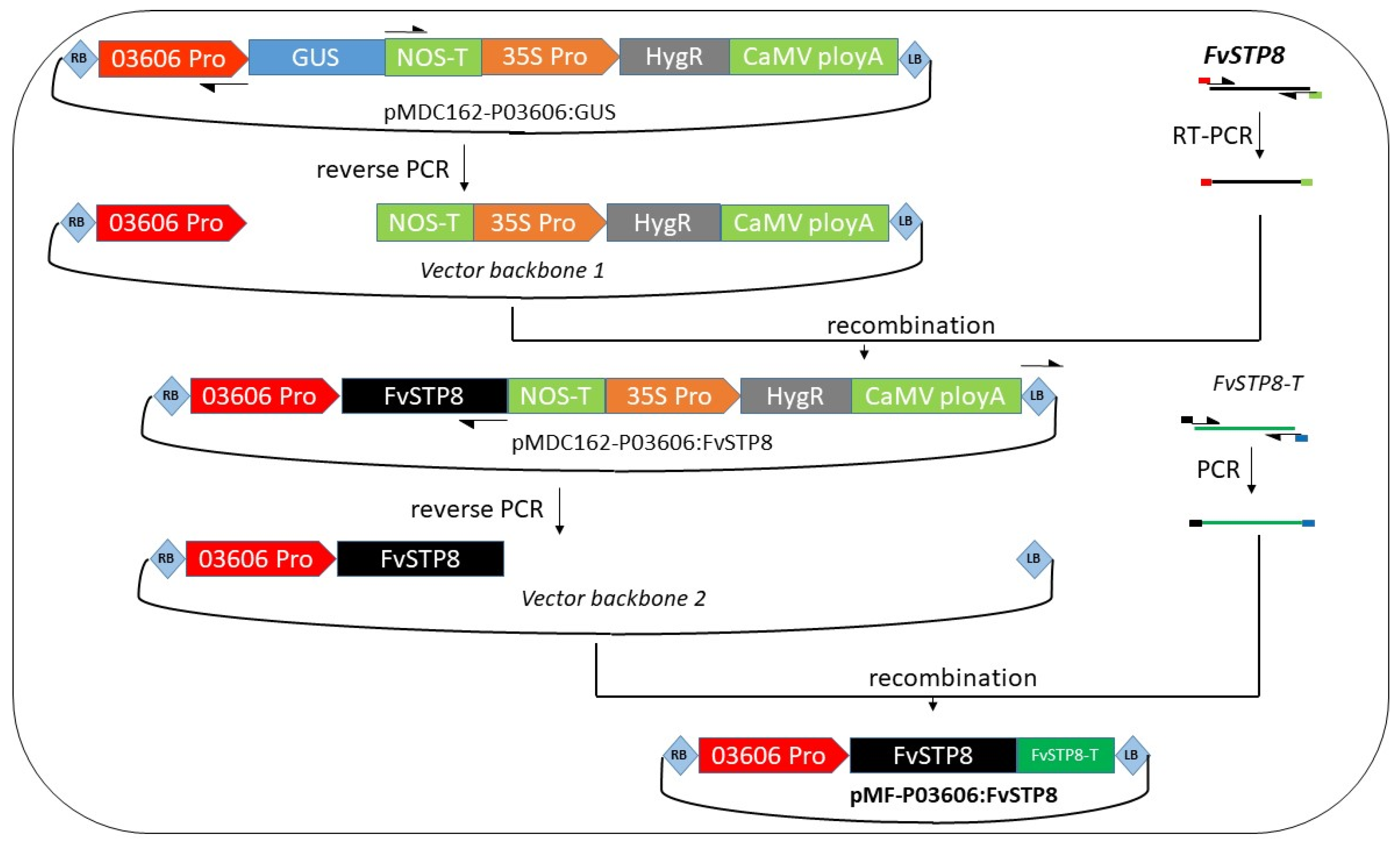
2.2. Generation of a Marker-Free Binary Vector with an Intragenic Expression Cassette
2.3. Development of Putative Intragenic Strawberry Plants via Agrobacterium-Mediated Transformation without Selection
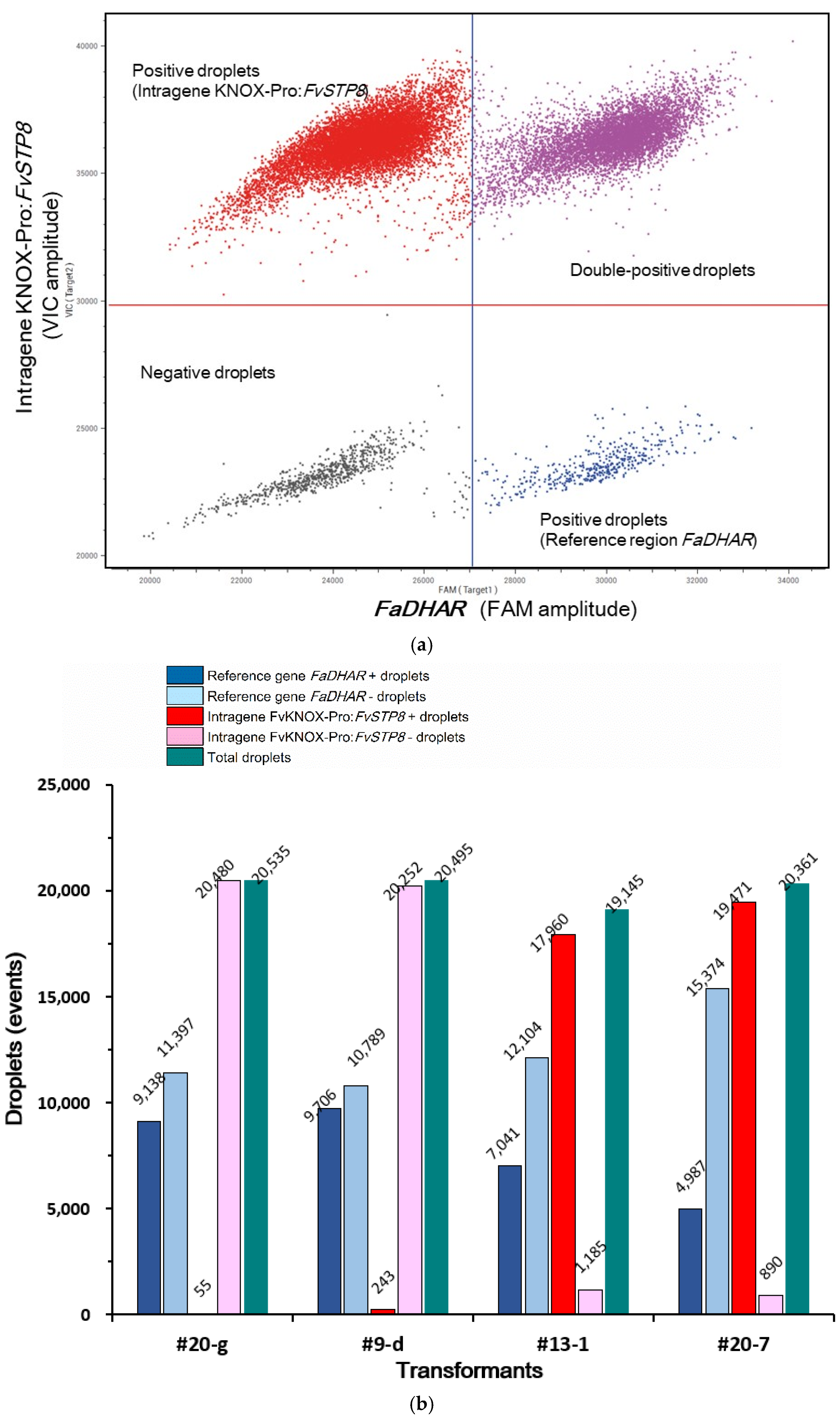
2.4. Selection of Transformants Using Two Rounds of PCR
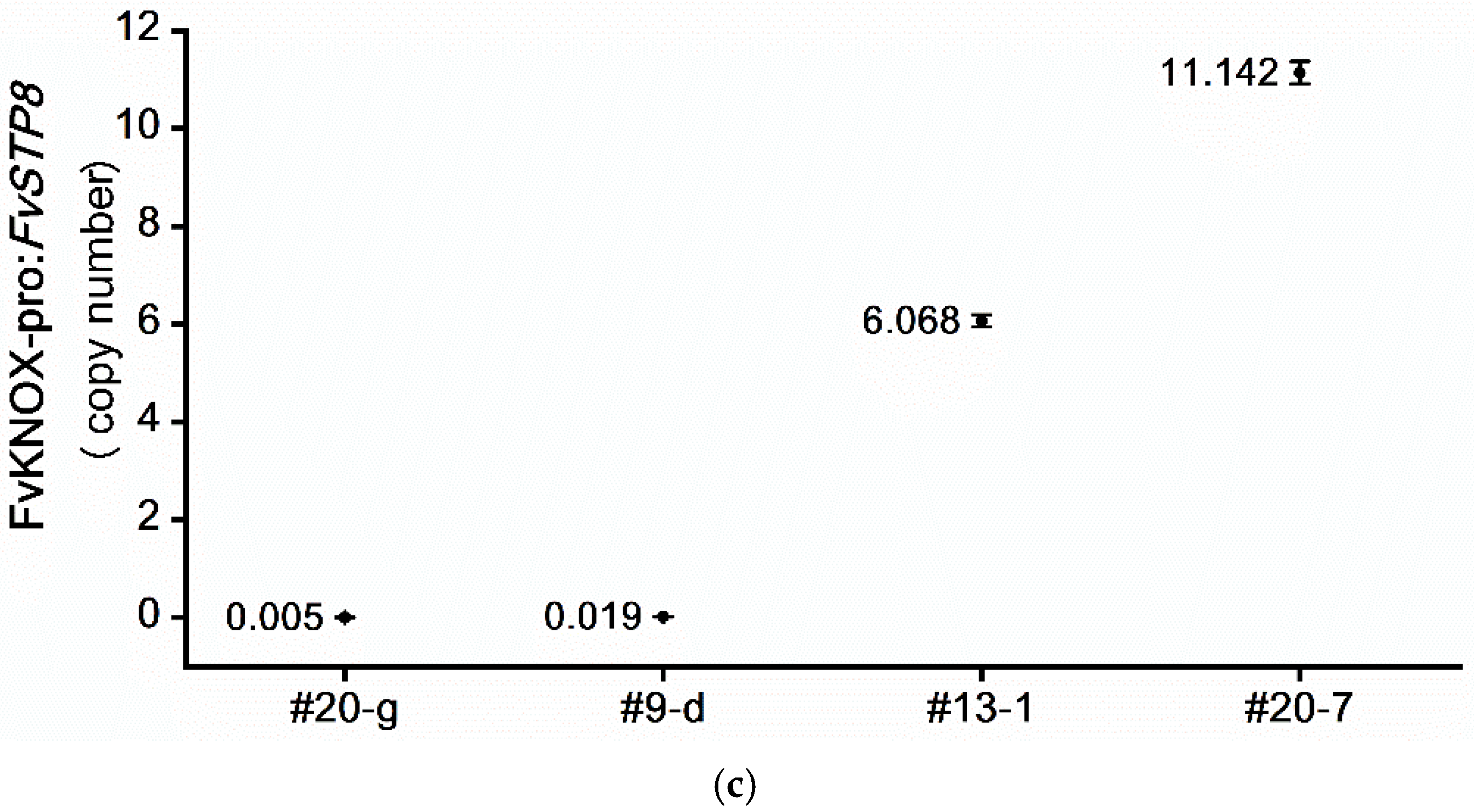
2.5. Determination of the Intragene Copy Number in Transformants Using ddPCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Agrobacterium Strain and Binary Vector
4.3. Explant Preparation and Strawberry Transformation
4.4. Direct PCR Analysis of Regenerated Shoots
4.5. DNA Purification and ddPCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. GigaScience 2018, 7, gix124. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Li, M.; Liu, Z.; Ai, X.; Li, Y. Reannotation of the cultivated strawberry genome and establishment of a strawberry genome database. Hortic. Res. 2021, 8, 41. [Google Scholar] [CrossRef]
- Whitaker, V.M.; Knapp, S.J.; Hardigan, M.A.; Edger, P.P.; Slovin, J.P.; Bassil, N.V.; Hytönen, T.; Mackenzie, K.K.; Lee, S.; Jung, S.; et al. A roadmap for research in octoploid strawberry. Hortic. Res. 2020, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Gaston, A.; Osorio, S.; Denoyes, B.; Rothan, C. Applying the Solanaceae Strategies to Strawberry Crop Improvement. Trends Plant Sci. 2020, 25, 130–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef] [PubMed]
- Lobato-Gómez, M.; Hewitt, S.; Capell, T.; Christou, P.; Dhingra, A.; Girón-Calva, P.S. Transgenic and genome-edited fruits: Background, constraints, benefits, and commercial opportunities. Hortic. Res. 2021, 8, 166. [Google Scholar] [CrossRef] [PubMed]
- Debnath, S.C.; Silva, J.A.T. Strawberry Culture In Vitro: Applications in Genetic Transformation and Biotechnology. Fruit Veg. Cereal Sci. Biotechnol. 2007, 1, 1–12. [Google Scholar]
- Park, J.-I.; Lee, Y.-K.; Chung, W.-I.; Lee, I.-H.; Choi, J.-H.; Lee, W.-M.; Ezura, H.; Lee, S.-P.; Kim, I.-J. Modification of sugar composition in strawberry fruit by antisense suppression of an ADP-glucose pyrophosphorylase. Mol. Breed. 2006, 17, 269–279. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.K.; Kim, I.J. Modulation of fruit softening by antisense suppression of endo-β-1,4-glucanase in strawberry. Mol. Breed. 2011, 27, 375–383. [Google Scholar] [CrossRef]
- Schaart, J.G.; Kjellsen, T.D.; Mehli, L.; Heggem, R.; Iversen, T.-H.; Schouten, H.J.; Krens, F.A. Towards the production of genetically modified strawberries which are acceptable to consumers. In Genomics, Transgenics, Molecular Breeding and Biotechnology of Strawberry; Husaini, A.M., Mercado, J.A., Eds.; Genes, Genomes and Genomics; Springer: Berlin/Heidelberg, Germany; London, UK, 2011; Volume 5, pp. 102–107. [Google Scholar]
- Gao, Q.; Luo, H.; Li, Y.; Liu, Z.; Kang, C. Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry. Plant Biotechnol. J. 2020, 18, 1550–1561. [Google Scholar] [CrossRef] [Green Version]
- Tuteja, N.; Verma, S.; Sahoo, R.K.; Raveendar, S.; Reddy, I.N. Recent advances in development of marker-free transgenic plants: Regulation and biosafety concern. J. Biosci. 2012, 37, 167–197. [Google Scholar] [CrossRef] [PubMed]
- de Vetten, N.; Wolters, A.M.; Raemakers, K.; van der Meer, I.; ter Stege, R.; Heeres, E.; Heeres, P.; Visser, R. A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nat. Biotechnol. 2003, 21, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Miroshnichenko, D.; Timerbaev, V.; Okuneva, A.; Klementyeva, A.; Sidorova, T.; Pushin, A.; Dolgov, S. Enhancement of resistance to PVY in intragenic marker-free potato plants by RNAi-mediated silencing of eIF4E translation initiation factors. Plant Cell Tissue Organ Cult. 2020, 140, 691–705. [Google Scholar] [CrossRef]
- Jo, K.R.; Kim, C.J.; Kim, S.J.; Kim, T.Y.; Bergervoet, M.; Jongsma, M.A.; Visser, R.G.; Jacobsen, E.; Vossen, J.H. Development of late blight resistant potatoes by cisgene stacking. BMC Biotechnol. 2014, 14, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, T.; Thistleton, J.; Higgins, T.J.; Howitt, C.; Ayliffe, M. Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell Tissue Organ Cult. 2014, 119, 647–659. [Google Scholar] [CrossRef]
- Schaart, J.G.; Krens, F.A.; Pelgrom, K.T.; Mendes, O.; Rouwendal, G.J. Effective production of marker-free transgenic strawberry plants using inducible site-specific recombination and a bifunctional selectable marker gene. Plant Biotechnol. J. 2004, 2, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Timerbaev, V.; Mitiouchkina, T.; Pushin, A.; Dolgov, S. Production of Marker-Free Apple Plants Expressing the Supersweet Protein Gene Driven by Plant Promoter. Front. Plant Sci. 2019, 10, 388. [Google Scholar] [CrossRef]
- Kleidon, J.; Brinin, A.; Paul, J.Y.; Harding, R.; Dale, J.; Dugdale, B. Production of selectable marker gene-free Cavendish banana (Musa spp.) using a steroid-inducible recombinase platform. Transgenic Res. 2020, 29, 81–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, C.; Sun, B.; Liu, Z.; Zhang, L.; Wei, X.; Zhou, Y.; Meng, Y.; Lai, Y.; Dai, Y.; Zhu, Z. An optimized double T-DNA binary vector system for improved production of marker-free transgenic tobacco plants. Biotechnol. Lett. 2020, 42, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.M. Transgenic organisms—Time for conceptual diversification? Nat. Biotechnol. 2003, 21, 227–228. [Google Scholar] [CrossRef] [PubMed]
- Rommens, C.M. All-native DNA transformation: A new approach to plant genetic engineering. Trends Plant Sci. 2004, 9, 457–464. [Google Scholar] [CrossRef]
- Rommens, C.M. Intragenic crop improvement: Combining the benefits of traditional breeding and genetic engineering. J. Agric. Food Chem. 2007, 55, 4281–4288. [Google Scholar] [CrossRef]
- Schouten, H.J.; Jacobsen, E. Cisgenesis and intragenesis, sisters in innovative plant breeding. Trends Plant Sci. 2008, 13, 260–261. [Google Scholar] [CrossRef]
- Molesini, B.; Pii, Y.; Pandolfini, T. Fruit improvement using intragenesis and artificial microRNA. Trends Biotechnol. 2012, 30, 80–88. [Google Scholar] [CrossRef]
- Holme, I.B.; Wendt, T.; Holm, P.B. Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol. J. 2013, 11, 395–407. [Google Scholar] [CrossRef]
- Espinoza, C.; Schlechter, R.; Herrera, D.; Torres, E.; Serrano, A.; Medina, C.; Arce-Johnson, P. Cisgenesis and intragenesis: New tools for improving crops. Biol. Res. 2013, 46, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, P. “Genetically Modified Lite” placates public but not activists: New technologies to manipulate plant genomes could help to overcome public concerns about GM crops. EMBO Rep. 2014, 15, 138–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricroch, A.E.; Hénard-Damave, M.C. Next biotech plants: New traits, crops, developers and technologies for addressing global challenges. Crit. Rev. Biotechnol. 2016, 36, 675–690. [Google Scholar] [CrossRef]
- Enfissi, E.M.A.; Drapal, M.; Perez-Fons, L.; Nogueira, M.; Berry, H.M.; Almeida, J.; Fraser, P.D. New plant breeding techniques and their regulatory implications: An opportunity to advance metabolomics approaches. J. Plant Physiol. 2021, 258–259, 153378. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Edenbrandt, A.K.; Vedel, S.E.; Andersen, M.M.; Landes, X.; Østerberg, J.T.; Falhof, J.; Olsen, L.I.; Christensen, S.B.; Sandøe, P.; et al. Are we ready for back-to-nature crop breeding? Trends Plant Sci. 2015, 20, 155–164. [Google Scholar] [CrossRef]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New Biotechnological Tools for the Genetic Improvement of Major Woody Fruit Species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef]
- Conner, A.J.; Barrell, P.J.; Baldwin, S.J.; Lokerse, A.S.; Cooper, P.A.; Erasmuson, A.K.; Nap, J.; Jacobs, J.M.E. Intragenic vectors for gene transfer without foreign DNA. Euphytica 2007, 154, 341–353. [Google Scholar] [CrossRef]
- Joshi, S.G.; Schaart, J.G.; Groenwold, R.; Jacobsen, E.; Schouten, H.J.; Krens, F.A. Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol. Biol. 2011, 75, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.L.; Karunairetnam, S.; Tomes, S.; Gleave, A.P. Developing apple intragenic transformation systems. Acta Hortic. 2013, 974, 109–115. [Google Scholar] [CrossRef]
- Waltz, E. USDA approves next-generation GM potato. Nat. Biotechnol. 2015, 33, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Holme, I.B.; Dionisio, G.; Brinch-Pedersen, H. A roadmap to modulated anthocyanin compositions in carrots. Plants 2021, 10, 472. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Głowacka, K.; Kromdijk, J.; Leonelli, L.; Niyogi, K.K.; Clemente, T.E.; Long, S.P. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 2016, 39, 908–917. [Google Scholar] [CrossRef]
- Xu, X.; Peng, C.; Wang, X.; Chen, X.; Wang, Q.; Xu, J. Comparison of droplet digital PCR with quantitative real-time PCR for determination of zygosity in transgenic maize. Transgenic Res. 2016, 25, 855–864. [Google Scholar] [CrossRef]
- Collier, R.; Dasgupta, K.; Xing, Y.; Hernandez, B.T.; Shao, M.; Rohozinski, D.; Kovak, E.; Lin, J.; de Oliveira, M.L.P.; Stover, E.; et al. Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. 2017, 90, 1014–1025. [Google Scholar] [CrossRef] [Green Version]
- Cassinari, K.; Quenez, O.; Joly-Hélas, G.; Beaussire, L.; Le Meur, N.; Castelain, M.; Goldenberg, A.; Guerrot, A.-M.; Brehin, A.-C.; Deleuze, J.-F.; et al. A Simple, Universal, and Cost-Efficient Digital PCR Method for the Targeted Analysis of Copy Number Variations. Clin. Chem. 2019, 65, 1153–1160. [Google Scholar] [CrossRef]
- Folta, K.M.; Dhingra, A.; Howard, L.; Stewart, P.J.; Chandler, C.K. Characterization of LF9, an octoploid strawberry genotype selected for rapid regeneration and transformation. Planta 2006, 224, 1058–1067. [Google Scholar] [CrossRef]
- Liu, H.; Xie, W.F.; Zhang, L.; Valpuesta, V.; Ye, Z.W.; Gao, Q.H.; Duan, K. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J Integr. Plant Biol. 2014, 56, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Shahan, R.; Li, D.; Liu, Z. Identification of genes preferentially expressed in wild strawberry receptacle fruit and demonstration of their promoter activities. Hortic. Res. 2019, 6, 50. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.T.; Ji, Y.; Liu, Y.; Tian, S.H.; Gao, Q.H.; Zou, X.H.; Yang, J.; Dong, C.; Tan, J.H.; Ni, D.A.; et al. The sugar transporter system of strawberry: Genome-wide identification and expression correlation with fruit soluble sugar-related traits in a Fragaria × ananassa germplasm collection. Hortic. Res. 2020, 7, 132. [Google Scholar] [CrossRef]
- Buttner, M. The monosaccharide transporter-like gene family in Arabidopsis. FEBS Lett. 2007, 581, 2318–2324. [Google Scholar] [CrossRef] [Green Version]
- Debnath, S.C. Zeatin overcomes thidiazuron-induced inhibition of shoot elongation and promotes rooting in strawberry culture in vitro. J. Hortic. Sci. Biotechnol. 2006, 81, 349–354. [Google Scholar] [CrossRef]
- Chen, L.; Li, W.; Katin-Grazzini, L.; Ding, J.; Gu, X.; Li, Y.; Gu, T.; Wang, R.; Lin, X.; Deng, Z.; et al. A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic. Res. 2018, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Rousseau-Gueutin, M.; Gaston, A.; Ainouche, A.; Ainouche, M.L.; Olbricht, K.; Staudt, G.; Richard, L.; Denoyes-Rothan, B. Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): New insights from phylogenetic analyses of low-copy nuclear genes. Mol. Phylogenet. Evol. 2009, 51, 515–530. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.-H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [Green Version]
- Gao, Q.; Tian, S.; Yang, J.; Duan, K.; Zhang, L.; Zou, X. ‘Shanghai Angel’, a New Strawberry Cultivar with High Quality and Disease Resistance. Acta Hortic. Sinica 2018, 45, 2741–2742. (In Chinese) [Google Scholar]
- Liu, Y. Evaluation of the Resistance to Anthracnose in Different Strawberry Cultivars and Construction of a Detection System for Latent Infection of Colletotrichum spp. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 3 June 2021; pp. 16–22. [Google Scholar]
- McCue, K.F.; Allen, P.V.; Shepherd, L.V.; Blake, A.; Maccree, M.M.; Rockhold, D.R.; Novy, R.G.; Stewart, D.; Davies, H.V.; Belknap, W.R. Potato glycosterol rhamnosyltransferase, the terminal step in triose side-chain biosynthesis. Phytochemistry 2007, 68, 327–334. [Google Scholar] [CrossRef]
- James, D.J.; Passey, A.J.; Barbara, D.J. Agrobacterium mediated transformation of the cultivated strawberry (Fragaria × ananassa Duch.) using disarmed binary vectors. Plant Sci. 1990, 69, 79–94. [Google Scholar] [CrossRef]
- Nehra, N.S.; Chibbar, R.N.; Kartha, K.K.; Datla, R.S.; Crosby, W.L.; Stushnoff, C. Agrobacterium—mediated trans formation of strawberry calli and recovery of transgenic plants. Plant Cell Rep. 1990, 9, 10–13. [Google Scholar] [CrossRef]
- Schestibratov, K.; Dolgov, S. Transgenic strawberry plants expressing a thaumatin II gene demonstrate enhanced resistance to Botrytis cinerea. Sci. Hortic. 2005, 106, 177–189. [Google Scholar] [CrossRef]
- Abdal-Aziz, S.A.; Pliego-Alfaro, F.; Quesada, M.A.; Mercado, J.A. Evidence of frequent integration of non-T-DNA vector backbone sequences in transgenic strawberry plant. J. Biosci. Bioeng. 2006, 101, 508–510. [Google Scholar] [CrossRef] [Green Version]
- Barceló, M.; El-Mansouri, I.; Mercado, J.A.; Quesada, M.A.; Alfaro, F.P. Regeneration and transformation via Agrobacterium tumefaciens of the strawberry cultivar Chandler. Plant Cell Tissue Organ Cult. 1998, 54, 29–36. [Google Scholar] [CrossRef]
- Ricci, A.; Sabbadini, S.; Prieto, H.; Padilla, I.M.; Dardick, C.; Li, Z.; Scorza, R.; Limera, C.; Mezzetti, B.; Perez-Jimenez, M.; et al. Genetic Transformation in Peach (Prunus persica L.): Challenges and Ways Forward. Plants 2020, 9, 971. [Google Scholar] [CrossRef]
- Raza, G.; Singh, M.B.; Bhalla, P.L. Somatic Embryogenesis and Plant Regeneration from Commercial Soybean Cultivars. Plants 2019, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Sabbadini, S.; Capriotti, L.; Molesini, B.; Pandolfini, T.; Navacchi, O.; Limera, C.; Ricci, A.; Mezzetti, B. Comparison of regeneration capacity and Agrobacterium-mediated cell transformation efficiency of different cultivars and rootstocks of Vitis spp. via organogenesis. Sci. Rep. 2019, 9, 582. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Q.; Davis, R.E. Transgene expression in strawberries driven by a heterologous phloem-specific promoter. Plant Cell Rep. 2004, 23, 224–230. [Google Scholar] [CrossRef]
- Wawrzyńczak, D.; Michalczuk, L.; Sowik, I. Modification in indole-3-acetic acid metabolism, growth and development of strawberry through transformation with maize IAA-glucose synthase gene (iaglu). Acta Physiol. Plant 2005, 27, 19–28. [Google Scholar] [CrossRef]
- Vellicce, G.R.; Ricci, J.; Hernández, L.; Castagnaro, A.P. Enhanced resistance to botrytis cinerea mediated by the transgenic expression of the chitinase gene ch5b in strawberry. Transgenic Res. 2006, 15, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Lunkenbein, S.; Coiner, H.; de Vos, C.H.R.; Schaart, J.G.; Boone, M.J.; Krens, F.A.; Schwab, W.; Salentijn, E.M.J. Molecular characterization of a stable antisense chalcone synthase phenotype in strawberry (Fragaria × ananassa). J. Agric. Food Chem. 2006, 54, 2145–2153. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.H.; Zhang, S.L. Factors influencing the efficiency of Agrobacterium mediated transformation in strawberry cultivar Toyonoka. J. Nucl. Agric. Sci. 2007, 21, 461–465. (In Chinese) [Google Scholar]
- Qin, Y.; da Silva, J.A.T.; Zhang, L.; Zhang, S. Transgenic strawberry: State of the art for improved traits. Biotechnol. Adv. 2008, 26, 219–232. [Google Scholar] [CrossRef]
- Holsters, M.; de Waele, D.; Depicker, A.; Messens, E.; van Montagu, M.; Schell, J. Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 1978, 163, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J. Optimization and Establishment of In Vitro Regeneration System and Genetic Transformation System of Different Strawberry Resources. Master’s Thesis, Shanghai Ocean University, Shanghai, China, May 2019. [Google Scholar]
- Oosumi, T.; Gruszewski, H.A.; Blischak, L.A.; Baxter, A.J.; Wadl, P.A.; Shuman, J.L.; Veilleux, R.E.; Shulaev, V. High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta 2006, 223, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ding, Z.; Wang, J.; Tian, S.; Duan, K.; Gao, Q. Bet v 1 potential allergens are involved in 10 anthracnose resistance of strawberry varieties. J. Berry Res. 2021, 11, 21–32. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H.J. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology; Krawetz, S., Ed.; Humana Press: Totowa, NJ, USA, 2000; pp. 365–386. [Google Scholar]
- Wang, K.; Li, H.; Xu, Y.; Shao, Q.; Yi, J.; Wang, R.; Cai, W.; Hang, X.; Zhang, C.; Cai, H.; et al. MFEprimer-3.0: Quality control for PCR primers. Nucleic Acids Res. 2019, 47, W610–W613. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Experiment a | No. of Initial Explants | No. of Shoots b | No. of PCR-Positive Shoots | Transformation Efficiency (%) c |
|---|---|---|---|---|
| I | 30 | 53 | 0 | 0 |
| II | 51 | 58 | 1 | 1.96% |
| III | 40 | 46 | 1 | 2.50% |
| IV | 38 | 84 | 2 | 5.26% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, K.; Zhao, Y.-J.; Li, Z.-Y.; Zou, X.-H.; Yang, J.; Guo, C.-L.; Chen, S.-Y.; Yang, X.-R.; Gao, Q.-H. A Strategy for the Production and Molecular Validation of Agrobacterium-Mediated Intragenic Octoploid Strawberry. Plants 2021, 10, 2229. https://doi.org/10.3390/plants10112229
Duan K, Zhao Y-J, Li Z-Y, Zou X-H, Yang J, Guo C-L, Chen S-Y, Yang X-R, Gao Q-H. A Strategy for the Production and Molecular Validation of Agrobacterium-Mediated Intragenic Octoploid Strawberry. Plants. 2021; 10(11):2229. https://doi.org/10.3390/plants10112229
Chicago/Turabian StyleDuan, Ke, Ying-Jie Zhao, Zi-Yi Li, Xiao-Hua Zou, Jing Yang, Cheng-Lin Guo, Si-Yu Chen, Xiu-Rong Yang, and Qing-Hua Gao. 2021. "A Strategy for the Production and Molecular Validation of Agrobacterium-Mediated Intragenic Octoploid Strawberry" Plants 10, no. 11: 2229. https://doi.org/10.3390/plants10112229
APA StyleDuan, K., Zhao, Y.-J., Li, Z.-Y., Zou, X.-H., Yang, J., Guo, C.-L., Chen, S.-Y., Yang, X.-R., & Gao, Q.-H. (2021). A Strategy for the Production and Molecular Validation of Agrobacterium-Mediated Intragenic Octoploid Strawberry. Plants, 10(11), 2229. https://doi.org/10.3390/plants10112229







