The Divergent Roles of the Rice bcl-2 Associated Athanogene (BAG) Genes in Plant Development and Environmental Responses
Abstract
1. Introduction
2. Materials and Methods
2.1. Database Searches for OsBAG Genes in Rice
2.2. Sequence and Structure Analysis
2.3. Phylogenetic Analyses and Motif Identification
2.4. Co-Expression Analysis of OsBAGs and OsCESAs in Rice
2.5. Expression Profile of OsBAGs in Responses to Environmental Stimulations
2.6. GO/KEGG Pathway Enrichment
2.7. Plasmid Construction and Plant Transformation
2.8. GUS Staining
3. Results
3.1. The Identification of the OsBAG Family in Rice
3.2. Structural and Phylogenetic Analyses of OsBAGs
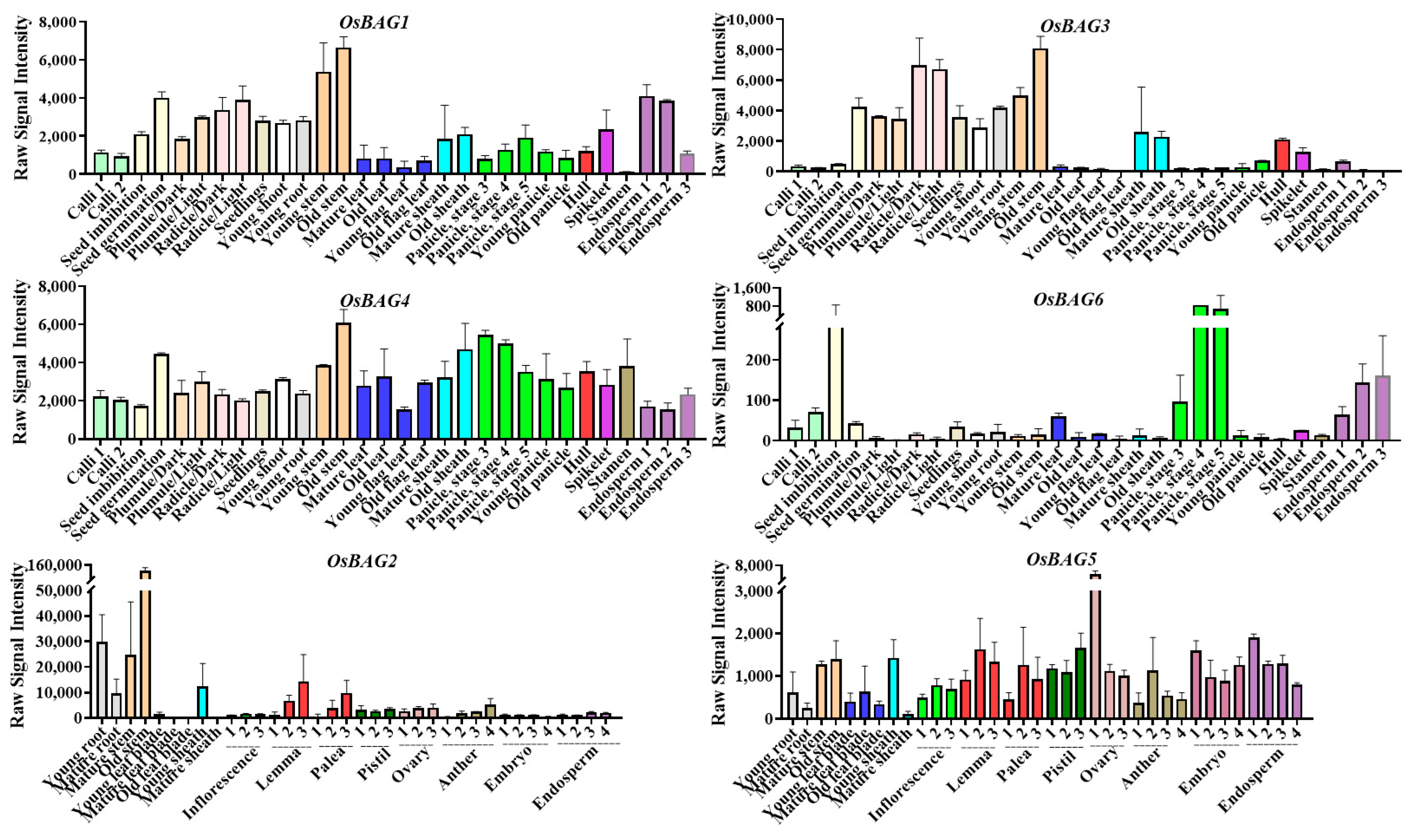
3.3. Expression Patterns of OSBAG Genes in Rice
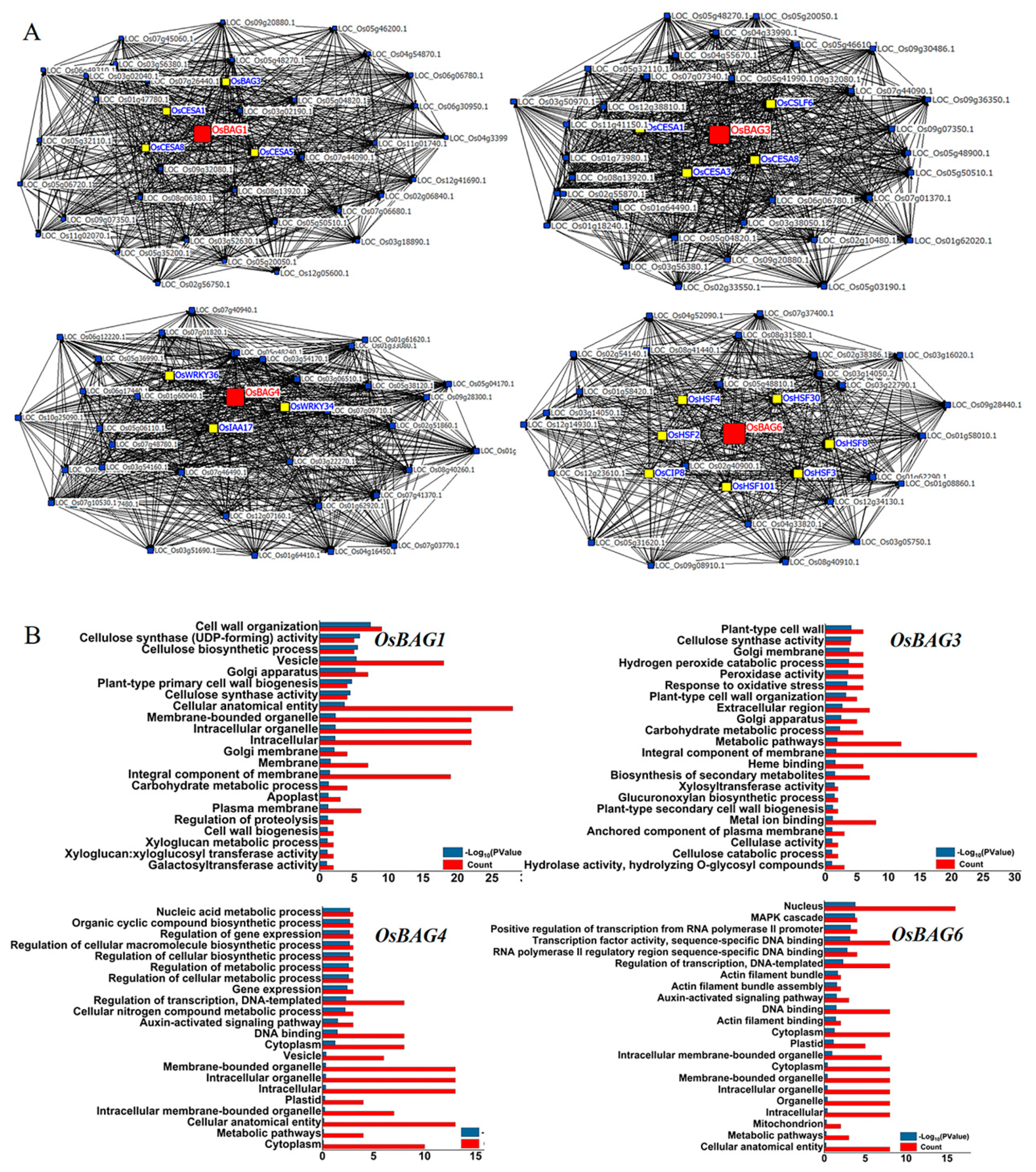
3.4. OsBAG Co-Expression Profiling and Functional Relevance Analysis
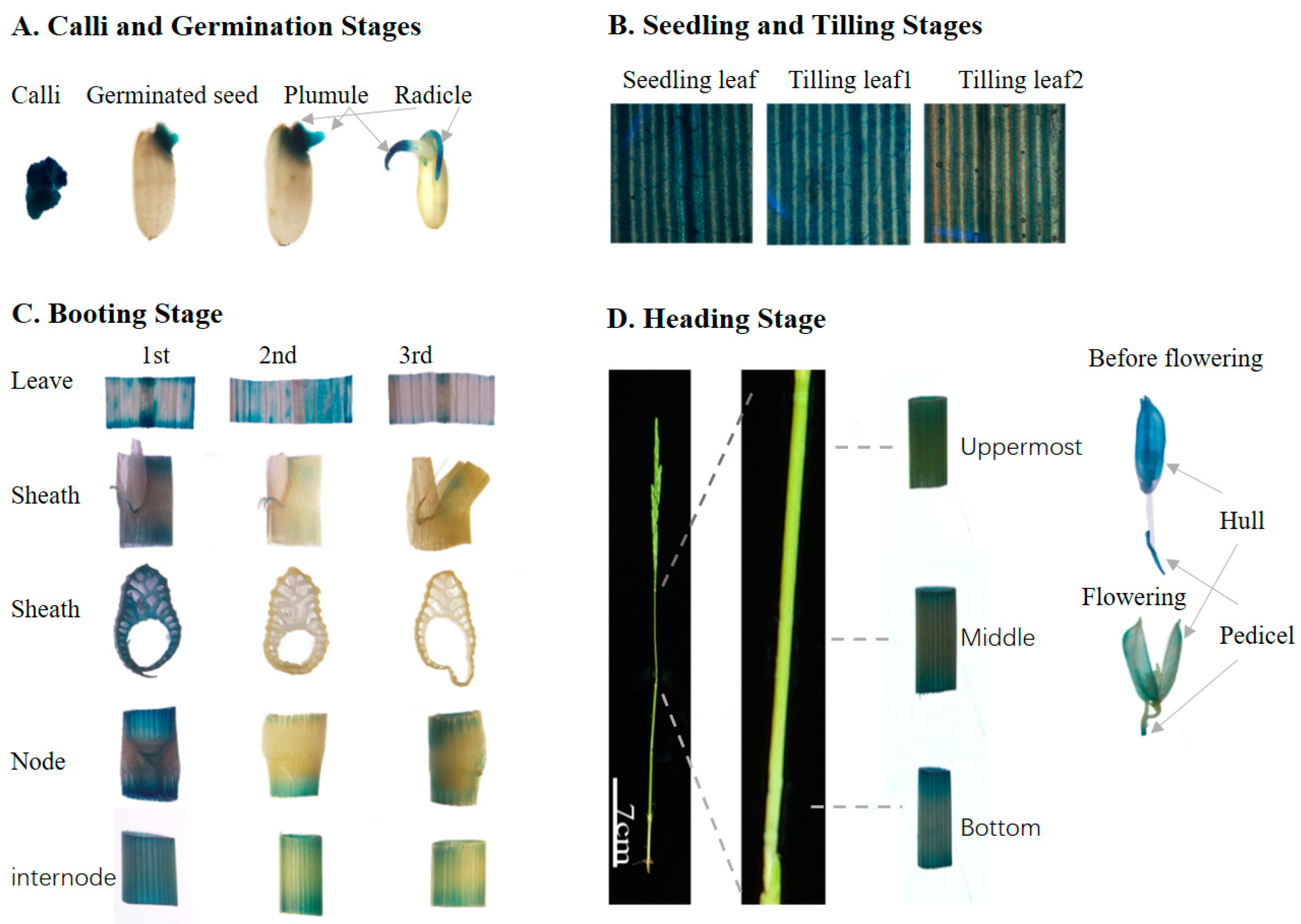
3.5. OsBAG3 Exhibited Young Tissues-Preferential GUS Staining Patterns, Consisting with the Transcriptional Profiling
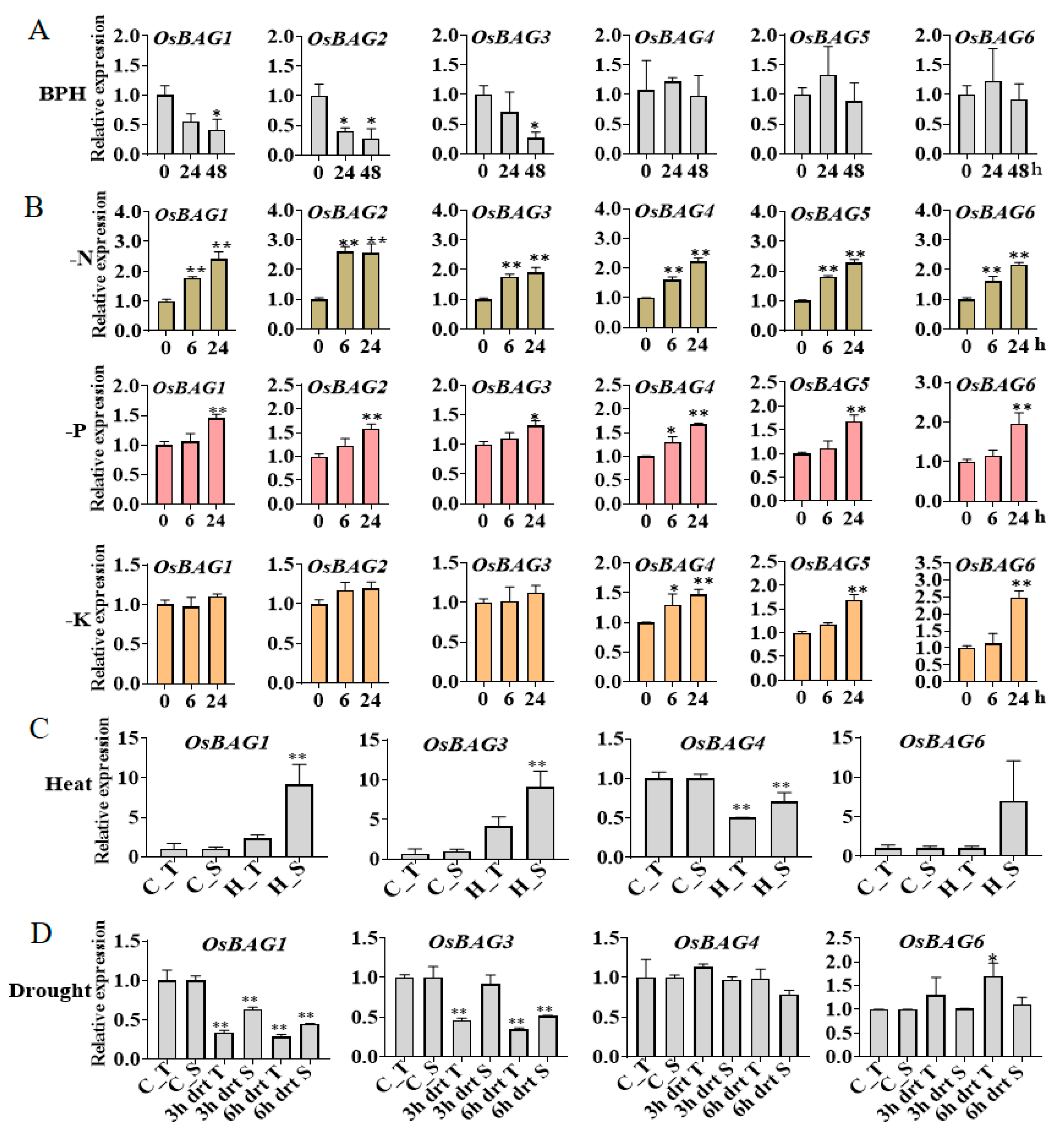
3.6. Expression Profiles of OsBAGs in Responses to Environmental Stimulations
4. Discussion
4.1. The Expression Levels of OsBAG1 and -3 Were in High Similarity but Subtle Differences during the Plant Development
4.2. The Divergent Roles of the Rice BAG Genes in Plant Development and in Responses to Environmental Stimulations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takayama, S.; Reed, J.C. Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 2001, 3, E237–E241. [Google Scholar] [CrossRef]
- Doukhanina, E.V.; Chen, S.; van der Zalm, E.; Godzik, A.; Reed, J.; Dickman, M.B. Identification and Functional Characterization of the BAG Protein Family in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 18793–18801. [Google Scholar] [CrossRef]
- Kabbage, M.; Dickman, M.B. The BAG proteins: A ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef]
- Takayama, S.; Sato, T.; Krajewski, S.; Kochel, K.; Irie, S.; Millan, J.A.; Reed, J.C. Cloning and functional analysis of BAG-1: A novel Bcl-2-binding protein with anti-cell death activity. Cell 1995, 80, 279–284. [Google Scholar] [CrossRef]
- Brive, L.; Takayama, S.; Briknarová, K.; Homma, S.; Ishida, S.K.; Reed, J.C.; Ely, K.R. The Carboxyl-Terminal Lobe of Hsc70 ATPase Domain Is Sufficient for Binding to BAG1. Biochem. Biophys. Res. Commun. 2001, 289, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Zhai, K.; Yang, D.; Yang, W.; Wu, J.; Liu, J.; Pan, W.; Wang, J.; Zhu, X.; Jian, Y.; et al. An E3 Ubiquitin Ligase-BAG Protein Module Controls Plant Innate Immunity and Broad-Spectrum Disease Resistance. Cell Host Microbe 2016, 20, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.S.; Beart, P.M.; Pascoe, C.J.; John, C.A.; Bernard, O. Human Bcl-2 protects against AMPA receptor-mediated apoptosis. J. Neurochem. 2000, 74, 1613–1620. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.H.; Jung, W.Y.; Kang, Y.H.; Kim, J.Y.; Kim, D.G.; Jeong, J.C.; Baek, D.W.; Jin, J.B.; Lee, J.Y.; Kim, M.O.; et al. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 2006, 13, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; He, C.; Zhang, H. The BAG-family proteins in Arabidopsis thaliana. Plant Sci. 2003, 165, 1–7. [Google Scholar] [CrossRef]
- Williams, B.; Kabbage, M.; Britt, R.; Dickman, M.B. AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. USA 2010, 107, 6088–6093. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xing, Y.; Chang, D.; Fang, S.; Cui, B.; Li, Q.; Wang, X.; Guo, S.; Yang, X.; Men, S.; et al. CaM/BAG5/Hsc70 signaling complex dynamically regulates leaf senescence. Sci. Rep. 2016, 6, 31889. [Google Scholar] [CrossRef]
- Li, Y.; Williams, B.; Dickman, M. Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 2017, 214, 695–705. [Google Scholar] [CrossRef]
- Williams, B.; Njaci, I.; Moghaddam, L.; Long, H.; Dickman, M.B.; Zhang, X.; Mundree, S. Trehalose Accumulation Triggers Autophagy during Plant Desiccation. PLoS Genet. 2015, 11, e1005705. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dickman, M. Processing of AtBAG6 triggers autophagy and fungal resistance. Plant Signal. Behav. 2016, 11, e1175699. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kabbage, M.; Liu, W.; Dickman, M.B. Aspartyl Protease-Mediated Cleavage of BAG6 Is Necessary for Autophagy and Fungal Resistance in Plants. Plant Cell 2016, 28, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liu, L.; Lin, Y.; Zu, Y.; Li, L.; Tang, Z. Ethylene Antagonizes Salt-Induced Growth Retardation and Cell Death Process via Transcriptional Controlling of Ethylene-, BAG- and Senescence-Associated Genes in Arabidopsis. Front. Plant Sci. 2016, 7, 696. [Google Scholar] [CrossRef] [PubMed]
- Ghag, S.B.; Shekhawat, U.K.S.; Ganapathi, T.R. Native cell-death genes as candidates for developing wilt resistance in transgenic banana plants. AoB Plants 2014, 6, u37. [Google Scholar] [CrossRef]
- Kobayashi, M.; Takato, H.; Fujita, K.; Suzuki, S. HSG1, a grape Bcl-2-associated athanogene, promotes floral transition by activating CONSTANS expression in transgenic Arabidopsis plant. Mol. Biol. Rep. 2012, 39, 4367–4374. [Google Scholar] [CrossRef]
- Li, X.; Yang, D.; Sun, L.; Li, Q.; Mao, B.; He, Z. The Systemic Acquired Resistance Regulator OsNPR1 Attenuates Growth by Repressing Auxin Signaling through Promoting IAA-Amido Synthase Expression. Plant Physiol. 2016, 172, 546–558. [Google Scholar] [CrossRef]
- Rana, R.M. Identification and characterization of the Bcl-2-associated athanogene (BAG) protein family in rice. Afr. J. Biotechnol. 2011, 11, 88–98. [Google Scholar]
- Wang, J.; Nan, N.; Li, N.; Liu, Y.; Wang, T.; Hwang, I.; Liu, B.; Xu, Z. A DNA Methylation Reader-Chaperone Regulator-Transcription Factor Complex ActivatesOsHKT1;5 Expression during Salinity Stress. Plant Cell 2020, 32, 3535–3558. [Google Scholar] [CrossRef]
- Hoang, T.M.L.; Moghaddam, L.; Williams, B.; Khanna, H.; Dale, J.; Mundree, S.G. Development of salinity tolerance in rice by constitutive-overexpression of genes involved in the regulation of programmed cell death. Front. Plant Sci. 2015, 6, 175. [Google Scholar] [CrossRef]
- Fu, S.; Li, L.; Kang, H.; Yang, X.; Men, S.; Shen, Y. Chronic mitochondrial calcium elevation suppresses leaf senescence. Biochem. Biophys. Res. Commun. 2017, 487, 672–677. [Google Scholar] [CrossRef]
- Ru-Hong, W.Z.L.L. Research Progress of BAG Family Proteins in Plants. J. Agric. Biotechnol. 2018, 26, 176–182. [Google Scholar]
- Guo Ay, Z.; Quin, C.X. GSDS: A Gene Structure Display Server; Yi Chuan: Beijing, China, 2007; Volume 29, pp. 1023–1026. [Google Scholar]
- Hooper, C.M.; Castleden, I.R.; Aryamanesh, N.; Jacoby, R.P.; Millar, A.H. Finding the Subcellular Location of Barley, Wheat, Rice and Maize Proteins: The Compendium of Crop Proteins with Annotated Locations (cropPAL). Plant Cell Physiol. 2016, 57, e9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, L.; Xie, W.; Chen, Y.; Tang, W.; Yang, J.; Ye, R.; Liu, L.; Lin, Y.; Xu, C.; Xiao, J.; et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010, 61, 752–766. [Google Scholar] [CrossRef]
- Usadel, B.R.; Obayashi, T.; Mutwil, M.; Giorgi, F.M.; Bassel, G.W.; Tanimoto, M.; Chow, A.; Steinhauser, D.; Persson, S.; Provart, N. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009, 32, 1633–1651. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lin, J.; Wang, X.; Li, Y.; Bi, F.; Cheng, L.; Huang, F.; Li, R.; Qiu, Y. Fine mapping, candidate genes analysis, and characterization of a brown planthopper (Nilaparvata lugens Stål) resistance gene in the rice variety ARC5984. Euphytica 2020, 216, 13. [Google Scholar] [CrossRef]
- Pabuayon, I.M.; Yamamoto, N.; Trinidad, J.L.; Longkumer, T.; Raorane, M.L.; Kohli, A. Reference genes for accurate gene expression analyses across different tissues, developmental stages and genotypes in rice for drought tolerance. Rice 2016, 9, 32. [Google Scholar] [CrossRef]
- Borah, P.; Sharma, E.; Kaur, A.; Chandel, G.; Mohapatra, T.; Kapoor, S.; Khurana, J.P. Analysis of drought-responsive signalling network in two contrasting rice cultivars using transcriptome-based approach. Sci. Rep. 2017, 7, 42131. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Suppl. 2), W316–W322. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of Total Cellular DNA from Plants, Algae and Fungi; Springer: Dordrecht, The Netherlands, 1994; pp. 183–190. [Google Scholar]
- Wu, C.; Li, X.; Yuan, W.; Chen, G.; Kilian, A.; Li, J.; Xu, C.; Li, X.; Zhou, D.; Wang, S.; et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003, 35, 418–427. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Zou, W.; Feng, Y.; Zhang, M.; Zhang, J.; Tu, F.; Xie, G.; Wang, L.; Wang, Y.; Klie, S.; et al. An integrated genomic and metabolomic framework for cell wall biology in rice. BMC Genom. 2014, 15, 596. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38 (Suppl. 1), D355–D360. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Arakawa, A.; Handa, N.; Ohsawa, N.; Shida, M.; Kigawa, T.; Hayashi, F.; Shirouzu, M.; Yokoyama, S. The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange. Structure 2010, 18, 309–319. [Google Scholar] [CrossRef]
- Coulson, M.; Robert, S.; Saint, R. Drosophilastarvin Encodes a Tissue-Specific BAG-Domain Protein Required for Larval Food Uptake. Genetics 2005, 171, 1799–1812. [Google Scholar] [CrossRef][Green Version]
- Moribe, Y.; Niimi, T.; Yamashita, O.; Yaginuma, T. Samui, a novel cold-inducible gene, encoding a protein with a BAG domain similar to silencer of death domains (SODD/BAG-4), isolated from Bombyx diapause eggs. Eur. J. Biochem. 2001, 268, 3432–3442. [Google Scholar] [CrossRef]
- Alberti, S.; Esser, C.; Hohfeld, J. BAG-1—A nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones 2003, 8, 225–231. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Dickman, M.B.; Wang, Z. Cytoprotective Co-chaperone BcBAG1 Is a Component for Fungal Development, Virulence, and Unfolded Protein Response (UPR) of Botrytis cinerea. Front. Microbiol. 2019, 10, 685. [Google Scholar] [CrossRef] [PubMed]
- Ficklin, S.P.; Luo, F.; Feltus, F.A. The Association of Multiple Interacting Genes with Specific Phenotypes in Rice Using Gene Coexpression Networks. Plant Physiol. 2010, 154, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Kim, Y.; Pham, T.T.M.; Song, S.I.; Kim, J.; Kang, K.Y.; An, G.; Jung, K.; Galbraith, D.W.; Kim, M.; et al. RiceArrayNet: A Database for Correlating Gene Expression from Transcriptome Profiling, and Its Application to the Analysis of Coexpressed Genes in Rice. Plant Physiol. 2009, 151, 16–33. [Google Scholar] [CrossRef] [PubMed]
- Takehisa, H.; Sato, Y.; Antonio, B.A.; Nagamura, Y. Global transcriptome profile of rice root in response to essential macronutrient deficiency. Plant Signal Behav. 2013, 8, e24409. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Antonio, B.; Namiki, N.; Motoyama, R.; Sugimoto, K.; Takehisa, H.; Minami, H.; Kamatsuki, K.; Kusaba, M.; Hirochika, H.; et al. Field transcriptome revealed critical developmental and physiological transitions involved in the expression of growth potential in japonica rice. BMC Plant Biol. 2011, 11, 10. [Google Scholar] [CrossRef]
- Gu, Y.; Kaplinsky, N.; Bringmann, M.; Cobb, A.; Carroll, A.; Sampathkumar, A.; Baskin, T.I.; Persson, S.; Somerville, C.R. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 12866–12871. [Google Scholar] [CrossRef]
- Persson, S.; Wei, H.; Milne, J.; Page, G.P.; Somerville, C.R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 2005, 102, 8633–8638. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Gao, H.; Yuan, M.; Xue, H. TheArabidopsis ARCP Protein, CSI1, Which Is Required for Microtubule Stability, Is Necessary for Root and Anther Development. Plant Cell 2012, 24, 1066–1080. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, C.; Mutwil, M.; Saxe, F.; Eder, M.; Nikoloski, Z.; Persson, S. Large-Scale Co-Expression Approach to Dissect Secondary Cell Wall Formation Across Plant Species. Front. Plant Sci. 2011, 2, 23. [Google Scholar] [CrossRef]
- Lee, H.K.; Hsu, A.K.; Sajdak, J.; Qin, J.; Pavlidis, P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004, 14, 1085–1094. [Google Scholar] [CrossRef]
- Stuart, J.M. A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. Science 2003, 302, 249–255. [Google Scholar] [CrossRef] [PubMed]


| Genes | TIGR Loci | Probsets | CDS/bp | KOME cDNA | Exons | Protein | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | PI | Domain | Molecular Weight (kDa) | Predicted Location (s) | ||||||
| OsBAG1 | LOC_Os09g35630.1 | Os.51065.1.S1_at | 1005 | AK105833 | 4 | 335 | 9.65 | Ubiquitin (PS50053), BAG (PS51035, PF02179) | 35.96 | Mitochondrion. |
| OsBAG2 | LOC_Os08g43270.1 | / | 951 | FP095381 | 2 | 317 | 9.61 | Ubiquitin (PS50053), BAG (PS51035, PF02179) | 34.7 | Cytosol. |
| OsBAG3 | LOC_Os06g03640.1 | Os.10179.1.S1_at | 1020 | AK065197 | 4 | 340 | 9.71 | Ubiquitin (PS50053), BAG (PS51035, PF02179) | 36.43 | Nucleus. |
| OsBAG4 | LOC_Os01g61500.1 | Os.20681.2.A1_at | 789 | AK070208 | 4 | 263 | 5.63 | Ubiquitin (PS50053), BAG (PS51035, PF02179) | 28.78 | Cytosol |
| OsBAG5 | LOC_Os02g48780.1 | / | 642 | AK119930 | 1 | 214 | 5.99 | IQ (PS50096), BAG (PS51035, PF02179) | 23.08 | Cytosol. |
| OsBAG6 | LOC_Os11g31060.1 | OsAffx.19095.1.S1_at | 1368 | FP100206 | 1 | 456 | 4.48 | IQ (PS50096), BAG (PS51035, PF02179) | 44.3 | Nucleus. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Li, J.; Liu, X.; Wei, X.; He, Z.; Hu, L.; Wang, J.; Duan, M.; Xie, G.; Wang, J.; et al. The Divergent Roles of the Rice bcl-2 Associated Athanogene (BAG) Genes in Plant Development and Environmental Responses. Plants 2021, 10, 2169. https://doi.org/10.3390/plants10102169
Zhou H, Li J, Liu X, Wei X, He Z, Hu L, Wang J, Duan M, Xie G, Wang J, et al. The Divergent Roles of the Rice bcl-2 Associated Athanogene (BAG) Genes in Plant Development and Environmental Responses. Plants. 2021; 10(10):2169. https://doi.org/10.3390/plants10102169
Chicago/Turabian StyleZhou, Hailian, Jiaying Li, Xueyuan Liu, Xiaoshuang Wei, Ziwei He, Lihua Hu, Jibin Wang, Mingzheng Duan, Guosheng Xie, Jihong Wang, and et al. 2021. "The Divergent Roles of the Rice bcl-2 Associated Athanogene (BAG) Genes in Plant Development and Environmental Responses" Plants 10, no. 10: 2169. https://doi.org/10.3390/plants10102169
APA StyleZhou, H., Li, J., Liu, X., Wei, X., He, Z., Hu, L., Wang, J., Duan, M., Xie, G., Wang, J., & Wang, L. (2021). The Divergent Roles of the Rice bcl-2 Associated Athanogene (BAG) Genes in Plant Development and Environmental Responses. Plants, 10(10), 2169. https://doi.org/10.3390/plants10102169







