Accelerating the Development of Heat Tolerant Tomato Hybrids through a Multi-Traits Evaluation of Parental Lines Combining Phenotypic and Genotypic Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phenotypic Evaluation of Parental Lines
2.2. Molecular Screening for Resistance Genes in Parental Lines
2.3. Genomic Characterization of Parental Lines
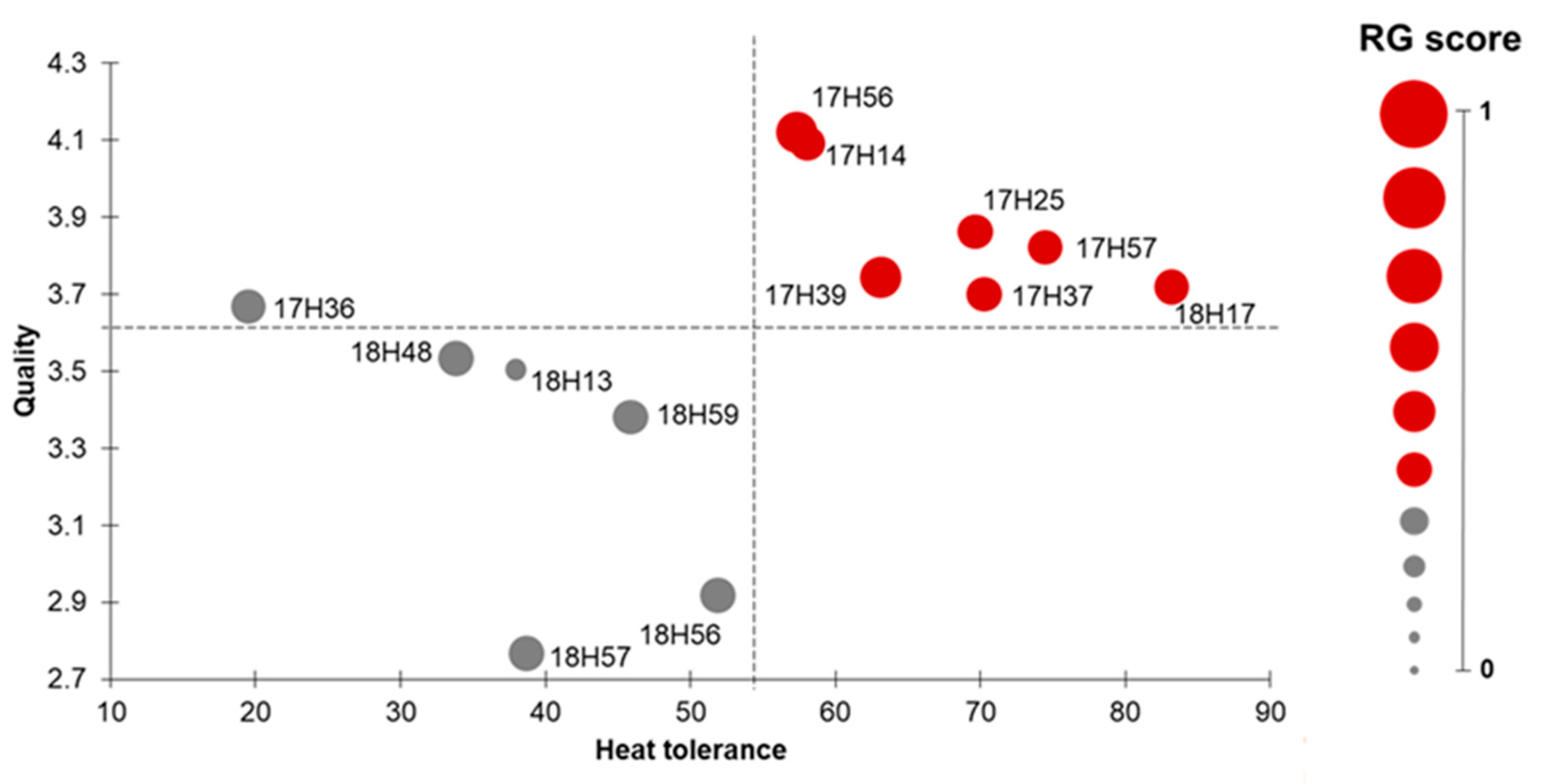
2.4. Multilevel Evaluation of F1 Hybrids
3. Materials and Methods
3.1. Plant Material
3.2. Phenotypic Evaluation
3.3. Statistical Analysis
3.4. Genomic Analysis
3.5. Marker Design and Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sato, S.; Peet, M.M.; Gardner, R.G. Altered flower retention and developmental patterns in nine tomato cultivars under elevated temperature. Sci. Hortic. 2004, 101, 95–101. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Fujita, M.; Ahmad, P.; Chandna, R.; Prasad, M.; Ozturk, M. Enhancing plant productivity under salt stress: Relevance of poly-omics. Salt Stress Plants 2013, 113–156. [Google Scholar] [CrossRef]
- Aliberti, A.; Olivieri, F.; Graci, S.; Rigano, M.M.; Barone, A.; Ruggieri, V. Genomic Dissection of a Wild Region in a Superior Solanum pennellii Introgression Sub-Line with High Ascorbic Acid Accumulation in Tomato Fruit. Genes 2020, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Farinetti, A.; Zurlo, V.; Manenti, A.; Coppi, F.; Mattioli, A.V. Mediterranean diet and colorectal cancer: A systematic review. Nutrition 2017, 43, 83–88. [Google Scholar] [CrossRef]
- Paupière, M.J.; van Haperen, P.; Rieu, I.; Visser, R.G.; Tikunov, Y.M.; Bovy, A.G. Screening for pollen tolerance to high temperatures in tomato. Euphytica 2017, 213, 130. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, R.S.; Kumar, L.; Shabani, F.; Picanço, M.C. Risk of spread of tomato yellow leaf curl virus (TYLCV) in tomato crops under various climate change scenarios. Agric. Syst. 2019, 173, 524–535. [Google Scholar] [CrossRef]
- Lapidot, M.; Polston, J.E. Resistance to Tomato yellow leaf curl virus in tomato. In Natural Resistance Mechanisms of Plants to Viruses; Springer: Berlin/Heidelberg, Germany, 2006; pp. 503–520. [Google Scholar]
- Trudgill, D.; Tzortzakakis, E. A comparative study of the thermal time requirements for embryogenesis in Meloidogyne javanica and M. incognita. Nematology 2005, 7, 313–315. [Google Scholar] [CrossRef] [Green Version]
- Panthee, D.R. ‘Mountain Crown’: Late Blight and Tomato mosaic virus-resistant Plum Hybrid Tomato and Its Parent, NC 1 Plum. HortScience 2020, 1, 1–2. [Google Scholar] [CrossRef]
- Prabhandakavi, P.; Pogiri, R.; Kumar, R.; Acharya, S.; Esakky, R.; Chakraborty, M.; Pinnamaneni, R.; Palicherla, S.R. Pyramiding Ty-1/Ty-3, Ty-2, ty-5 and ty-6 genes into tomato hybrid to develop resistance against tomato leaf curl viruses and recurrent parent genome recovery by ddRAD sequencing method. J. Plant Biochem. Biotechnol. 2020, 30, 462–476. [Google Scholar] [CrossRef]
- Vijeth, S.; Dhaliwal, M.S.; Jindal, S.K.; Sharma, A. Evaluation of tomato hybrids for resistance to leaf curl virus disease and for high-yield production. Hortic.Environ. Biotechnol. 2018, 59, 699–709. [Google Scholar] [CrossRef]
- Hamilton, J.P.; Sim, S.C.; Stoffel, K.; Van Deynze, A.; Buell, C.R.; Francis, D.M. Single nucleotide polymorphism discovery in cultivated tomato via sequencing by synthesis. Plant Genome 2012, 5, 17–29. [Google Scholar] [CrossRef]
- Bauchet, G.; Grenier, S.; Samson, N.; Segura, V.; Kende, A.; Beekwilder, J.; Cankar, K.; Gallois, J.L.; Gricourt, J.; Bonnet, J. Identification of major loci and genomic regions controlling acid and volatile content in tomato fruit: Implications for flavor improvement. New Phytol. 2017, 215, 624–641. [Google Scholar]
- Ruggieri, V.; Calafiore, R.; Schettini, C.; Rigano, M.M.; Olivieri, F.; Frusciante, L.; Barone, A. Exploiting genetic and genomic resources to enhance heat-tolerance in tomatoes. Agronomy 2019, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Poland, J.A.; Rife, T.W. Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivieri, F.; Calafiore, R.; Francesca, S.; Schettini, C.; Chiaiese, P.; Rigano, M.M.; Barone, A. High-Throughput Genotyping of Resilient Tomato Landraces to Detect Candidate Genes Involved in the Response to High Temperatures. Genes 2020, 11, 626. [Google Scholar]
- Sacco, A.; Ruggieri, V.; Parisi, M.; Festa, G.; Rigano, M.M.; Picarella, M.E.; Mazzucato, A.; Barone, A. Exploring a tomato landraces collection for fruit-related traits by the aid of a high-throughput genomic platform. PLoS ONE 2015, 10, e0137139. [Google Scholar] [CrossRef] [Green Version]
- Ruggieri, V.; Francese, G.; Sacco, A.; D’Alessandro, A.; Rigano, M.M.; Parisi, M.; Milone, M.; Cardi, T.; Mennella, G.; Barone, A. An association mapping approach to identify favourable alleles for tomato fruit quality breeding. BMC Plant Biol. 2014, 14, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tranchida-Lombardo, V.; Aiese Cigliano, R.; Anzar, I.; Landi, S.; Palombieri, S.; Colantuono, C.; Bostan, H.; Termolino, P.; Aversano, R.; Batelli, G. Whole-genome re-sequencing of two Italian tomato landraces reveals sequence variations in genes associated with stress tolerance, fruit quality and long shelf-life traits. DNA Res. 2018, 25, 149–160. [Google Scholar] [CrossRef] [Green Version]
- Scarano, A.; Olivieri, F.; Gerardi, C.; Liso, M.; Chiesa, M.; Chieppa, M.; Frusciante, L.; Barone, A.; Santino, A.; Rigano, M.M. Selection of tomato landraces with high fruit yield and nutritional quality under elevated temperatures. J. Sci. Food Agric. 2020, 100, 2791–2799. [Google Scholar] [CrossRef]
- Gimenez, E.; Salinas, M.; Manzano-Agugliaro, F. Worldwide research on plant defense against biotic stresses as improvement for sustainable agriculture. Sustainability 2018, 10, 391. [Google Scholar] [CrossRef] [Green Version]
- Beckles, D.M. Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit. Postharvest Biol. Technol. 2012, 63, 129–140. [Google Scholar] [CrossRef]
- Kader, A.; Lyons, J.; Morris, L. Postharvest responses of vegetables to preharvest field temperature. HortScience 1974, 9, 523–527. [Google Scholar]
- Janda, T.; Khalil, R.; Tajti, J.; Pál, M.; Darkó, É. Responses of young wheat plants to moderate heat stress. Acta Physiol. Plant. 2019, 41, 137. [Google Scholar] [CrossRef] [Green Version]
- Gorovits, R.; Moshe, A.; Ghanim, M.; Czosnek, H. Recruitment of the host plant heat shock protein 70 by Tomato yellow leaf curl virus coat protein is required for virus infection. PLoS ONE 2013, 8, e70280. [Google Scholar] [CrossRef] [PubMed]
- Schuster, D.J.; Mann, R.S.; Toapanta, M.; Cordero, R.; Thompson, S.; Cyman, S.; Shurtleff, A.; Morris, R.F., II. Monitoring neonicotinoid resistance in biotype B of Bemisia tabaci in Florida. Pest Manag. Sci. Former. Pestic. Sci. 2010, 66, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Foolad, M.R.; Panthee, D.R. Marker-assisted selection in tomato breeding. Crit. Rev. Plant Sci. 2012, 31, 93–123. [Google Scholar] [CrossRef]
- Jung, J.; Kim, H.J.; Lee, J.M.; Oh, C.S.; Lee, H.-J.; Yeam, I. Gene-based molecular marker system for multiple disease resistances in tomato against Tomato yellow leaf curl virus, late blight, and verticillium wilt. Euphytica 2015, 205, 599–613. [Google Scholar] [CrossRef]
- Kim, M.; Park, Y.; Lee, J.; Sim, S.-C. Development of molecular markers for Ty-2 and Ty-3 selection in tomato breeding. Sci. Hortic. 2020, 265, 109230. [Google Scholar] [CrossRef]
- Santos, D.; da Silva, P.M.; Abrantes, I.; Maleita, C. Tomato Mi-1.2 gene confers resistance to Meloidogyne luci and M. ethiopica. Eur. J. Plant Pathol. 2020, 156, 571–580. [Google Scholar] [CrossRef]
- Ren, Z.; You, Z.; Munir, S.; Zhang, Y.; Li, H.; Zhang, J.; Wang, T.; Zheng, W.; Ye, Z. Development of a highly specific co-dominant marker for genotyping the Ph-3 (tomato late blight resistance) locus by comparing cultivated and wild ancestor species. Mol. Breed. 2019, 39, 1–8. [Google Scholar] [CrossRef]
- Vela-Avitúa, S.; Meuwissen, T.H.; Luan, T.; Ødegård, J. Accuracy of genomic selection for a sib-evaluated trait using identity-by-state and identity-by-descent relationships. Genet. Sel. Evol. 2015, 47, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, C.M.; Paytuvi-Gallart, A.; Di Donato, A.; Sundesha, V.; Andolfo, G.; Aiese Cigliano, R.; Sanseverino, W.; Ercolano, M.R. PRGdb 3.0: A comprehensive platform for prediction and analysis of plant disease resistance genes. Nucleic Acids Res. 2017, 46, D1197–D1201. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Cui, J.; Hou, X.; Yang, G.; Xiao, Y.; Han, L.; Meng, J.; Luan, Y. Sl-lncRNA15492 interacts with Sl-miR482a and affects Solanum lycopersicum immunity against Phytophthora infestans. Plant J. 2020, 103, 1561–1574. [Google Scholar] [CrossRef]
- Chellappan, P.; Qiyue, M.; Reza, S.; Stewart, K.S.; Hutton, S.F.; Scott, J.W.; Zhangjun, F.; Kai-Shu, L. Comprehensive transcriptome analysis and functional characterization of PR-5 for its involvement in tomato Sw-7 resistance to tomato spotted wilt tospovirus. Sci. Rep. 2019, 9, 7673. [Google Scholar]
- Catanzariti, A.M.; Do, H.T.; Bru, P.; de Sain, M.; Thatcher, L.F.; Rep, M.; Jones, D.A. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR 1 and SERK 3/BAK 1. Plant J. 2017, 89, 1195–1209. [Google Scholar] [CrossRef] [Green Version]
- Devran, Z.; Kahveci, E.; Hong, Y.; Studholme, D.J.; Tör, M. Identifying molecular markers suitable for Frl selection in tomato breeding. Theor. Appl. Genet. 2018, 131, 2099–2105. [Google Scholar] [CrossRef] [Green Version]
- Solieman, T.; El-Gabry, M.; Abido, A. Heterosis, potence ratio and correlation of some important characters in tomato (Solanum lycopersicum L.). Sci. Hortic. 2013, 150, 25–30. [Google Scholar] [CrossRef]
- Tamta, S.; Singh, J. Heterosis in tomato for growth and yield traits. Int. J. Veg. Sci. 2018, 24, 169–179. [Google Scholar] [CrossRef]
- Yadav, S.K.; Singh, B.; Baranwal, D.; Solankey, S. Genetic study of heterosis for yield and quality components in tomato (Solanum lycopersicum). Afr. J. Agric. Res. 2013, 8, 5585–5591. [Google Scholar]
- Avdikos, I.D.; Tagiakas, R.; Tsouvaltzis, P.; Mylonas, I.; Xynias, I.N.; Mavromatis, A.G. Comparative Evaluation of Tomato Hybrids and Inbred Lines for Fruit Quality Traits. Agronomy 2021, 11, 609. [Google Scholar] [CrossRef]
- Kumari, S.; Sharma, M.K. Exploitation of heterosis for yield and its contributing traits in tomato, Solanum lycopersicum L. Int. J. Farm Sci. 2011, 1, 45–55. [Google Scholar]
- Hannan, M.; Ahmed, M.; Razvy, M.; Karim, R.; Khatun, M.; Haydar, A.; Hossain, M.; Roy, U. Heterosis and correlation of yield and yield components in tomato (Lycopersicon esulentum Mill.). Am.-Eurasian J. Sci. Res. 2007, 2, 146–150. [Google Scholar]
- George, B.; Kaur, C.; Khurdiya, D.; Kapoor, H. Antioxidants in tomato (Lycopersium esculentum) as a function of genotype. Food Chem. 2004, 84, 45–51. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, s13742-015. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene | Mutated Genotype (no. SNPs) | Predicted Effect | Protein Function |
|---|---|---|---|
| Solyc01g014520 | E42 | missense_variant | Receptor-like protein kinase |
| Solyc01g074010 | E11, E42 | missense_variant | Protein kinase domain |
| Solyc01g080880 | E20, E42, PDVIT | missense_variant | Protein kinase domain |
| Solyc01g087200 | E42, E55 | missense_variant | Disease resistance protein |
| Solyc02g070000 | E11, E36, E42, E55 | missense_variant | Leucine-rich receptor-like protein kinase family protein |
| Solyc02g078780 | PDVIT | missense_variant | Protein STRUBBELIG-RECEPTOR FAMILY 3 |
| Solyc02g093100 | E7, E11, E36, E42, E45, E48, E55, PDVIT | missense_variant | Leucine-rich repeat protein kinase family protein |
| Solyc04g007030 | E11, E36, E42 | missense_variant | Disease resistance protein |
| Solyc04g008650 | E7. E11, E20, E36, E42, E45 | missense_variant | Inactive leucine-rich repeat receptor-like serinethreonine-protein kinase |
| Solyc04g014400 | E11, E36, E42, E55 | missense_variant | LRR receptor-like serine/threonine-protein kinase GSO1 |
| Solyc04g015130 | E42, E55, PDVIT | missense_variant | Protein kinase G11A |
| Solyc04g049400 | E42, E55, PDVIT | missense_variant | Protein kinase domain |
| Solyc04g054200 | E42, E55, PDVIT | missense_variant | Leucine-rich receptor-like protein kinase family protein |
| Solyc04g057930 | E11 (2), E42 (2), E55 (2) E109, PDVIT (2) | missense_variant | Pkinase domain-containing protein/Usp domain-containing protein |
| Solyc04g074270 | E11 (3), PDVIT (3) | missense_variant | Outer arm dynein light chain 1 |
| Solyc04g082510 | E55 (2) | missense_variant; stop_gained | Protein kinase |
| Solyc04g082620 | E55 (2) | missense_variant | Serine/threonine-protein kinase/endoribonuclease IRE1a |
| Solyc05g009740 | E7, E11 (2), E20, E45, E109 (2) | missense_variant | Disease resistance protein |
| Solyc05g009800 | E11, E109 | missense_variant | Leucine-rich repeat receptor-like serine/threonine-protein kinase |
| Solyc05g013280 | LA3120 (2) | missense_variant | Pseudomonas resistance |
| Solyc05g050700 | E11, E36, E42, E109, PDVIT | missense_variant | Leucine-rich repeat protein |
| Solyc05g051050 | E11, E36, E42, E109 | missense_variant | Shaggy-related protein kinase theta |
| Solyc05g053010 | E11, E109, PDVIT | missense_variant | Lectin receptor kinase |
| Solyc05g053930 | E55 | missense_variant | Protein kinase APK1B, chloroplastic |
| Solyc05g054340 | E55, PDVIT | missense_variant | Plant resistance protein |
| Solyc06g036470 | E55 | missense_variant | G-type lectin S-receptor-like serine/threonine-protein kinase |
| Solyc06g060690 | E11, E55, PDVIT | stop_gained | Protein kinase superfamily protein |
| Solyc06g068920 | E7, E20, E36, E42, E48 | stop_lost | Protein kinase domain |
| Solyc06g072340 | E11, E36, E45, E55, E109 | missense_variant | Protein kinase domain |
| Solyc07g007140 | E11, E42, E48, E55, E109 | missense_variant | MAP kinase kinase kinase 44 |
| Solyc07g053010 | E42, PDVIT | missense_variant | Disease resistance protein |
| Solyc07g053300 | E42 | missense_variant | ABC transporter G family member 10 |
| Solyc07g053910 | E55 | missense_variant | Protein kinase domain |
| Solyc07g055670 | E11, E42, E45, E48, E55, PDVIT | missense_variant | Lectin-domain receptor-like kinase |
| Solyc07g062040 | E42, E55 | stop_gained | Receptor-like serine/threonine-protein kinase |
| Solyc08g081210 | E20, E36, E42, PDVIT | missense_variant | MAP kinase kinase kinase 66 |
| Solyc09g005080 | E36, PDVIT | missense_variant | Verticillium resistance |
| Solyc09g007110 | E36 (5), E45 (5), E48 (5), PDVIT (5) | missense_variant | Leucine-rich receptor-like protein kinase family protein |
| Solyc09g011320 | E11, E109 | missense_variant | Serine/threonine-protein kinase |
| Solyc09g074240 | E11, E36, E109, PDVIT | missense_variant | Protein kinase domain |
| Solyc09g091580 | E11, E55 | missense_variant | Protein kinase domain |
| Solyc09g091990 | E42, E48, PDVIT | missense_variant | Receptor like protein kinase S.2 |
| Solyc10g005140 | E55 | missense_variant | probably inactive receptor-like protein kinase At2g46850 |
| Solyc10g084390 | E55 | missense_variant | Protein kinase superfamily protein |
| Solyc11g007280 | E42 | missense_variant | Pleiotropic drug resistance protein 2 |
| Solyc11g011080 | E11 (3), E36 (3), E109 (3) | missense_variant | Disease resistance protein (TIR-NBS-LRR class) |
| Solyc11g011090 | E11 (2), E36, E109 (2) | missense_variant | Disease resistance protein (TIR-NBS-LRR class) |
| Solyc11g011180 | E11, E36, E109 | missense_variant | Lrr receptor-like serinethreonine-protein kinase gso1 |
| Solyc11g013880 | E11 | missense_variant | G-type lectin S-receptor-like serine/threonine-protein kinase |
| Solyc11g018690 | E11, E42, PDVIT | missense_variant | ABC transporter G family member 25 |
| Solyc11g020230 | E11, PDVIT | missense_variant | Serine/threonine-protein kinase-like protein CCR4 |
| Solyc11g020280 | E11, PDVIT | missense_variant | Receptor-like protein kinase |
| Solyc11g033270 | E11, PDVIT | missense_variant | MAP kinase kinase kinase 82 |
| Solyc11g042990 | E11, PDVIT | missense_variant | Kinase protein |
| Solyc11g056680 | E11, PDVIT | missense_variant | Leucine-rich repeat receptor-like protein |
| Solyc12g016220 | E55 | missense_variant | Disease resistance protein |
| Solyc12g021280 | E11, E36, E45, E48, E55 | missense_variant | Serine/threonine-protein kinase STN7, chloroplastic |
| Genotype | Heat Tolerance | Fruit Quality 3 | Genetic Distance 4 | Variability In Prg Database 5 | Resistance Allele | |
|---|---|---|---|---|---|---|
| FS 1 | YP 2 | |||||
| E7 | HH | M | M | L | L | Mi-1.2 |
| E11 | M | M | M | M | H | Mi-1.2, Ph-3, Ve-1 |
| E20 | M | M | M | L | L | Mi-1.2 |
| E36 | H | H | M | L | M | Mi-1.2, Ph-3 |
| E42 | M | M | M | H | H | Mi-1.2, Ve-1 |
| E45 | M | M | H | L | L | Mi-1.2, Ph-3 |
| E48 | M | M | M | L | L | Mi-1.2, Ph-3 |
| E55 | H | M | L | M | M | Mi-1.2, Ph-3, Ty-3 |
| E103 | HH | L | H | - | - | Mi-1.2, Tm-2 |
| E109 | M | M | M | M | L | Mi-1.2, Ph-3, Ve-1 |
| E111 | M | L | H | - | - | Mi-1.2, Ph-3 |
| LA2662 | H | H | L | L | - | Mi-1.2 |
| LA3120 | H | H | L | L | L | Mi-1.2 |
| PDLUC | - | - | H | - | - | Mi-1.2, Ph-3, Ty-3 |
| PDVIT | H | M | H | H | H | Mi-1.2, Tm-2, Ty-3 |
| Hybrid | Trait | |||||
|---|---|---|---|---|---|---|
| TNF | FW | YP | TSSC | TA | TSSC/TA | |
| 17H14 | −1.37 | −21.44 | 8.00 | 7.16 | 14.41 | −6.02 |
| 17H25 | 85.10 | 9.34 | 127.76 | 7.03 | 20.13 | −15.17 |
| 17H36 | −30.35 | −10.65 | −24.21 | 7.50 | −0.98 | −2.93 |
| 17H37 | 49.28 | 15.08 | 76.14 | 2.22 | −12.74 | 15.17 |
| 17H39 | 128.27 | 27.05 | 183.09 | −1.12 | −16.92 | 16.92 |
| 17H56 | 69.83 | 22.42 | 120.76 | 5.49 | −17.80 | 35.21 |
| 17H57 | 136.32 | 18.49 | 192.65 | 3.63 | −12.12 | 20.04 |
| 18H13 | −11.71 | 8.12 | −5.52 | 5.88 | 16.54 | −13.52 |
| 18H17 | 79.27 | 4.99 | 75.77 | 12.48 | 32.23 | −16.01 |
| 18H48 | 9.60 | 12.70 | 22.04 | 9.45 | 2.72 | 18.94 |
| 18H56 | 80.34 | 13.33 | 126.21 | 0.87 | −6.18 | 9.95 |
| 18H57 | −4.79 | −31.29 | −8.24 | 1.32 | −20.79 | 22.71 |
| 18H59 | −6.97 | −24.50 | 47.79 | 7.52 | 20.64 | −8.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olivieri, F.; Graci, S.; Francesca, S.; Rigano, M.M.; Barone, A. Accelerating the Development of Heat Tolerant Tomato Hybrids through a Multi-Traits Evaluation of Parental Lines Combining Phenotypic and Genotypic Analysis. Plants 2021, 10, 2168. https://doi.org/10.3390/plants10102168
Olivieri F, Graci S, Francesca S, Rigano MM, Barone A. Accelerating the Development of Heat Tolerant Tomato Hybrids through a Multi-Traits Evaluation of Parental Lines Combining Phenotypic and Genotypic Analysis. Plants. 2021; 10(10):2168. https://doi.org/10.3390/plants10102168
Chicago/Turabian StyleOlivieri, Fabrizio, Salvatore Graci, Silvana Francesca, Maria Manuela Rigano, and Amalia Barone. 2021. "Accelerating the Development of Heat Tolerant Tomato Hybrids through a Multi-Traits Evaluation of Parental Lines Combining Phenotypic and Genotypic Analysis" Plants 10, no. 10: 2168. https://doi.org/10.3390/plants10102168
APA StyleOlivieri, F., Graci, S., Francesca, S., Rigano, M. M., & Barone, A. (2021). Accelerating the Development of Heat Tolerant Tomato Hybrids through a Multi-Traits Evaluation of Parental Lines Combining Phenotypic and Genotypic Analysis. Plants, 10(10), 2168. https://doi.org/10.3390/plants10102168








