1. Introduction
Alfalfa (
Medicago sativa L.) is one of the most widely grown and valuable perennial forage legumes, with an estimated global cropping area of over 30 million hectares. This popularity stems from the high nutritional value, palatability, environmental adaptability, and biomass yield of alfalfa, as well as its low fertilizer requirements due to its ability to fix nitrogen through symbiosis with rhizobia [
1]. The demand for ruminant products such as meat and milk is expected to grow substantially in coming years due to our expanding population and affluence, and high levels of forage crop production will therefore be a necessity [
2]. While alfalfa is relatively drought tolerant compared to many other crop species as a result of the typical presence of a deep tap root, its production depends upon irrigation in many growing regions, and it consumes a particularly high amount of water due to its long growing season and dense canopy [
3]. Unfortunately, there is a progressively limited supply of water for irrigation [
4], as well as an escalation in the severity and frequency of drought events due to climate change [
5], which will negatively impact alfalfa productivity [
6]. As such, there is a vital need to develop alfalfa cultivars that use water more efficiently and/or exhibit improved drought resilience compared to current varieties in a timely manner [
7,
8].
Plants respond to drought stress using a variety of physiological, cellular, and molecular processes, with the aim of enhancing their ability to withstand or recover from such challenging conditions. At the molecular level, drought stress leads to an increase in the production of reactive oxygen species (ROS) that act as important signaling molecules in the abiotic stress response. However, when present at levels above a particular threshold, they can be detrimental to plant cells due to resulting lipid peroxidation, as well as the degradation of nucleic acids and proteins [
9]. In order to minimize such damage, plants tend to enhance the production/activity of various enzymatic and non-enzymatic antioxidants under water deficit as a means of scavenging ROS and maintaining redox homeostasis [
10]. ROS can also influence the production of certain phytohormones, which then often function through an interplay with the ROS signaling cascade [
11]. Abscisic acid (ABA) in particular is known to be a key signaling hormone under drought stress in plants, where its accumulation leads to stomatal closure and the regulation of numerous transcription factors [
12]. The differential activity of these ABA-responsive transcription factors, as well as the transcriptional regulation of many genes that are modulated in ABA-independent pathways, then leads to an increased accumulation of various products that contribute to stress tolerance, including protective proteins such as late embryogenesis abundant (LEA) proteins, osmoprotectants such as proline, glycine betaine, and trehalose, and the aforementioned antioxidant system components [
13]. Although this general framework for the cascade of events that occurs following plant exposure to drought stress has been elucidated, the complex regulatory processes that coordinate these responses have yet to be established in full at the genetic level, and precise mechanisms can differ between plant species and genotypes. Above and beyond drought tolerance mechanisms, plants can also make use of drought avoidance, which typically entails a short life cycle and/or developmental plasticity, and drought escape, which tends to involve enhanced water uptake and/or reduced water loss [
13].
Until fairly recently, the enhancement of drought resilience has not been a major focus of alfalfa breeding efforts, and the scope of genetic variation and mechanisms responsible for this trait are not fully known [
14]. While conventional breeding programs have begun to focus on the improvement of this trait in alfalfa [
2,
15], and a growing number of studies are dedicated towards the enhancement of drought tolerance in alfalfa using a transgenic approach [
16,
17,
18,
19,
20], such efforts are complicated by a lack of understanding of the exact mechanisms by which alfalfa senses, reacts, and adapts to water deficit.
Medicago sativa subsp.
falcata, which can be found in diploid or tetraploid form, is a close relative of tetraploid
M. sativa subsp.
sativa. While the
sativa subspecies is the most commonly cultivated subspecies of alfalfa, current-day varieties have been bred using introgressions from, and hybridization with, other subspecies, including
falcata, and thus the delineation between subspecies is not necessarily quite so obvious [
2]. In any case,
M. sativa subsp.
falcata is well known to exhibit superior cold tolerance compared to the
sativa subspecies [
21,
22], and a small number of studies have also shown it to be more resilient to other abiotic stresses such as drought [
22,
23,
24,
25]. While
M. sativa subsp.
falcata germplasm has been used for many years in alfalfa breeding programs as a means of harnessing its enhanced abiotic stress tolerance, it typically exhibits relatively low productivity and persistence [
15,
26,
27], which has made its use in this context challenging.
The drought resilience of the
falcata subspecies has been suggested to result from several physiological and biochemical characteristics, including reduced stomatal density and conductance, delayed leaf senescence, increased root length, and a greater accumulation of carbohydrate osmoprotectants (raffinose, myo-inositol, and galactinol) and flavonoid antioxidants [
24,
25]. In addition, various studies have demonstrated that the heterologous expression of certain genes from
M. sativa subsp.
falcata genotypes, including those encoding Universal Stress Protein 1 (
MfUSP1) [
19], galactinol synthase (
MfGolS1) [
16],
myo-inositol phosphate synthase (
MfMIPS1) [
28], and late embryogenesis abundant protein 3 (
MfLEA3) [
20], in tobacco led to enhanced tolerance to osmotic or dehydration stress in transgenic lines. Although comparative transcriptional responses between a drought-sensitive
sativa (Chilean) and a drought-tolerant
falcata (Wisfal) variety have been assessed under drought stress using microarray analysis previously [
24], this type of evaluation has not been extended to other accessions/genotypes as of yet. Therefore, we sought to carry out further physiological, biochemical, and transcriptomic comparisons of distinct
falcata and
sativa accessions exhibiting differential levels of drought tolerance to provide new insight into the molecular mechanisms driving the resilience of
falcata to water-deficient environmental conditions.
3. Discussion
Previously, several
falcata accessions were shown to exhibit superior drought tolerance or water use efficiency compared to the
sativa subspecies; a phenomenon that has been suggested to be related to characteristics such as differences in root morphology, reduced stomatal density and conductance, delayed leaf senescence under drought, and increased RSO and (iso)flavonoid accumulation [
22,
24,
25,
26]. However, relatively few
falcata accessions have been examined in-depth in terms of drought tolerance, and only a single
falcata accession (Wisfal) has been comparatively assessed at the transcriptional level in this context thus far [
24]. As such, we sought to compare various physiological, biochemical, and transcriptional characteristics in ‘sativa’ (Beaver) and ‘falcata’ (PI 641381) genotypes as a means of furthering our knowledge regarding the mechanisms driving improved resilience to water deficit in alfalfa.
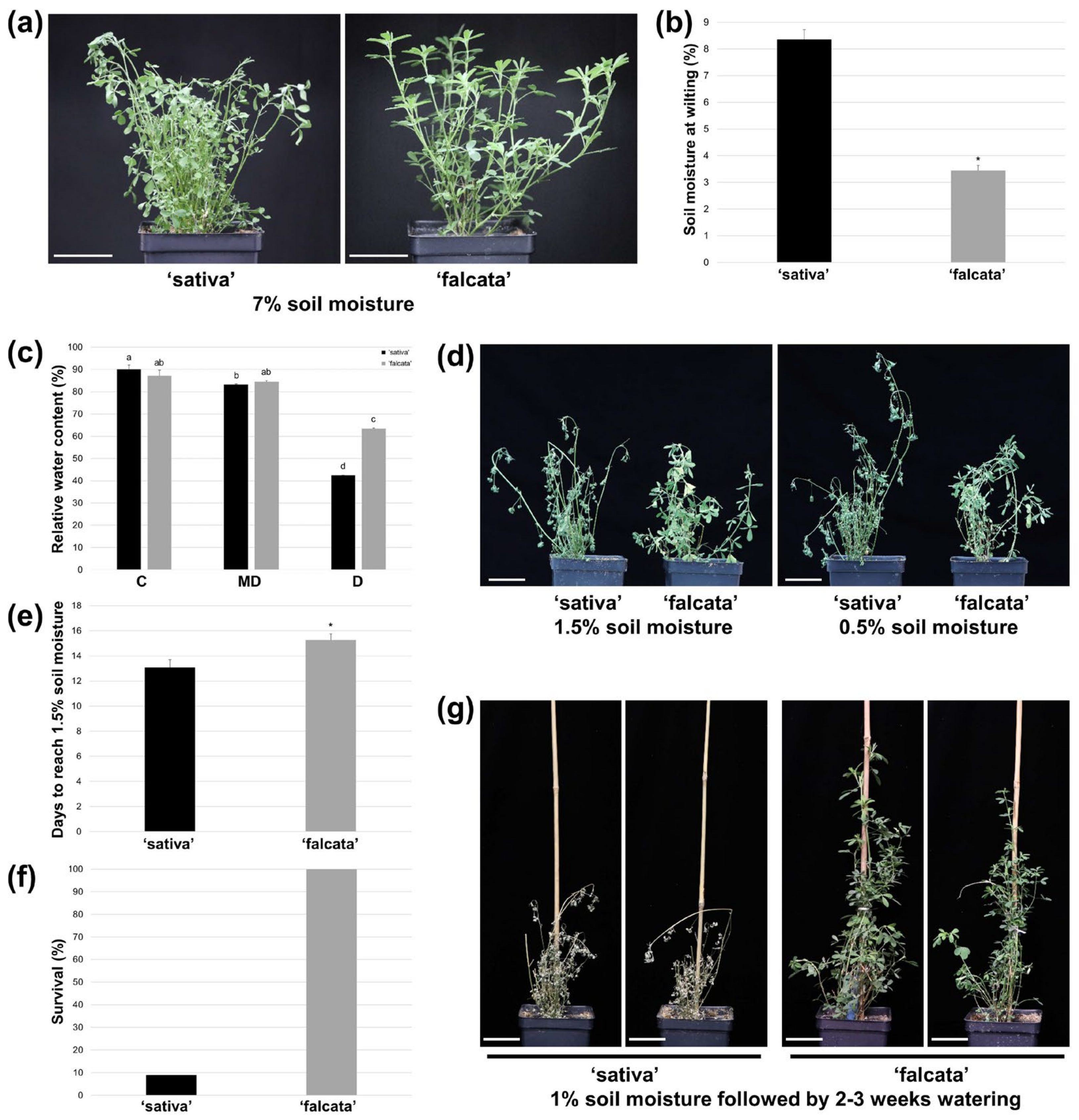
In the current study, ‘falcata’ was found to exhibit superior drought tolerance compared to ‘sativa’, as evidenced by a significant reduction in the soil moisture level at which plants began to wilt (
Figure 1a–b), and enhanced survival following severe drought (
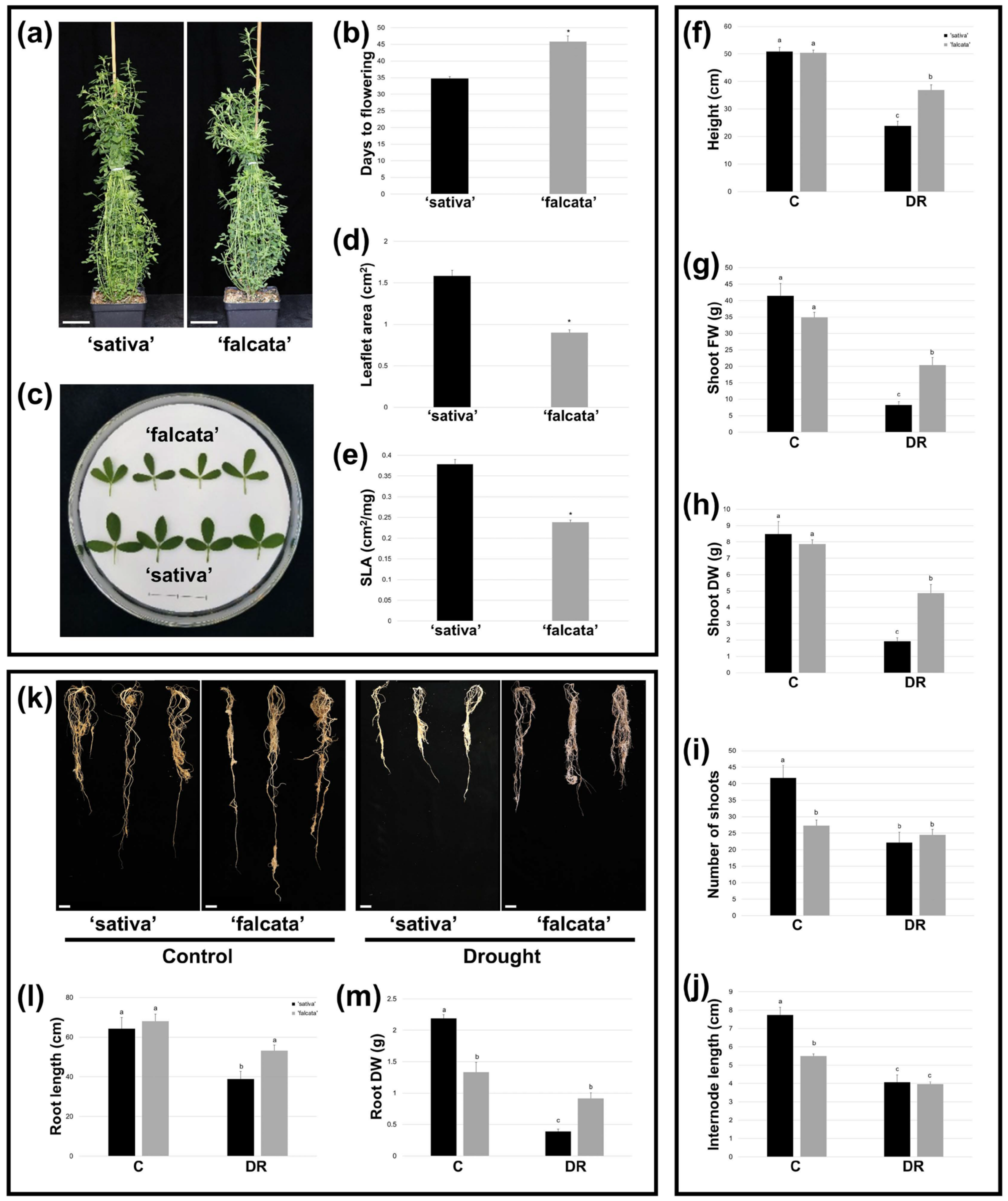
Figure 1f–g). In addition, while both ‘sativa’ and ‘falcata’ exhibited decreases in growth following drought recovery, which is a typical response of alfalfa [
29,
30], reductions were far more substantial in ‘sativa’ (
Figure 2f–h). Given the mounting limitations in water resources, minimizing yield loss under drought conditions will be a crucial component of attaining both agricultural and environmental sustainability in alfalfa production [
31,
32].
Drought-induced senescence is commonplace among drought-sensitive plants and is typically correlated with a reduction in leaf chlorophyll content [
24], which can lead to a decrease in photosynthetic capacity and eventually the death of the plant. Indeed, the ability to retain chlorophyll, and thus photosynthesis, under water deficit has been suggested to be a key contributor to drought tolerance in
falcata genotypes assessed previously [
24,
25]. In the present study, we did not note any changes in leaf chlorophyll content between drought and well-watered conditions in either genotype (
Figure S1b), and light-saturated photosynthetic rates declined under water deficit to a similar extent in both genotypes (
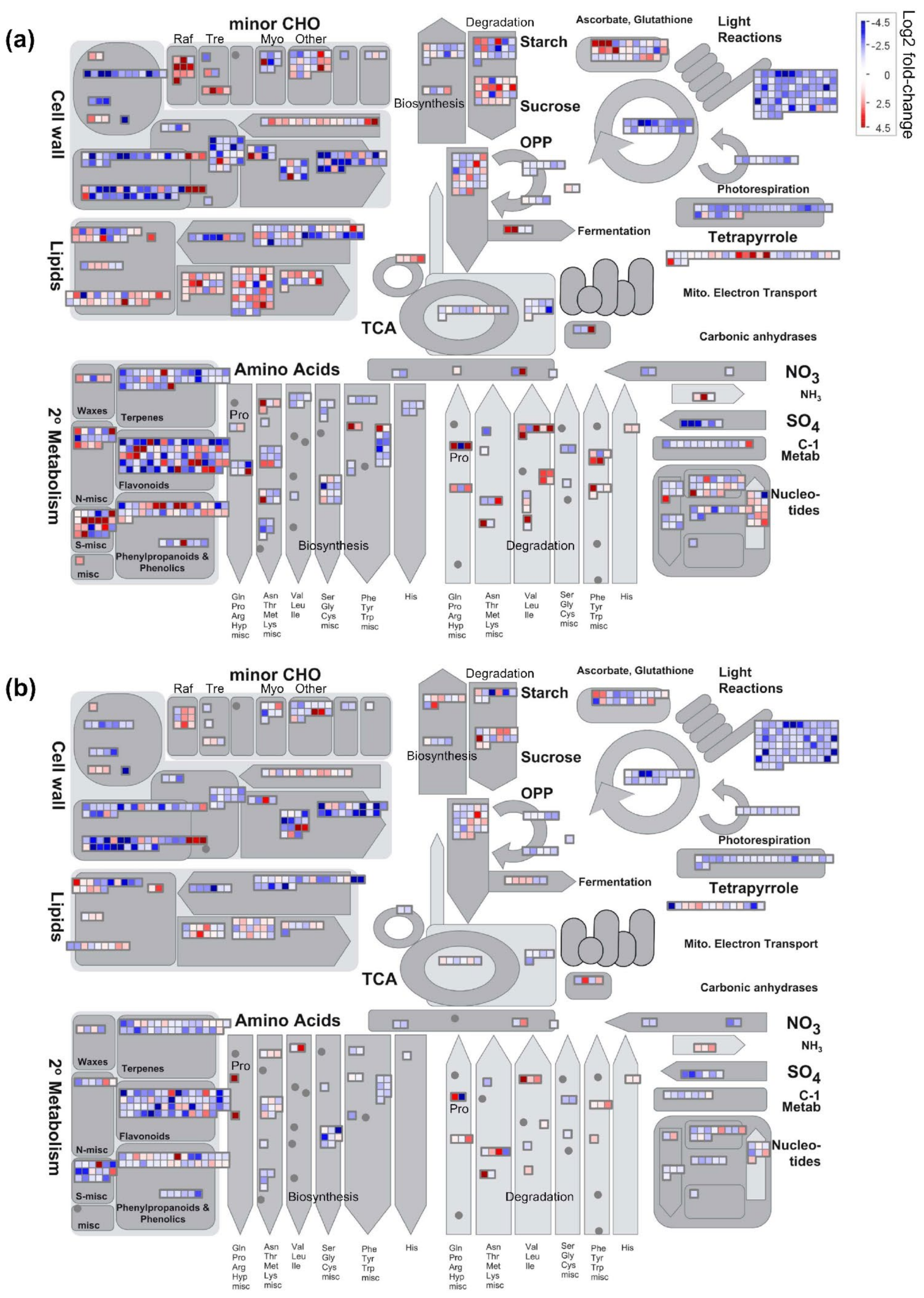
Figure S1h), which was consistent with the down-regulation of a multitude of genes involved in the photosynthetic process (
Figure 4). This implies that unlike previously studied
falcata genotypes, chlorophyll content was not correlated with photosynthetic rate in this study, and neither trait appeared to contribute to the improvement of drought tolerance in ‘falcata’.
The fact that ‘falcata’ plants wilted at a lower volumetric soil moisture content than ‘sativa’ and leaves retained a higher level of turgidity under severe drought stress (
Figure 1) suggests that ‘falcata’ is better able to maintain a high water status under stress, thus avoiding dehydration and allowing for the continuation of metabolic function for a longer period of time. This corresponds with our finding that ‘sativa’ leaves exhibited a greater number of DEGs under drought compared to well-watered conditions than ‘falcata’ (
Figure S4a), which resembles the outcome of a previous comparative microarray assessment of drought-tolerant Wisfal
falcata and drought-sensitive Chilean
sativa shoots [
24]. However, PAGE analysis of our RNA-Seq data also indicated that up-regulated genes within the ‘response to stress’ category were significantly enriched in ‘falcata’ compared to ‘sativa’ under drought (
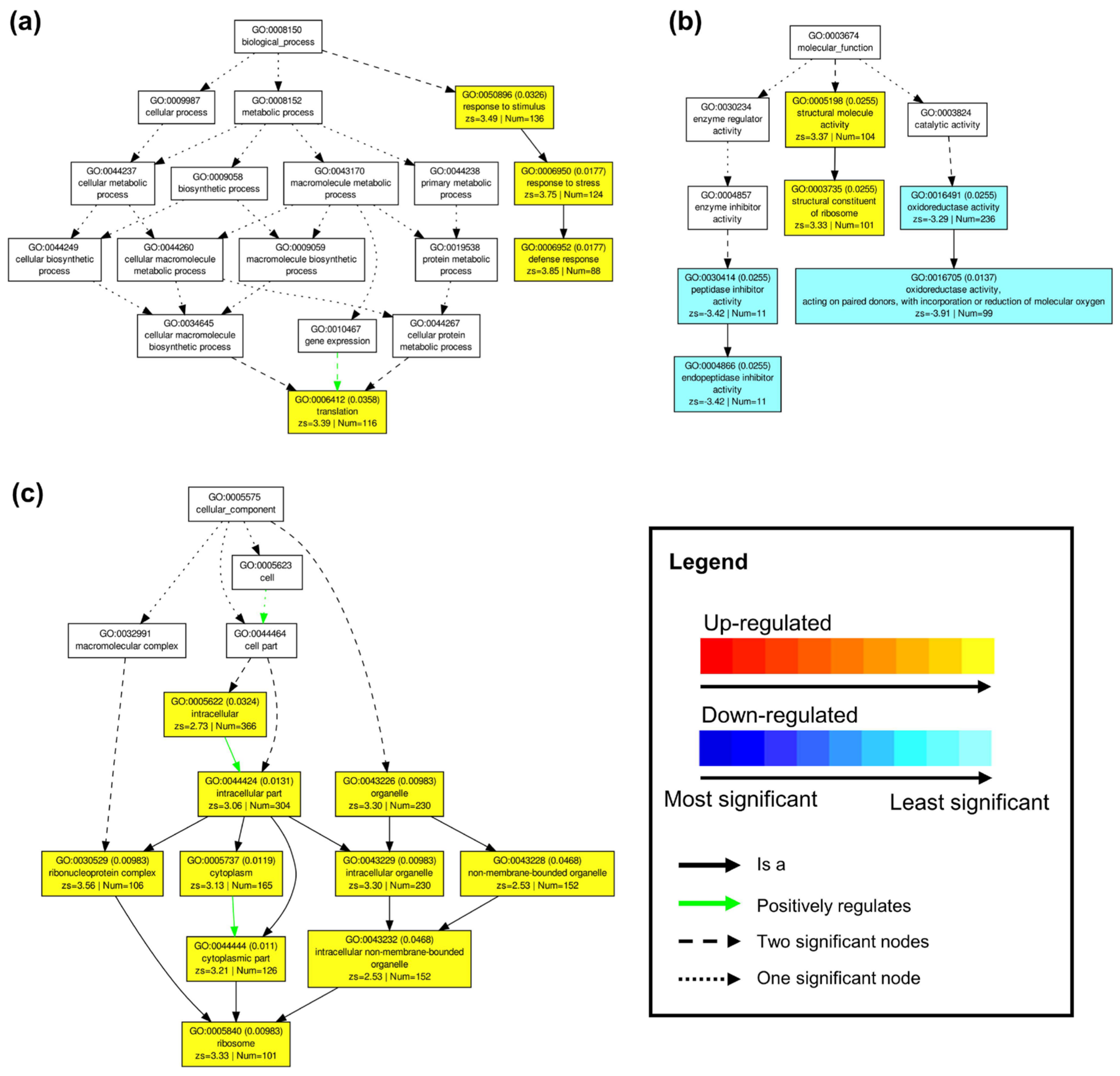
Figure 3a). This signifies that while ‘sativa’ may have undergone an overall greater transcriptional response under drought than ‘falcata’, the latter exhibited the preferential regulation of genes known to be specifically involved in stress response.
One way in which plants can avoid cellular dehydration under drought stress is through the superior capture of soil moisture [
33], at least under certain water deficit scenarios. Accordingly, several
falcata genotypes have been shown previously to exhibit higher root-to-shoot ratios under drought conditions than their
sativa counterparts [
24,
25]. In the current study, root DW was lower in ‘falcata’ than ‘sativa’ under well-watered conditions. However, only ‘sativa’ exhibited a significant reduction in root length and DW following drought recovery (
Figure 2k–m), which corresponds with the greater overall growth penalties incurred in this genotype. Therefore, although we did not examine root morphology in fine detail in this study, root size did not appear to be a main component of the improved drought tolerance in ‘falcata’.
Another manner in which plant water status can be maintained under water-limited conditions is through a reduction in water loss. Given that ‘falcata’ took approximately 2 days longer to reach a soil moisture level of 1.5% (from approximately 50% soil moisture) than ‘sativa’ following drought treatment (
Figure 1e), it is possible that this characteristic played a greater role in maintaining cellular hydration than enhanced soil moisture capture through the roots. Reductions in stomatal density and conductance are traits that have been associated with reduced water loss and are often linked to increased drought tolerance in many plant species, including alfalfa [
24,
25,
30]. In the present study, we observed an increase in stomatal density in ‘falcata’ compared to ‘sativa’, but a decrease in stomatal width and area (
Figure S1c–g), which corresponds with the lack of difference in stomatal conductance and transpiration rate on a per area basis between genotypes (
Figure S1i–j). This is in stark contrast to a selection of
falcata genotypes assessed previously, whereby they were found to exhibit decreases in stomatal density and/or stomatal conductance compared to
sativa genotypes [
24,
25,
26]. Although ABA levels, which can contribute to drought resilience in part through stomatal-related changes [
12], were not examined in the current study, few differences were noted between genotypes with respect to the expression of ABA biosynthetic genes (
File S2). Taken together, this implies that ABA-independent pathways and non-stomatal traits may be more important than ABA-dependent pathways in terms of eliciting superior drought tolerance in ‘falcata’.
Water loss can also be curtailed through non-stomatal leaf characteristics, such as a small leaf size and low SLA, and these traits are thus also associated with superior drought tolerance in plants [
30,
34,
35]. In line with this, ‘falcata’ plants bore leaves with a significantly lower area and SLA than ‘sativa’ under well-watered conditions (
Figure 2c–e). While we did not observe any difference between genotypes in detached leaf water loss assays (
Figure S1a), which suggests that ‘falcata’ leaves were not inherently better at minimizing water loss than ‘sativa’ on a per weight basis, it is possible that the leaf characteristics of ‘falcata’ may be contributing to an overall improvement in transpiration efficiency on a whole-plant level. Such a phenomenon could also feasibly have been a factor in the slower rate of soil moisture utilization observed in these plants.
An increase in the production of compatible solutes, such as proline, glycine betaine, and certain soluble sugars including trehalose and raffinose, within a plant under drought conditions can also have a positive impact on cellular hydration by augmenting osmotic pressure in the cytoplasm of plant cells, thus protecting enzymes and cell membranes under drought stress [
36,
37]. Proline accumulation did not appear to be a differentiating factor in drought resilience between ‘falcata’ and ‘sativa’ (
Figure 4,
Figure 5b and
Figure S2a), which is consistent with previous findings in Wisfal
falcata and Chilean
sativa [
24]. However, single genes involved in glycine betaine and trehalose biosynthesis, respectively, were up-regulated in ‘falcata’ compared to ‘sativa’ under well-watered conditions (
Figure 5b,
File S2). Similarly,
GolS1, which encodes a galactinol synthase that catalyzes the production of galactinol that is utilized for the subsequent biosynthesis of RSO and elicits enhanced tolerance to drought, salinity, and cold tolerance when over-expressed in tobacco [
16], was also up-regulated in ‘falcata’ compared to ‘sativa’ under drought conditions (
File S2). Wisfal
falcata was found previously to possess increased levels of several minor carbohydrates with known functions as osmoprotectants, including raffinose and myo-inositol, compared to Chilean
sativa under both well-watered and water-limited conditions [
24]. Together, these findings hint at the possibility that the accumulation of particular carbohydrates may be a general drought response mechanism across multiple
falcata genotypes. Although total soluble carbohydrate levels were found to increase similarly under drought conditions in both ‘sativa’ and ‘falcata’ in the current study (
Figure S2b), these changes do not necessarily reflect alterations in specific types of soluble carbohydrate, and it is thus feasible that an increase in the levels of these, and potentially other osmoprotectants may be a contributing factor to the enhanced ability of ‘falcata’ to maintain cell turgor under drought conditions.
In addition to a reduction in cellular hydration, drought stress also typically leads to an increase in the production of ROS such as superoxide (O
2-) and hydrogen peroxide (H
2O
2). These ROS have important signaling functions in drought response, particularly through their function as secondary messengers that contribute to the coordination of specific physiological, molecular, and metabolic events [
38]. However, above a certain threshold they have detrimental effects on cells and their components [
36], and as such, plants typically increase the transcriptional levels/enzymatic activities of various enzymatic and non-enzymatic antioxidant systems under stress as a means of scavenging and detoxifying ROS [
30,
39]. The enzymatic antioxidant activity of SOD, APX, and CAT did not appear to contribute to the increased drought resilience of ‘falcata’ in this study (
Figure S2c–e), and instead, both SOD and CAT activity increased to a greater extent in ‘sativa’ than ‘falcata’ under drought treatment. This implies that in line with the fact that ‘falcata’ was able to maintain a higher RWC than ‘sativa’ under the same intensity of water deficit, ROS levels may have been less impacted in the former genotype and there was thus less of a need to increase enzymatic antioxidant activity under these conditions. Interestingly, the transcriptional changes in genes encoding enzymatic antioxidants did not appear to follow this same pattern overall (
Figures S10a,b and S12a, File S2). This was not wholly unexpected since the expression patterns of genes encoding antioxidant enzymes have not always been found to be congruent with fluctuations in enzyme activity under drought stress [
40,
41,
42], which may be related to post-translational regulatory mechanisms that have yet to be fully elucidated [
43,
44].
Non-enzymatic antioxidants, such as flavonoids (Baskar et al. 2018) and isoprenoids including carotenoids [
45], tocopherols [
46], and a variety of other terpenoids [
47], also function to reduce ROS levels during abiotic stress. In line with this, shoot flavonoids were found previously to be elevated in Wisfal
falcata compared to Chilean
sativa under both control and water-deficient conditions, with a concomitant increase in the propensity for the up-regulation of genes involved in their biosynthesis in the
falcata genotype under drought compared to well-watered conditions [
24]. However, a similar trend was not observed in the present study (
Figure 4,
File S2), and instead, a relatively large proportion of genes involved in the flavonoid biosynthetic pathway were expressed at lower levels in ‘falcata’ than ‘sativa’ under drought stress (
Figure S12a). In agreement with our findings, a drought-tolerant alfalfa genotype was shown previously to exhibit a reduction in flavonoid levels under drought stress, whereas levels were not altered significantly in a drought-sensitive cultivar [
32]. As such, it is possible that the role of these metabolites in the abiotic stress response may differ between species/genotypes [
48,
49], and that different alfalfa genotypes may employ distinct mechanisms to withstand drought stress.
In the case of terpenoids, no obvious distinction was noted in the differential expression of genes involved in the metabolism of carotenoids or tocopherols between ‘sativa’ and ‘falcata’ (
File S2). In contrast, we noted the significant up-regulation of eight genes involved in the metabolism of other terpenoids in ‘falcata’ compared to ‘sativa’ under well-watered conditions, while no down-regulated genes in this category were present (
Figure 5c). Higher levels of metabolites involved in terpenoid biosynthesis were found in a drought-tolerant alfalfa genotype relative to a sensitive genotype under well-watered conditions previously [
32], which is analogous to what we observed in the current study. As there has been a relative paucity of research attempting to unravel the role of non-carotenoid and non-tocopherol terpenoids in plants, it is possible that these specialized metabolites may provide an as of yet undeciphered mechanism contributing to drought tolerance in alfalfa. Therefore, while carotenoids and tocopherols do not likely contribute to the differential drought response observed between genotypes in the present study, further examination will be necessary to definitively determine whether baseline terpenoid levels/composition are involved.
Differential levels of heat shock proteins, LEAs, and aquaporins have also been found to contribute to drought tolerance in plants [
50]. Aquaporins function to regulate the movement of water across cellular membranes [
51], and at least certain members of this family tend to be down-regulated in response to drought in plants, which may reduce water loss during dehydration [
52]. In contrast, heat shock proteins and LEAs (including dehydrins), which function, at least in part, as molecular chaperones to maintain protein stability and cellular homeostasis, are both typically up-regulated in response to drought stress [
53,
54]. Genes encoding all three types of protein have been shown to follow this trend at the transcriptional level in Wisfal
falcata and Chilean
sativa, with many being more responsive to drought in
sativa than
falcata [
24]. While we observed a similar pattern in the current study, there were several exceptions (
Figure S13). Of particular note was a gene encoding LEA3, which was significantly up-regulated in ‘falcata’ compared to ‘sativa’ under control, but not drought, conditions (
Figure S13b). Previously, the over-expression of
MfLEA3 (derived from a
falcata accession) in tobacco enhanced tolerance to drought, cold, and high light, possibly due in part to a concomitant decrease in ROS accumulation [
20]. Therefore, it is possible that higher levels of baseline expression of this gene may play a role in the superior drought tolerance observed in ‘falcata’ compared to ‘sativa’ via a protective effect on as of yet unidentified proteins.
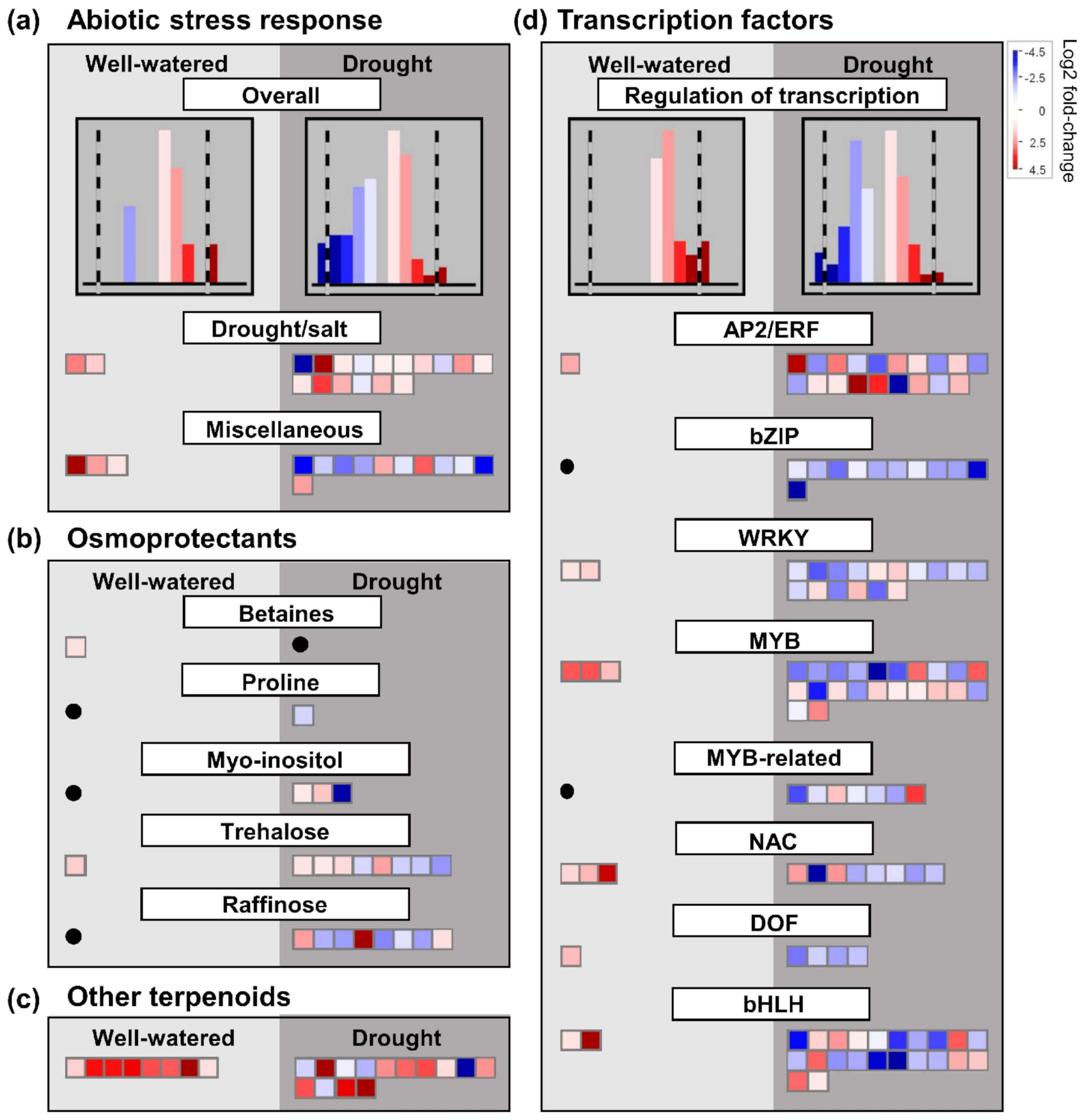
Alterations in the expression of genes encoding particular transcription factors have also been shown to be important for the regulation of drought stress response [
55]. In the present study, we found that overall, genes encoding transcription factors were more highly regulated in ‘sativa’ than ‘falcata’ under drought compared to control conditions (
Figure S11). Intriguingly, we also found that despite the greater overall response of ‘sativa’ under drought in this context, all differentially expressed genes encoding abiotic stress-related transcription factors assessed in this study were up-regulated in ‘falcata’ compared to ‘sativa’ under well-watered conditions (
Figure 5d), which implies that ‘falcata’ might possess higher baseline levels of these transcription factors than ‘sativa’. It is noteworthy that, in certain cases, drought tolerance can be attributed to the enhanced expression of genes, including a subset of transcription factors, prior to the onset of drought, thus rendering the plant quicker to respond [
56]. Accordingly,
NAC3, which was shown previously to positively regulate cold tolerance in
M. truncatula [
57], was up-regulated in ‘falcata’ compared to ‘sativa’ under control but not drought conditions (
File S2).
Finally, we also found that the expression of
USP1 was significantly higher in ‘falcata’ than ‘sativa’ under both well-watered and drought conditions (
File S2). The heterologous expression of a
falcata homolog of this gene in tobacco was previously shown to enhance tolerance to various types of abiotic stress, including osmotic stress, at least in part by lowering ROS accumulation [
19]. Although the precise mechanism by which this gene elicits improvements in abiotic stress tolerance remains to be determined, it is possible that its increased expression may also influence drought response in ‘falcata’.
In conclusion, unlike the small number of
falcata genotypes assessed previously [
24,
25,
26], ‘falcata’ PI 641381 did not appear to elicit its superior drought resilience through alterations in stomatal-related traits, root size, or delayed senescence under drought. While ‘falcata’ may make use of increased RSO accumulation, as evidenced by the up-regulation of
GolS1 in ‘falcata’ compared to ‘sativa’ under drought conditions, which resembles one possible factor behind improved drought tolerance in the Wisfal
falcata genotype [
24], it also appears to utilize a unique suite of additional mechanisms to achieve drought resilience. These putative mechanisms include the presence of smaller, thicker leaves and an increase in baseline transcriptional levels of a number of genes under well-watered conditions, which could feasibly allow a more rapid response to drought. This suggests that different
falcata accessions/genotypes may make use of distinct mechanisms to enhance their ability to thrive under drought conditions. While the majority of studies to date have focused on common drought response mechanisms in particular alfalfa accessions/cultivars, little effort has been directed towards elucidating differences between drought-tolerant genotypes thus far. However, such discrepancies have been found to occur in other plant species [
58,
59], highlighting the complexity of drought tolerance mechanisms in general. As such, deciphering the molecular and regulatory processes driving the superior ability of
falcata genotypes to withstand adverse conditions will provide knowledge and molecular tools to improve alfalfa, and potentially also other crops, in the future.
4. Materials and Methods
4.1. Plant Growth Conditions
Seeds of tetraploid
M. sativa subsp.
falcata accession PI 641381 (‘falcata’; determined to exhibit superior resilience to drought stress in preliminary experiments) were obtained from the United States Department of Agriculture—Agricultural Research Service Germplasm Resources Information Network (
https://www.ars-grin.gov/Pages/Collections; accessed on 16 October 2017). This accession was derived from a population grown in Russia at a latitude of 56°. Seeds of
M. sativa subsp.
sativa cv. Beaver (‘sativa’), which is relatively susceptible to drought and is typically grown under irrigation [
60], were provided by Dr. Surya Acharya (Agriculture and Agri-Food Canada, Lethbridge Research and Development Centre, Lethbridge, Canada). Due to the outcrossing nature of
M. sativa, all assessments were carried out using biological replicates (details are provided in specific subsections below, as well as in figure legends) of a single genotype of each accession/cultivar derived from vegetative stem cuttings for each treatment, respectively.
All plants were grown in Cornell mix in 4” pots under greenhouse conditions with supplemental light with a 16 h/8 h photoperiod, and a day/night temperature of approximately 20/15 °C. Pots were rotated daily to prevent microclimate effects and plants were cut back to approximately 5 cm at least twice prior to assessments. Drought treatment involved withholding water and daily monitoring of volumetric soil moisture content using a ML3 ThetaKit soil moisture meter (Hoskin Scientific Ltd., Burnaby, Canada), as well as stress symptoms, such as dry shoots and wilted leaves. All pots were adjusted to a soil moisture content of 50% prior to the experiment. All physiological, biochemical, and transcriptomic analyses were carried out at volumetric soil moisture contents of approximately 50% (control treatment) and 7–8% (drought treatment; when ‘sativa’ plants first began exhibiting signs of dehydration stress). All growth measurements were carried out using well-watered plants (approximately 50% volumetric soil moisture content) and in certain cases also following drought recovery (2–3 weeks of re-watering after allowing volumetric soil moisture contents to reach approximately 4% (aboveground measurements) or 1% (root measurements)).
4.2. Assessment of Growth Characteristics
Between ten and eleven vegetative stem cuttings for each treatment (drought recovery and control) were generated from a single genotype of ‘sativa’ and ‘falcata’, respectively, in order to assess growth characteristics and penalties in each case. Flowering time was defined as the number of days following cutting to the appearance of the first open flower in well-watered plants.
To evaluate growth penalties following drought recovery, aboveground and belowground plant growth characteristics were assessed. For aboveground evaluations, measurements were carried out 35–37 days after cutting under well-watered control conditions (soil moisture levels were maintained at approximately 50%) and following drought recovery (soil moisture content was allowed to reach approximately 4%, after which time plants were re-watered for 2 weeks). Plant height was derived from the length of the longest shoot, internode length consisted of the mean value of the longest internode on the three longest shoots of each plant, and the number of shoots comprised the total number of primary, secondary, and tertiary shoots per plant. Aboveground FW was evaluated by weighing all aboveground tissue immediately following harvest, while DW was determined following drying at 65 °C for at least 1 week.
Root assessments were carried out 54 days after cutting under well-watered conditions (control) and following drought recovery, which comprised allowing soil moisture content to reach approximately 1%, followed by re-watering for 3 weeks. At the time of evaluation, plants were removed from their pots and the roots were washed thoroughly. Root length was determined from the longest root on each plant. Root DW measurements were determined by drying at 65 °C for at least 1 week prior to weighing.
For evaluation of survival following drought treatment, water was withheld until volumetric soil water content reached approximately 1%, after which time plants were re-watered normally for 2–3 weeks and plant survival was assessed by determining the percentage of plants that regenerated.
4.3. Leaf Characteristics and Stomatal Measurements
Leaf length, leaf width, leaf area, and SLA were measured 21 days after cutting on five biological replicate plants of each genotype, with three leaves assessed per plant, using the middle leaflet of the third fully expanded trifoliate leaf from the shoot tip. Leaf area was resolved using the Petiole plant leaf area meter app (version 2.0.1;
https://play.google.com/store/apps/details?id=com.petioleapp.petiole&hl=en_CA&gl=US; accessed on 29 April 2019) and leaf dry weight was then determined after drying at 80 °C for 24 h. Specific leaf area was established by dividing leaf area by dry weight in each case.
Stomatal density, length, width, and area were measured using middle leaflets of third trifoliate leaves (from the shoot tip) from three biological replicate 35-day-old plants. Stomatal density was determined by applying clear nail polish to the abaxial side of leaflets (four to five leaflets from each of the three biological replicate plants), which were then allowed to dry for 10–15 min. The leaf imprints were then peeled off with the help of transparent tape, and slides were prepared. The resulting slides were visualized with an EVOS FL Auto Imaging System (Thermo Fisher Scientific, Waltham, MA, USA) under 20× magnification and stomata were counted. Values were then converted to the number of stomata per mm2 in each case. Stomatal length, width, and area were assessed on six to seven randomly selected stomata from each of the three biological replicate plants (twenty stomata total for each genotype) using the EVOS FL Auto Cell Imaging System software (Thermo Fisher Scientific, Waltham, MA, USA).
4.4. Measurement of Relative Water Content and Detached Leaf Water Loss
Water deficit was estimated by measuring the RWC of first fully expanded trifoliate leaves from ten biological replicates of each genotype, as described previously [
61], when soil moisture content reached approximately 50% (well-watered), 20% (mild drought), and 7% (drought). In brief, the fresh weight (FW) of trifoliate leaves was recorded immediately after harvesting, turgid weights (TW) were resolved after submersing petioles in water in an enclosed Eppendorf tube for 3–4 h, and dry weights (DW) were established by drying turgid leaves at 80 °C overnight. RWC (%) was then calculated as [(FW − DW)/(TW − DW)] × 100.
Detached leaf water loss assays were carried out on five biological replicate plants of ‘sativa’ and ‘falcata’ by weighing a fully expanded trifoliate leaf from each plant immediately upon harvest (Winitial), placing the leaf in the open air on a benchtop, and then weighing every 30 min for 180 min. The rate of water loss as a percentage was calculated as (Winitial − Wat particular time/Winitial) × 100.
4.5. Biochemical Assessments
All biochemical assessments were conducted using first fully expanded trifoliate leaves from nine to ten biological replicates of each genotype in each treatment. In all instances, leaf tissue was freeze-dried prior to carrying out assays. The control treatment comprised plants with soil moisture levels maintained at approximately 50%, while drought-treated samples were harvested when soil moisture levels reached approximately 7% (when ‘sativa’ plants were beginning to show signs of drought stress).
Total soluble sugar content was assessed using the Plant Soluble Sugar Content Assay Kit according to the manufacturer’s instructions (MyBioSource Inc., San Diego, CA, USA). Two technical replicates were carried out for each sample. Proline content was determined as described previously [
62] with minor modifications. In brief, approximately 50 mg (FW) leaf tissue was freeze dried and ground using a TissueLyzer II (Qiagen Inc., Toronto, ON, Canada), after which time 1 mL 3% aqueous sulphosalicylic acid was added to each tube. The tissue was further homogenized and incubated at room temperature for 3 h, and then centrifuged at 1500×
g for 10 min. Following centrifugation, 600 μL of sample supernatant (or various concentrations of L-proline standard) was added to 600 μL glacial acetic acid and 600 μL ninhydrin reagent (0.025 g/mL ninhydrin, 0.6 mL/mL glacial acetic acid, 2. 4M H
3PO
4). Reactions were incubated at 100 °C for 45 min, then cooled on ice for 30 min. To each tube, 1.2 mL toluene was added, and reactions were vortexed and then centrifuged at 1000×
g for 5 min. Proline content was determined in triplicate in microplate format using toluene as a blank in a Synergy Mx Multi-Mode Microplate Reader spectrophotometer (BioTek Instruments Inc., Winooski, VT, USA) at 520 nm.
4.6. Evaluation of Antioxidant Activity
Antioxidant assays were carried out using a single first fully expanded trifoliate leaf from nine to ten biological replicates of ‘sativa’ and ‘falcata’ plants under well-watered (approximately 50% soil water content) and drought (approximately 7% soil water content) conditions. Tissue was immediately frozen in liquid nitrogen, ground in a TissueLyzer for 1 min at 30 Hz, and placed back in liquid nitrogen. To each sample, 1.5 mL extraction buffer (0.15 M potassium phosphate buffer, pH 7.8 and 1 mM EDTA) was added and samples were vortexed for 30 s. Subsequently, the samples were centrifuged at 12,000×
g at 4 °C for 20 min, and the supernatant was transferred to a fresh tube. Extracts were used for all subsequent assays, which were carried out in triplicate. Protein concentration was determined using a Bradford assay [
63], with 4 μL extract and the Quickstart Protein Assay according to the manufacturer’s microassay protocol, with bovine serum albumin as a standard (Bio-Rad Laboratories Ltd., Hercules, CA, USA).
CAT activity was evaluated as described previously [
64] with minor modifications. Initially, the spectrophotometer (SmartSpec Plus; Bio-Rad Laboratories Ltd., Hercules, CA, USA) was blanked at 240 nm with 50 mM potassium phosphate buffer (pH 7.0). For each assay, 33 μL sample extract was added to 967 μL CAT reaction mixture (50 mM potassium phosphate buffer, pH 7.0, 10 mM H
20
2) in a 1 mL cuvette and the absorbance was immediately measured at 240 nm (A
sampleT0). A second reading was carried out after precisely 3 min (A
sampleT3). CAT activity (nM H
20
2 · min
−1 · mg protein
−1) was calculated as (A
sampleT0 − A
sampleT3)/(e × d × t × C), where e corresponds to 39.4 M
−1 (extinction coefficient of H
20
2), d corresponds to 1 cm (cuvette path length), t corresponds to 3 min (incubation time), and C corresponds to the amount of protein (mg) within the 33 μL of extract used for analysis.
APX activity was determined as described previously [
65] with minor modifications. To a 1 mL cuvette, 967 μL APX reaction mixture (50 mM potassium phosphate buffer, pH 7.0, 0.5 mM ascorbic acid, 0.1 mM EDTA) was mixed with 33 μL sample extract and 5 μL 200 mM H
20
2. APX reaction buffer was used in place of sample extract for blanks. Absorbances were read immediately at 290 nm (A
sampleT0), and a second reading was taken after precisely 3 min (A
sampleT3). APX activity (μM ascorbate · min
−1 · mg protein
−1) was calculated as (A
sampleT0 − A
sampleT3)/(e × d × t × C), where e corresponds to 2.8 mM
−1 · cm
−1 (extinction coefficient of H
20
2), d corresponds to 1 cm (cuvette path length), t corresponds to 3 min (incubation time), and C corresponds to the amount of protein (mg) within the 33 μL of extract used for analysis.
SOD activity was assessed as described previously [
66] with minor modifications to allow for microplate format. Briefly, each reaction consisted of 5 μL of sample extract and 195 μL SOD reaction mixture (50 mM potassium phosphate buffer, pH 7.8, 2 mM EDTA, 9.9 mM L-methionine, 55 μM nitroblue tetrazolium, 0.025% Triton X-100 and 1 μM riboflavin). Every reaction was replicated under light (approximately 80 μmol m
−2 s
−1) and dark conditions, and was incubated for 10 min at room temperature in each case. Absorbance was then measured with a microplate spectrophotometer (Synergy Mx Multi-Mode Microplate Reader; BioTek Instruments Inc., Winooski, VT, USA) at 560 nm with extraction buffer as a blank (A
blank). Sample absorbance (A
sample) was calculated as absorbance in the dark subtracted from absorbance in the light. SOD activity (SOD units/mg protein) was calculated as [(A
blank − A
sample)/(A
blank × 0.5)]/mg protein.
4.7. Chlorophyll- and Photosynthesis-Related Measurements
Chlorophyll content in the leaves of ten to eleven biological replicates of ‘sativa’ and ‘falcata’ genotypes was determined using a CCM-200 Chlorophyll Content Meter (Hoskin Scientific Ltd., Burlington, ON, Canada). The middle leaflet of third fully expanded trifoliate leaves was used for measurements in each case, with the values obtained from three leaves averaged for each biological replicate. Leaves were assessed under well-watered conditions (approximately 50% soil moisture content) and when drought-treated ‘sativa’ plants were just beginning to display symptoms of stress (soil moisture content of approximately 8%).
Stomatal conductance (gs), transpiration rate (E), and light-saturated photosynthetic rate (Asat) were measured with an LI-6800 (LI-COR Inc., Lincoln, NE, USA). The centre leaflet of a first fully expanded dark green trifoliate leaf was used for measurements, and all observations were carried out between 12:45 pm and 3:45 pm in the greenhouse. Leaves were evaluated under well-watered conditions (approximately 50% soil moisture content) and under drought treatment (average soil moisture content of approximately 7%). Four biological replicates were utilized for ‘sativa’ under drought conditions and eleven biological replicates were used for the remaining groups. The number of ‘sativa’ drought-treated plants assessed was lower than other groups due to the lack of viable tissue in a proportion of plants as a result of drought stress on the day testing was carried out. Within the chamber, light intensity was maintained at 1500 μmol m−2 s−1, relative humidity at 65%, air temperature at 22 °C, and CO2 level at 410 μmol CO2/mol air. Each leaflet was allowed 3 min to stabilize within the chamber prior to assessment. All three measurements were adjusted for leaf area, which was determined using the Petiole app (version 2.0.1) as described in a previous section.
4.8. RNA-Seq Data Analysis
4.8.1. Sequencing and Annotation
Leaf tissue was harvested from control (soil moisture content of approximately 50%) and drought-treated plants (soil moisture content of approximately 7%), flash frozen in liquid nitrogen, and stored at −80 °C. Tissue was harvested from four biological replicates of ‘sativa’ and ‘falcata’ under each treatment, respectively. Total RNA was extracted from ground tissue using the Spectrum Plant Total RNA Kit according to the manufacturer’s instructions (Sigma-Aldrich Corp., St. Louis, MO, USA). RNA integrity was confirmed by resolving a small aliquot on a 1% agarose gel and using a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA). A stranded mRNA library was prepared using 250 ng of total RNA and the NEBNext® system (New England Biolabs Ltd., Whitby, ON, Canada), and sequencing was carried out using an Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA) with 100 bp paired-end reads by a third party (Genome Québec Centre d’Expertise et de Services, Montreal, QC, Canada). The resulting raw RNA-Seq data was analyzed as described previously [
67] with minor modifications. Briefly, raw reads were trimmed using the ‘sickle’ script in Linux with default parameters, and read quality was assessed using the FASTQC tool (
https://www.bioinformatics.babraham.ac.uk/projects/fastqc/; accessed on 30 May 2020). High-quality filtered reads were mapped to the
Medicago truncatula genome (Mt4.0 v2;
http://www.medicagogenome.org/downloads; accessed on 31 May 2020; a close diploid relative of alfalfa with a well-annotated genome sequence) using Tophat2 [
68].
DEGs were identified and normalized to FPKM using Cuffdiff [
69]. Genes with a false discovery rate (FDR) [
70] of less than 0.05 were considered DEGs (
File S1). PCA was performed using total exon read counts, which were obtained using the featureCounts program [
71], followed by analysis using freely available R-software (v4.0). Venn diagrams were generated using freely available software (
http://bioinformatics.psb.ugent.be/webtools/Venn/; accessed on 15 June 2021). The sequence data generated in this study are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (BioProject accession number PRJNA765383).
4.8.2. GO Term Enrichment and Pathway Analysis
SEA of either up-regulated or down-regulated DEGs observed between control and drought conditions for both ‘sativa’ and ‘falcata’ were carried out using AgriGO v2.0 (
http://systemsbiology.cau.edu.cn/agriGOv2/; accessed on 13 January 2021) [
72] with
M. truncatula as the reference species. In both cases, the hypergeometric statistical test was used, along with the Yekutieli (FDR under dependency) multi-test adjustment method, with a significance level of 0.05. SEACOMPARE was conducted by inputting SEA results. PAGE was carried out by inputting all up-regulated and down-regulated DEGs observed between ‘sativa’ and ‘falcata’ when grown either under control or drought conditions, along with log2 fold-change expression values (expression values of 0 were artificially set to 0.001 in order to provide numerical log2 fold-changes), using the same program with the Hochberg (FDR) multi-test adjustment method and a significance level of 0.05. Heat maps were generated using the freely available Morpheus tool (
https://software.broadinstitute.org/morpheus/; accessed on 20 May 2021). Visualization of DEG-associated pathways between ‘sativa’ and ‘falcata’ under both well-watered and control conditions was performed using MapMan V3.6 software (
https://mapman.gabipd.org/) with the
M. truncatula genome as a reference sequence (Mt4.0 v2).
4.9. Validation of RNA-Seq Results
Total RNA that was extracted from leaf tissues for RNA-Seq analysis was utilized for qRT-PCR validation. First-strand cDNA synthesis was carried out using the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) and quantitative real-time RT-PCR assays were conducted using an appropriate dilution of each cDNA template along with PerfeCTa SYBR Green Supermix (VWR International LLC, Mississauga, ON, Canada) in a final reaction volume of 10 µL. Assays were accomplished on a Quantstudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) using primers designed to anneal to a region of coding sequence for ten genes selected based on their up- or down-regulation in RNA-Seq analyses (see
Table S2 for primer sequences used for qRT-PCR assays). A 183 nt region of the constitutively expressed actin-depolymerizing factor (
ADF) gene, which has been shown previously to act as a highly stable reference gene for qRT-PCR across developmental stages and environmental conditions (including water stress) in alfalfa [
73], was amplified as an internal control. Thermal parameters for amplification were as follows: initial denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 45 s. Dissociation curves were generated to confirm the presence of a single amplification product in each case. Levels of gene expression were established using the standard curve method and Applied Biosystems
TM analysis software v4.0 (Thermo Fisher Scientific, Waltham, MA, USA), with the expression of each target gene comprising mean values of four biological replicates (three technical replicates each) normalized to that of the internal control. Log2 fold-change values between control and drought samples were calculated for comparison to RNA-Seq values.
4.10. Statistical Analyses
For the majority of multi-variate comparisons, the observed response variables were modeled using the GLIMMIX procedure in SAS (SAS version 9.4, SAS Institute Inc., Cary, NC, USA) with one-thousand iterations at multiple levels of iteration (MAXOPT = 1000 and NLOPTIONS MAXITER = 1000). The normal distribution of the response was not assumed and therefore the models were “generalized.” The best-fitting distribution from the exponential family of distributions (e.g., gamma, inverse Gaussian, lognormal, shifted-t, normal, and exponential) was selected for each variable, based on the model fit statistics, that is, the Bayesian information criterion (BIC). The models were “mixed” due to the inclusion of fixed factors (Genotype, Growth_conditions, and Genotype × Growth_conditions) and random factors (Biological_replicate). Variance homogeneity was not assumed and models of variance heterogeneity were tested and selected based on the BIC of the models. Bonferroni’s method was used to adjust for multiple comparisons. For assessments that involved comparisons between genotypes under a single treatment, as well as for evaluations of photosynthetic rate, stomatal conductance, and transpiration rate, 2-tailed Student’s t-tests assuming unequal variance were used for statistical analysis. Differences were considered significant at p ≤ 0.05.