The Soybean High Density ‘Forrest’ by ‘Williams 82’ SNP-Based Genetic Linkage Map Identifies QTL and Candidate Genes for Seed Isoflavone Content
Abstract
1. Introduction
2. Results and Discussion
2.1. The SNP-Based Genetic Map
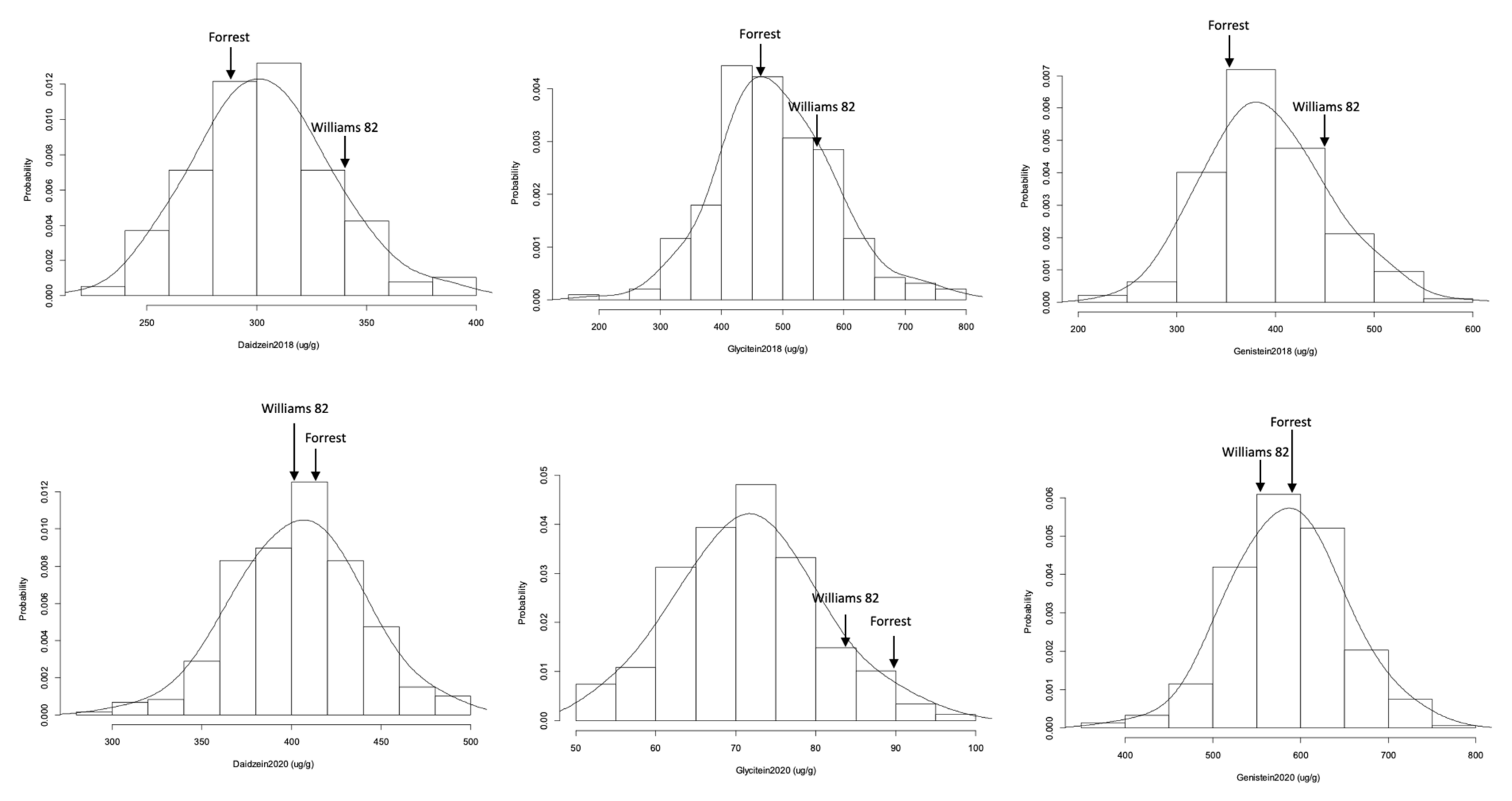
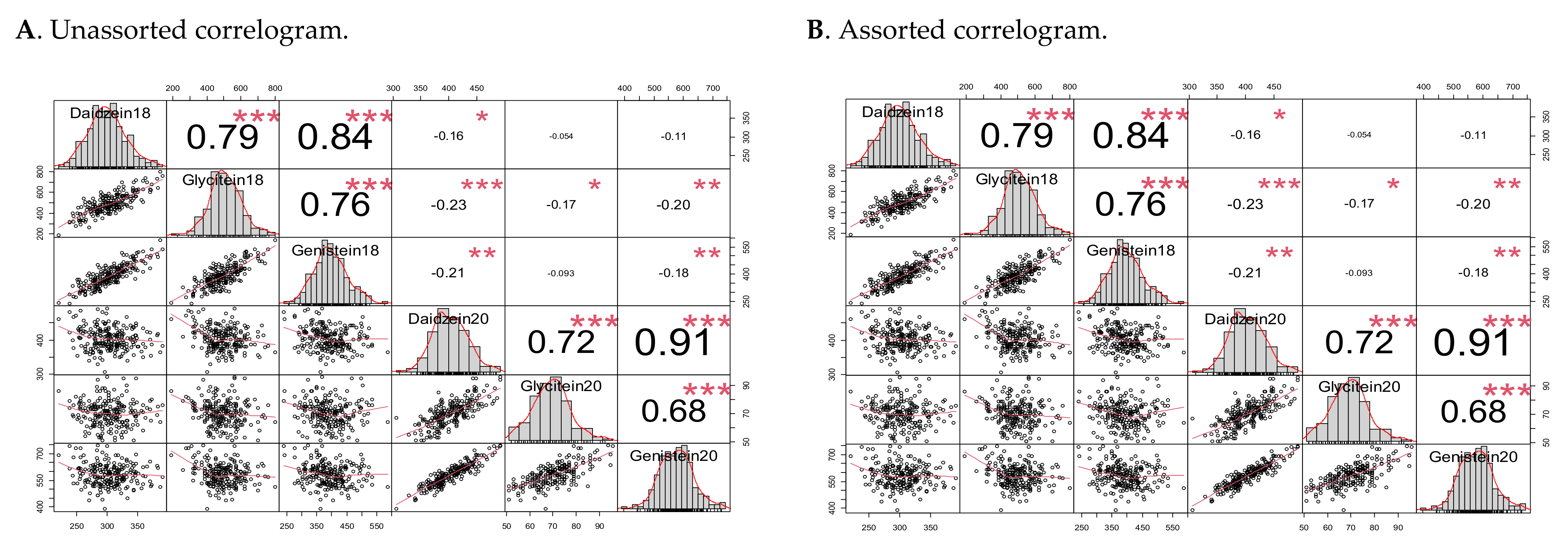
2.2. Isoflavone Contents Frequency Distribution, Heritability, and Correlation
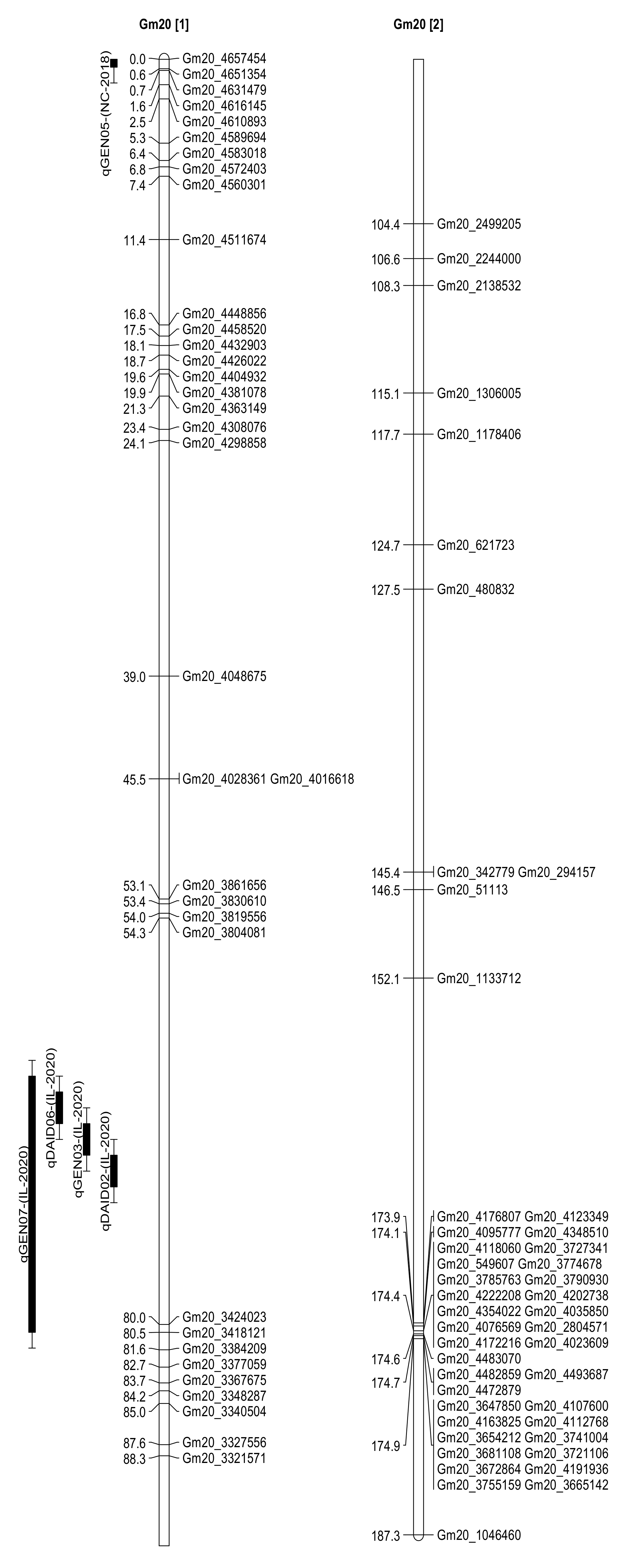
2.3. Seed Isoflavone Contents QTL
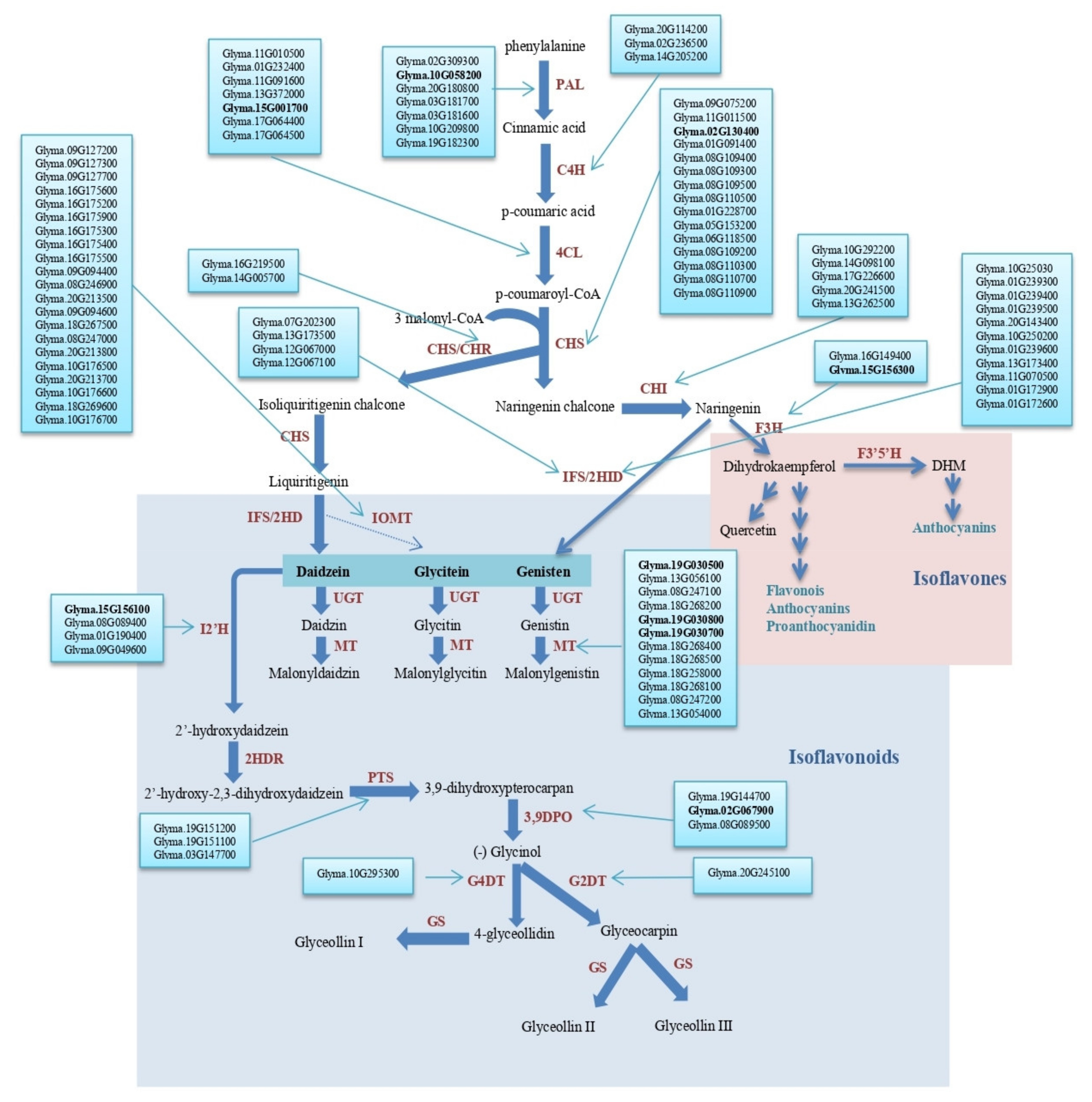
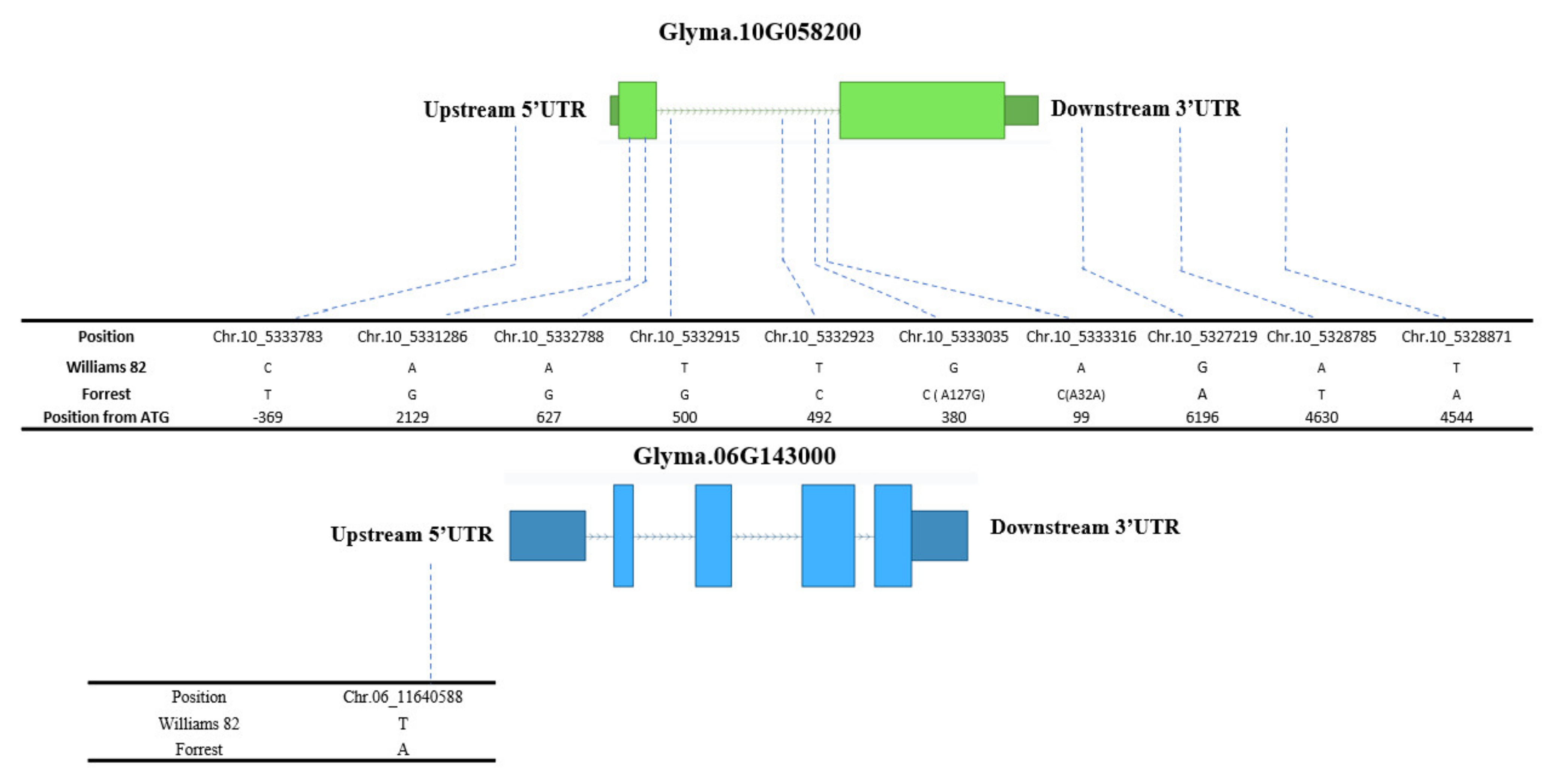
2.4. Seed Isoflavone Candidate Genes
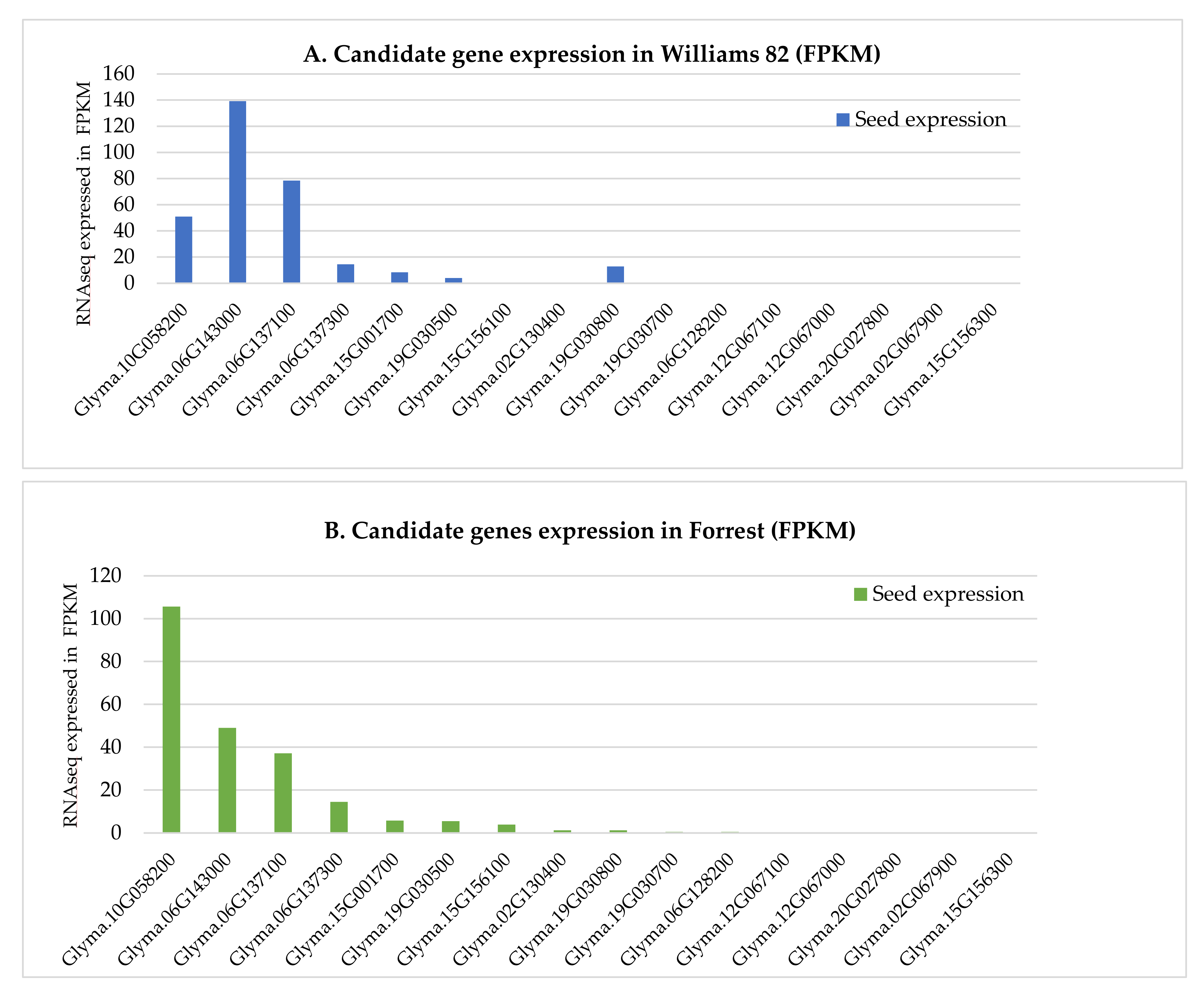
2.5. Expression Analysis
2.6. Conclusions
3. Materials and Methods
3.1. Plant Material and Growth Conditions
3.2. Isoflavone Quantification
3.3. DNA Isolation, SNP Genotyping, and Genetic Map Construction
3.4. Isoflavone QTL Detection and Statistical Analysis
3.5. Isoflavone Candidate Genes Identification
3.6. Expression Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cavallini, D.C.U.; Bedani, R.; Bomdespacho, L.Q.; Vendramini, R.C.; Rossi, E.A. Effects of probiotic bacteria, isoflavones and simvastatin on lipid profile and atherosclerosis in cholesterol-fed rabbits: A randomized double-blind study. Lipids Health Dis. 2009, 8, 1. [Google Scholar] [CrossRef]
- Cederroth, C.R.; Zimmermann, C.; Nef, S. Soy, phytoestrogens and their impact on reproductive health. Mol. Cell. Endocrinol. 2012, 355, 192–200. [Google Scholar] [CrossRef]
- Kwon, Y. Effect of soy isoflavones on the growth of human breast tumors: Findings from preclinical studies. Food Sci. Nutr. 2014, 2, 613–622. [Google Scholar] [CrossRef]
- Setchell, K.D.R.; Cassidy, A. Dietary Isoflavones: Biological Effects and Relevance to Human Health. J. Nutr. 1999, 129, 758S–767S. [Google Scholar] [CrossRef]
- Cederroth, C.R.; Nef Soy, S. phytoestrogens and metabolism: A review. Mol. Cell. Endocrinol. 2009, 304, 30–42. [Google Scholar] [CrossRef]
- Jiang, Q.; Payton-Stewart, F.; Elliott, S.; Driver, J.; Rhodes, L.V.; Zhang, Q.; Zheng, S.; Bhatnagar, D.; Boue, S.M.; Collins-Burow, B.M.; et al. Effects of 7-O Substitutions on Estrogenic and Anti-Estrogenic Activities of Daidzein Analogues in MCF-7 Breast Cancer Cells. J. Med. Chem. 2010, 53, 6153–6163. [Google Scholar] [CrossRef]
- Hamilton-Reeves, J.M.; Banerjee, S.; Banerjee, S.K.; Holzbeierlein, J.M.; Thrasher, J.B.; Kambhampati, S.; Keighley, J.; Veld-huizen, P.V. Short-Term Soy Isoflavone Intervention Patients with Localized Prostate Cancer: A Randomized, Double-Blind, Placebo-Controlled Trial. PLoS ONE 2013, 8, e68331. [Google Scholar] [CrossRef] [PubMed]
- Fritz, H.; Seely, D.; Flower, G.; Skidmore, B.; Fernandes, R.; Vadeboncoeur, S.; Kennedy, D.; Cooley, K.; Wong, R.; Sagar, S.; et al. Soy, Red Clover, and Isoflavones and Breast Cancer: A Systematic Review. PLoS ONE 2013, 8, e81968. [Google Scholar] [CrossRef]
- Chen, M.; Rao, Y.; Zheng, Y.; Wei, S.; Li, Y.; Guo, T.; Yin, P. Association between Soy Isoflavone Intake and Breast Cancer Risk for Pre- and Post-Menopausal Women: A Meta-Analysis of Epidemiological Studies. PLoS ONE 2014, 9, e89288. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Bhatt, S.; Chang, L.M.; Sampson, H.A.; Masilamani, M. Isoflavones, Genistein and Daidzein, Regulate Mucosal Immune Response by Suppressing Dendritic Cell Function. PLoS ONE 2012, 7, e47979. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Kogiso, M. Soy isoflavones and immunity. J. Med. Investig. 2008, 55, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Stacey, G.; Yu, O. Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci. 2007, 12, 282–285. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, G.-J.; Song, T.T.; Murphy, P.A.; Hendrich, S. Urinary disposition of the soybean isoflavones daidzein, genistein and glycitein differs among humans with moderate fecal isoflavone degradation activity. J. Nutr. 1999, 129, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Thigpen, J.E.; Setchell, K.D.R.; Ahlmark, K.B.; Locklear, J.; Spahr, T.; Cavines, G.F.; Goelz, M.F.; Haseman, J.K.; Newbold, R.R.; Forsyth, D.B. Phytoestrogen content of purified, open- and closed-formula laboratory animal diets. Lab. Anim. Sci. 1999, 49, 530–536. [Google Scholar] [PubMed]
- Kassem, M.A.; Meksem, K.; Iqbal, M.J.; Njiti, V.N.; Banz, W.J.; Winters, T.A.; Wood, A.J.; Lightfoot, D.A. Definition of Soybean Genomic Regions That Control Seed Phytoestrogen Amounts. J. Biomed. Biotechnol. 2004, 2004, 52–60. [Google Scholar] [CrossRef]
- Kassem, M.A.; Shultz, J.; Meksem, K.; Cho, Y.; Wood, A.; Iqbal, M.J.; Lightfoot, D.A. An updated ‘Essex’ by ‘Forrest’ linkage map and first composite interval map of QTL underlying six soybean traits. Theor. Appl. Genet 2006, 113, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Gonzalez, J.J.; Wu, X.; Zhang, J.; Lee, J.-D.; Ellersieck, M.; Shannon, J.G.; Yu, O.; Nguyen, H.T.; Sleper, D.A. Genetic control of soybean seed isoflavone content: Importance of statistical model and epistasis in complex traits. Theor. Appl. Genet. 2009, 119, 1069–1083. [Google Scholar] [CrossRef]
- Liang, H.Z.; Yu, Y.L.; Wang, S.F.; Lian, Y.; Wang, T.G.; Wei, T.L.; Gong, P.T.; Liu, X.Y.; Fang, X.J.; Zhang, M.C. QTL Mapping of Isoflavone, Oil and Protein Contents in Soybean (Glycine max L. Merr.). Agric. Sci. China 2010, 9, 1108–1116. [Google Scholar] [CrossRef]
- Gutierrez-Gonzalez, J.J.; Vuong, T.D.; Zhong, R.; Yu, O.; Lee, J.-D.; Shannon, G.; Ellersieck, M.; Nguyen, H.T.; Sleper, D.A. Major locus and other novel additive and epistatic loci involved in modulation of isoflavone concentration in soybean seeds. Theor. Appl. Genet. 2011, 123, 1375–1385. [Google Scholar] [CrossRef]
- Smallwood, C.J.; Nyinyi, C.N.; Kopsell, D.A.; Sams, C.E.; West, D.R.; Chen, P.; Kantartzi, S.K.; Cregan, P.B.; Hyten, D.L.; Pantalone, V.R. Detection and Confirmation of Quantitative Trait Loci for Soybean Seed Isoflavones. Crop. Sci. 2014, 54, 595–606. [Google Scholar] [CrossRef]
- Zhao, G.; Jiang, Z.; Li, D.; Han, Y.; Hu, H.; Wu, L.; Wang, Y.; Gao, Y.; Teng, W.; Li, Y.; et al. Molecular loci associated with seed isoflavone content may underlie resistance to soybean pod borer (Leguminivora glycinivorella). Plant Breed. 2015, 134, 78–84. [Google Scholar] [CrossRef]
- Akond, A.; Liu, S.; Kantartzi, S.K.; Meksem, K.; Bellaloui, N.; Lightfoot, D.A.; Yuan, J.; Wang, D.; Anderson, J.; Kassem, M.A. A SNP Genetic Linkage Map Based on the ‘Hamilton’ by ‘Spencer’ Recombinant Inbred Line (RIL) Population of Soybean [Glycine max (L.) Merr.] Identified QTL for Seed Isoflavone Contents. Plant Breed. 2015, 134, 580–588. [Google Scholar] [CrossRef]
- Watanabe, S.; Yamada, R.; Kanetake, H.; Kaga, A.; Anai, T. Identification and characterization of a major QTL underlying soybean isoflavone malonylglycitin content. Breed. Sci. 2019, 69, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Teng, W.; Wang, Y.; Zhao, X.; Wu, L.; Li, D.; Li, W. Unconditional and conditional QTL underlying the genetic interrelationships between soybean seed isoflavone, and protein or oil contents. Plant Breed. 2015, 134, 300–309. [Google Scholar] [CrossRef]
- Murphy, S.E.; Lee, E.A.; Woodrow, L.; Seguin, P.; Kumar, J.; Rajcan, I.; Ablett, G.R. Genotype × Environment Interaction and Stability for Isoflavone Content in Soybean. Crop. Sci. 2009, 49, 1313–1321. [Google Scholar] [CrossRef]
- Zhang, H.J.; Li, J.W.; Liu, Y.J.; Jiang, W.Z.; Du, X.L.; Li, L.; Li, X.W.; Su, L.T.; Wang, Q.Y.; Wang, Y. Quantitative trait loci analysis of individual and total isoflavone contents in soybean seeds. J. Genet. 2014, 93, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kamala, S.; Tian, R.; Du, H.; Li, W.; Kong, Y.; Zhang, C. Identification and validation of quantitative trait loci controlling seed isoflavone content across multiple environments and backgrounds in soybean. Mol. Breed. 2018, 38, 8. [Google Scholar] [CrossRef]
- Wu, D.; Li, D.; Zhao, X.; Zhan, Y.; Teng, W.; Qiu, L.; Zheng, H.; Li, W.; Han, Y. Identification of a candidate gene associated with isoflavone content in soybean seeds using genome-wide association and linkage mapping. Plant J. 2020, 104, 950–963. [Google Scholar] [CrossRef]
- Brown, A.V.; Conners, S.I.; Huang, W.; Wilkey, A.P.; Grant, D.; Weeks, N.T.; Cannon, S.B.; Graham, M.A.; Nelson, R.T. A new decade and new data at SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2020, 49, D1496–D1501. [Google Scholar] [CrossRef]
- Kassem, M.A. Two Decades of QTL Mapping of Isoflavone in Soybean Seed. In Soybean Seed Composition: Protein, Oil, Fatty Acids, Amino Acids, Sugars, Mineral Nutrients, and Isoflavone, 1st ed.; Kassem, M.A., Ed.; Springer Nature: Basingstoke, UK, 2021. [Google Scholar]
- Grant, D.; Nelson, R.T.; Cannon, S.B.; Shoemaker, R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010, 38, D843–D846. [Google Scholar] [CrossRef]
- Yang, C.; Yan, J.; Jiang, S.; Li, X.; Min, H.; Wang, X.; Hao, D. Resequencing 250 Soybean Accessions: New Insights into Genes Associated with Agronomic Traits and Genetic Networks. bioRxiv 2021. [Google Scholar] [CrossRef]
- Akond, M.; Liu, S.; Schoener, L.; Anderson, J.A.; Kantartzi, S.K.; Meksem, K.; Song, Q.; Wang, D.; Wen, Z.; Lightfoot, D.A.; et al. A SNP-Based Genetic Linkage Map of Soybean Using the SoyS—NP6K Illumina Infinium BeadChip Genotyping Array. Plant Genet. Genom. Sci. 2013, 1, 80–89. [Google Scholar] [CrossRef][Green Version]
- Kassem, M.A.; Ramos, L.; Leandro, L.; Mbofung, G.; Hyten, D.L.; Kantartzi, S.K.; Grier, R.L.; Njiti, V.N.; Cianzio, S.; Meksem, K. The ‘PI 438489B’ by ‘Hamilton’ SNP-Based Genetic Linkage Map of Soybean [Glycine max (L.) Merr.] Identified Quantitative Trait Loci that Underlie Seedling SDS Resistance. J. Plant Genome Sci. 2012, 1, 18–30. [Google Scholar] [CrossRef]
- Azam, M.; Zhang, S.; Abdelghany, A.M.; Shaibu, A.S.; Feng, Y.; Li, Y.; Tian, Y.; Hong, H.; Li, B.; Sun, J. Seed isoflavone profiling of 1168 soybean accessions from major growing ecoregions in China. Food Res. Int. 2020, 130, 108957. [Google Scholar] [CrossRef]
- Hsiao, Y.-H.; Ho, C.-T.; Pan, M.-H. Bioavailability and health benefits of major isoflavone aglycones and their metabolites. J. Funct. Foods 2020, 74, 104164. [Google Scholar] [CrossRef]
- Zhang, J.; Ge, Y.; Han, F.; Li, B.; Yan, S.; Sun, J.; Wang, L. Isoflavone Content of Soybean Cultivars from Maturity Group 0 to VI Grown in Northern and Southern China. J. Am. Oil Chem. Soc. 2014, 91, 1019–1028. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Yin, X.; Bellaloui, N.; McClure, M.A.; Mengistu, A. Soybean seed isoflavones respond differentially to phosphorus applications in low and high phosphorus soils. Nutr. Cycl. Agroecosyst. 2019, 113, 217–230. [Google Scholar] [CrossRef]
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5; Department of Statistics, NCSU: Raleigh, NC, USA, 2012; Available online: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 10 March 2021).
- Pei, R.; Zhang, J.; Tian, L.; Zhang, S.; Han, F.; Yan, S.; Wang, L.; Li, B.; Sun, J. Identification of novel QTL associated with soy-bean isoflavone content. Crop. J. 2018, 6, 244–252. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Fliegmann, J.; Furtwängler, K.; Malterer, G.; Cantarello, C.; Schüler, G.; Ebel, J.; Mithöfer, A. Flavone synthase II (CYP93B16) from soybean (Glycine max L.). Phytochemistry 2010, 71, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.L.; Han, Y.P.; Teng, W.L.; Li, Y.G.; Bin Li, W. QTL underlying the resistance to soybean aphid (Aphis glycines Matsumura) through isoflavone-mediated antibiosis in soybean cultivar ‘Zhongdou 27’. Theor. Appl. Genet. 2011, 123, 1459–1465. [Google Scholar] [CrossRef] [PubMed]
- Kassem, M.A. QTL that Control Seed Protein, Oil, and Fatty Acids Contents. In Soybean Seed Composition: Protein, Oil, Fatty Acids, Amino Acids, Sugars, Mineral Nutrients, and Isoflavone, 1st ed.; Kassem, M.A., Ed.; Springer Nature: Basingstoke, UK, 2021. [Google Scholar]
- Phytozome. Available online: https://phytozome.jgi.doe.gov/pz/portal.html# (accessed on 5 July 2021).
- Hartwig, E.E.; Epps, J.M. Registration of ‘Forrest’ soybeans. Crop. Sci. 1973, 13, 287. [Google Scholar] [CrossRef]
- Bernard, R.L.; Cremeens, C.R. Registration of Williams 82 soybean. Crop. Sci. 1988, 28, 1027–1028. [Google Scholar] [CrossRef]
- Shultz, J.L.; Kurunam, D.; Shopinski, K.; Iqbal, M.J.; Kazi, S.; Zobrist, K.; Bashir, R.; Yaegashi, S.; Lavu, N.; Afzal, A.J.; et al. The soybean genome database (SoyGD): A browser for display of duplicated, polyploid, regions and sequence tagged sites on the integrated physical and genetic maps of Glycine max. Nucleic Acids Res. 2006, 34, D758–D765. [Google Scholar] [CrossRef]
- Wu, X.; Vuong, T.D.; Leroy, J.A.; Shannon, J.G.; Sleper, D.A.; Nguyen, H.T. Selection of a core set of RILs from Forrest × Williams 82 to develop a framework map in soybean. Theor. Appl. Genet. 2011, 122, 1179–1187. [Google Scholar] [CrossRef][Green Version]
- Weather Spark. Available online: https://weatherspark.com (accessed on 5 July 2021).
- Bellaloui, N.; Mengistu, A.; Fisher, D.K.; Abel, C.A. Soybean Seed Composition Constituents as Affected by Drought andPhomopsisin Phomopsis Susceptible and Resistant Genotypes. J. Crop. Improv. 2012, 26, 428–453. [Google Scholar] [CrossRef]
- Vuong, T.D.; Sleper, D.A.; Shannon, J.G.; Nguyen, H.T. Novel quantitative trait loci for broad-based resistance to soybean cyst nematode (Heterodera glycines Ichinohe) in soybean PI 567516C. Theor. Appl. Genet. 2010, 121, 1253–1266. [Google Scholar] [CrossRef]
- Song, Q.; Yan, L.; Quigley, C.; Fickus, E.; Wei, H.; Chen, L.; Dong, F.; Araya, S.; Liu, J.; Hyten, D.; et al. Soybean BARCSoySNP6K: An assay for soybean genetics and breeding research. Plant J. 2020, 104, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. Joinmap 4.0 Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Plant Res Intl.: Wageningen, The Netherlands, 2006. [Google Scholar]
- Pilet-Nayel, M.L.; Muehlbauer, F.J.; McGee, R.J.; Kraft, J.M.; Baranger, A.; Coyne, C.J. Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor. Appl. Genet. 2002, 106, 28–39. [Google Scholar] [CrossRef] [PubMed]
- R Software. Available online: https://www.r-project.org (accessed on 10 March 2021).
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTL. J. Heredity 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Lakhssassi, N.; Lopes-Caitar, V.S.; Knizia, D.; Cullen, M.A.; Badad, O.; El Baze, A.; Zhou, Z.; Embaby, M.G.; Meksem, J.; Lakhssassi, A.; et al. TILLING-by-Sequencing+ Reveals the Role of Novel Fatty Acid Desaturases (GmFAD2-2s) in Increasing Soybean Seed Oleic Acid Content. Cells 2021, 10, 1245. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.1–11.14.19. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Lakhssassi, N.; Zhou, Z.; Liu, S.; Piya, S.; Patil, G.B.; Cullen, M.A.; El Baz, A.; Badad, O.; Embaby, M.G.; Meksem, J.; et al. Soybean TILLING-by-Sequencing+ reveals the role of novel GmSACPD members in the unsaturated fatty acid biosynthesis while maintaining healthy nodules. J. Exp. Bot. 2020, 71, 6969–6987. [Google Scholar] [CrossRef] [PubMed]




| Chr. | No. of SNP Markers | Length (cM) | Average Marker Density (cM) | Maximum Gap (cM) |
|---|---|---|---|---|
| 1 | 110 | 190.1 | 1.73 | 48.7 |
| 2 | 161 | 308.3 | 1.91 | 59.0 |
| 3 | 92 | 173.9 | 1.89 | 22.9 |
| 4 | 71 | 214.9 | 3.03 | 57.4 |
| 5 | 138 | 167.2 | 1.21 | 38.1 |
| 6 | 114 | 253.7 | 2.23 | 43.4 |
| 7 | 117 | 224.0 | 1.91 | 18.4 |
| 8 | 71 | 211.1 | 2.97 | 45.3 |
| 9 | 109 | 179.2 | 1.64 | 62.6 |
| 10 | 100 | 216.5 | 2.17 | 48.5 |
| 11 | 95 | 168.9 | 1.78 | 41.3 |
| 12 | 73 | 192.6 | 2.64 | 31.5 |
| 13 | 156 | 265.7 | 1.70 | 57.7 |
| 14 | 50 | 158.9 | 3.18 | 25.8 |
| 15 | 94 | 219.1 | 2.33 | 68.4 |
| 16 | 95 | 169.0 | 1.78 | 46.7 |
| 17 | 79 | 185.4 | 2.35 | 46.5 |
| 18 | 144 | 153.7 | 1.07 | 23.1 |
| 19 | 125 | 190.4 | 1.52 | 53.3 |
| 20 | 81 | 187.3 | 2.31 | 25.7 |
| Totals | 2075 | 4029.9 | Av. = 1.94 | Av. = 43.2 |
| Trait | Mean | Range | CV | SE | Skewness | Kurtosis | p Value (p > 0.05) |
|---|---|---|---|---|---|---|---|
| Daidzein 2018 | 303.22 | 171 | 10.11 | 2.23 | 0.26 | 3.11 | 0.99 |
| Glycitein2018 | 490.47 | 610 | 19.37 | 7.01 | 0.29 | 3.65 | 0.98 |
| Genistein2018 | 391 | 348 | 15.47 | 4.4 | 0.3 | 3.04 | 0.99 |
| Daidzein 2020 | 14.48 | 8.08 | 13.72 | 0.42 | −0.08 | 3.17 | 0.99 |
| Glycitein2020 | 71.79 | 46 | 12.59 | 0.53 | 0.178 | 2.94 | 0.99 |
| Genistein2020 | 584.88 | 383 | 10.94 | 3.73 | −0.02 | 3.31 | 0.99 |
| Response: Daidzein | ||||
|---|---|---|---|---|
| Df | Sum Sq | Mean Seq | H2 | |
| Line | 301 | 541,800 | 1800 | 0.428 |
| Year | 1 | 974,711 | 974,711 | |
| Line: Year | 181 | 186,226 | 1029 | |
| Residuals | 0 | 0 | NA | |
| Response: Glycitein | ||||
| Df | Sum Sq | Mean Seq | H2 | |
| Line | 301 | 5,086,274 | 16,898 | 0.724 |
| Year | 1 | 16,033,506 | 16,033,506 | |
| Line: Year | 181 | 843,473 | 4660 | |
| Residuals | 0 | 0 | NA | |
| Response: Genistein | ||||
| Df | Sum Sq | Mean Seq | H2 | |
| Line | 301 | 1,922,339 | 6387 | 0.425 |
| Year | 1 | 3,630,207 | 3,630,207 | |
| Line: Year | 181 | 668,735 | 3695 | |
| Residuals | 0 | 0 | NA |
| 2A. QTL Identified in Spring Lake, NC (2018) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Trait | QTL | Chr. | Marker | Interval (cM) | LOD | R2 (%) | Additive Effect | Environment |
| Daidzein | qDAID01 | 5 | Gm05_1705841 | 160.41 | 2.01 | 6.07 | −7.61 | Spring Lake, NC |
| Daidzein | 5 | Gm05_9012813 | 166.51 | 2.12 | 4.18 | −6.35 | Spring Lake, NC | |
| Daidzein | 5 | Gm05_9097414 | 166.71 | 2.19 | 4.32 | −6.46 | Spring Lake, NC | |
| Daidzein | 5 | Gm05_8916450 | 166.81 | 2.11 | 4.16 | −6.34 | Spring Lake, NC | |
| Genistein | qGEN03 | 5 | Gm05_1705841 | 152.41 | 2.06 | 9.37 | −18.72 | Spring Lake, NC |
| Genistein | 5 | Gm05_9012813 | 166.51 | 2.27 | 4.22 | −12.53 | Spring Lake, NC | |
| Genistein | 5 | Gm05_9097414 | 166.71 | 2.36 | 4.39 | −12.79 | Spring Lake, NC | |
| Genistein | 5 | Gm05_8916450 | 166.81 | 2.28 | 4.25 | −12.57 | Spring Lake, NC | |
| Glycitein | qGLY02 | 5 | Gm05_1705841 | 146.41 | 2.01 | 9.07 | −29.89 | Spring Lake, NC |
| Genistein | qGEN01 | 6 | Gm06_5014399 | 64.41 | 2.58 | 8.52 | 21.15 | Spring Lake, NC |
| Daidzein | qDAID02 | 6 | Gm06_5014399 | 62.41 | 2.06 | 7.55 | 10.35 | Spring Lake, NC |
| Daidzein | 6 | Gm06_3941524 | 78.21 | 2.02 | 7.24 | 8.67 | Spring Lake, NC | |
| Genistein | qGEN04 | 6 | Gm06_5014399 | 60.41 | 2.26 | 9.06 | 22.94 | Spring Lake, NC |
| Genistein | 6 | Gm06_3941524 | 70.21 | 2.11 | 3.98 | 13.86 | Spring Lake, NC | |
| Genistein | qGEN02 | 12 | Gm12_915327 | 179.21 | 2.56 | 4.8 | −16.87 | Spring Lake, NC |
| Genistein | 12 | Gm12_1064727 | 179.41 | 2.58 | 4.85 | −16.93 | Spring Lake, NC | |
| Genistein | 12 | Gm12_1229101 | 179.71 | 2.95 | 5.51 | −17.79 | Spring Lake, NC | |
| Genistein | 12 | Gm12_1374970 | 179.91 | 2.95 | 5.5 | −17.78 | Spring Lake, NC | |
| Genistein | 12 | Gm12_1433336 | 180.61 | 2.85 | 5.33 | −17.4 | Spring Lake, NC | |
| Glycitein | qGLY01 | 12 | Gm12_553862 | 177.31 | 2.76 | 5.82 | −26.95 | Spring Lake, NC |
| Glycitein | 12 | Gm12_915327 | 179.21 | 2.58 | 4.79 | −27.05 | Spring Lake, NC | |
| Glycitein | 12 | Gm12_1064727 | 179.41 | 2.59 | 4.82 | −27.12 | Spring Lake, NC | |
| Glycitein | 12 | Gm12_1229101 | 179.71 | 2.83 | 5.23 | −27.76 | Spring Lake, NC | |
| Glycitein | 12 | Gm12_1374970 | 179.91 | 2.83 | 5.23 | −27.76 | Spring Lake, NC | |
| Glycitein | 12 | Gm12_1433336 | 180.61 | 2.7 | 5.02 | −27.03 | Spring Lake, NC | |
| Genistein | qGEN05 | 12 | Gm12_553862 | 171.31 | 2.03 | 5.74 | −15.18 | Spring Lake, NC |
| Genistein | 12 | Gm12_975837 | 178.71 | 2.47 | 4.64 | −16.47 | Spring Lake, NC | |
| Genistein | 12 | Gm12_1632399 | 181.31 | 2.32 | 4.38 | −15.76 | Spring Lake, NC | |
| Glycitein | qGLY03 | 12 | Gm12_553862 | 169.31 | 2.22 | 5.63 | −23.71 | Spring Lake, NC |
| Glycitein | 12 | Gm12_975837 | 178.71 | 2.46 | 4.58 | −26.29 | Spring Lake, NC | |
| Glycitein | 12 | Gm12_1632399 | 181.31 | 2.2 | 4.1 | −24.52 | Spring Lake, NC | |
| Daidzein | qDAID03 | 19 | Gm19_4552537 | 109.51 | 2.14 | 4.17 | 6.92 | Spring Lake, NC |
| Glycitein | qGLY04 | 19 | Gm19_3010363 | 35.31 | 2.04 | 3.83 | 19.46 | Spring Lake, NC |
| Genistein | qGEN06 | 20 | Gm20_4657454 | 0.01 | 2.03 | 3.75 | 11.84 | Spring Lake, NC |
| 2B. QTL Identified in Carbondale, IL (2020) | ||||||||
| Trait | QTL | Chr. | Marker | Interval (cM) | LOD | R2 (%) | Additive Effect | Environment |
| Daidzein | qDAID03 | 2 | Gm02_2282900 | 24.01 | 2.01 | 10.61 | −11.55 | Carbondale, IL |
| Genistein | qGEN04 | 4 | Gm04_4461164 | 190.31 | 2.26 | 3.96 | −12.97 | Carbondale, IL |
| Genistein | qGEN01 | 10 | Gm10_4670275 | 130.81 | 2.6 | 3.52 | −12.38 | Carbondale, IL |
| Genistein | 10 | Gm10_4035277 | 130.91 | 2.61 | 3.53 | −12.39 | Carbondale, IL | |
| Daidzein | qDAID04 | 10 | Gm10_4670275 | 130.81 | 2.18 | 2.97 | −6.11 | Carbondale, IL |
| Daidzein | 10 | Gm10_4035277 | 130.91 | 2.19 | 2.97 | −6.12 | Carbondale, IL | |
| Genistein | qGEN05 | 10 | Gm10_4670275 | 128.81 | 2.15 | 3.2 | −11.75 | Carbondale, IL |
| Genistein | 10 | Gm10_4035277 | 132.91 | 2.37 | 3.39 | −12.11 | Carbondale, IL | |
| Daidzein | qDAID01 | 12 | Gm12_9193994 | 53.21 | 2.56 | 4.07 | 7.18 | Carbondale, IL |
| Daidzein | 12 | Gm12_1430950 | 61.71 | 3.99 | 5.61 | 8.49 | Carbondale, IL | |
| Daidzein | 12 | Gm12_1423120 | 62.31 | 4.12 | 5.78 | 8.62 | Carbondale, IL | |
| Daidzein | 12 | Gm12_1539402 | 63.01 | 4.53 | 6.34 | 9.01 | Carbondale, IL | |
| Daidzein | 12 | Gm12_1678702 | 63.11 | 4.59 | 6.45 | 9.1 | Carbondale, IL | |
| Daidzein | 12 | Gm12_3052701 | 64.11 | 4.89 | 6.82 | 9.42 | Carbondale, IL | |
| Daidzein | 12 | Gm12_2097199 | 64.41 | 4.4 | 6.18 | 8.97 | Carbondale, IL | |
| Daidzein | 12 | Gm12_2432082 | 65.31 | 4.26 | 5.97 | 8.77 | Carbondale, IL | |
| Daidzein | 12 | Gm12_1547239 | 65.51 | 3.77 | 5.31 | 8.27 | Carbondale, IL | |
| Daidzein | 12 | Gm12_1428801 | 65.91 | 3.36 | 4.75 | 7.87 | Carbondale, IL | |
| Genistein | qGEN02 | 12 | Gm12_9193994 | 55.21 | 2.5 | 4.24 | 13.58 | Carbondale, IL |
| Genistein | 12 | Gm12_1430950 | 61.71 | 3.78 | 5.28 | 15.31 | Carbondale, IL | |
| Genistein | 12 | Gm12_1423120 | 62.31 | 3.76 | 5.25 | 15.2 | Carbondale, IL | |
| Genistein | 12 | Gm12_1539402 | 63.01 | 4.39 | 6.1 | 16.4 | Carbondale, IL | |
| Genistein | 12 | Gm12_1678702 | 63.11 | 4.38 | 6.1 | 16.4 | Carbondale, IL | |
| Genistein | 12 | Gm12_3052701 | 64.11 | 4.66 | 6.46 | 16.99 | Carbondale, IL | |
| Genistein | 12 | Gm12_2097199 | 64.41 | 4.05 | 5.65 | 15.64 | Carbondale, IL | |
| Genistein | 12 | Gm12_2432082 | 65.31 | 3.83 | 5.35 | 15.25 | Carbondale, IL | |
| Genistein | 12 | Gm12_1547239 | 65.51 | 3.38 | 4.74 | 14.39 | Carbondale, IL | |
| Genistein | 12 | Gm12_1428801 | 65.91 | 3.16 | 4.44 | 13.96 | Carbondale, IL | |
| Daidzein | qDAID05 | 12 | Gm12_9193994 | 51.21 | 2.03 | 2.91 | 6.05 | Carbondale, IL |
| Daidzein | 12 | Gm12_1428801 | 73.91 | 2.31 | 4.8 | 7.83 | Carbondale, IL | |
| Genistein | qGEN06 | 12 | Gm12_1428801 | 71.91 | 2.37 | 4.4 | 13.77 | Carbondale, IL |
| Glycitein | qGLY01 | 15 | Gm15_756303 | 212.31 | 2.07 | 2.86 | −1.53 | Carbondale, IL |
| Glycitein | 15 | Gm15_2072075 | 218.41 | 2.24 | 3.1 | −1.6 | Carbondale, IL | |
| Glycitein | 15 | Gm15_2021199 | 218.81 | 2.06 | 2.84 | −1.53 | Carbondale, IL | |
| Daidzein | qDAID02 | 20 | Gm20_3804081 | 70.31 | 2.55 | 6.13 | −8.81 | Carbondale, IL |
| Genistein | qGEN03 | 20 | Gm20_3804081 | 68.31 | 2.65 | 6.72 | −16.81 | Carbondale, IL |
| Daidzein | qDAID06 | 20 | Gm20_3804081 | 66.31 | 2.22 | 5.58 | −8.41 | Carbondale, IL |
| Genistein | qGEN07 | 20 | Gm20_3804081 | 64.31 | 2 | 5.08 | −14.62 | Carbondale, IL |
| Genistein | 20 | Gm20_3424023 | 80.01 | 2.44 | 3.3 | −11.8 | Carbondale, IL | |
| Genistein | 20 | Gm20_3418121 | 80.51 | 2.1 | 2.85 | −10.96 | Carbondale, IL | |
| (A). | |||||||
|---|---|---|---|---|---|---|---|
| Environment | Trait | QTL | Chr. | Gene | Start | End | Distance (cM) |
| 2018 CIM QTL with LOD Scores > 2.5 | |||||||
| Spring Lake, NC | Genistein | qGEN01 | 6 | Glyma.06G128200 | 10,543,911 | 10,545,747 | 5.52 cM |
| Glyma.06G137100 | 11,225,188 | 11,228,664 | 6.21 cM | ||||
| Glyma.06G137300 | 11,237,072 | 11,239,469 | 6.22 cM | ||||
| Glyma.06G143000 | 11,642,031 | 11,644,022 | 6.62 cM | ||||
| Spring Lake, NC | Genistein | qGEN02 | 12 | Glyma.12G067000 | 4,909,073 | 4,911,905 | 3.47 cM |
| Glyma.12G067100 | 4,919,960 | 4,922,998 | 3.48 cM | ||||
| Spring Lake, NC | Glycitein | qGLY01 | 12 | Glyma.12G067000 | 4,909,073 | 4,911,905 | 3.47 cM |
| Glyma.12G067100 | 4,919,960 | 4,922,998 | 3.48 cM | ||||
| 2018 CIM QTL with LOD Scores 2.0 < LOD < 2.5 | |||||||
| Spring Lake, NC | Daidzein | qDAID01 | 5 | - | - | - | - |
| Spring Lake, NC | Daidzein | qDAID02 | 6 | Glyma.06G128200 | 10,543,911 | 10,545,747 | 5.52 cM |
| Spring Lake, NC | Daidzein | qDAID03 | 19 | Glyma.19G030500 | 3,779,017 | 3,781,453 | 0.77 cM |
| Glyma.19G030700 | 3,794,404 | 3,796,426 | 0.75 cM | ||||
| Glyma.19G030800 | 3,799,941 | 3,801,335 | 0.75 cM | ||||
| Spring Lake, NC | Genistein | qGEN03 | 5 | - | |||
| Spring Lake, NC | Genistein | qGEN04 | 6 | Glyma.06G128200 | 10,543,911 | 10,545,747 | 5.52 cM |
| Spring Lake, NC | Genistein | qGEN05 | 12 | Glyma.12G067000 | 4,909,073 | 4,911,905 | 3.27 cM |
| Glyma.12G067100 | 4,919,960 | 4,922,998 | 3.28 Cm | ||||
| Spring Lake, NC | Genistein | qGEN06 | 20 | Glyma.20G027800 | 3,179,955 | 3,183,453 | 1.47 cM |
| Spring Lake, NC | Glycitein | qGLY02 | 5 | - | - | - | - |
| Spring Lake, NC | Glycitein | qGLY03 | 12 | Glyma.12G067000 | 4,909,073 | 4,911,905 | 3.27 cM |
| Glyma.12G067100 | 4,919,960 | 4,922,998 | 3.28 Cm | ||||
| Spring Lake, NC | Glycitein | qGLY04 | 19 | Glyma.19G030500 | 3,779,017 | 3,781,453 | 0.76 cM |
| Glyma.19G030700 | 3,794,404 | 3,796,426 | 0.78 cM | ||||
| Glyma.19G030800 | 3,799,941 | 3,801,335 | 0.78 cM | ||||
| (B). | |||||||
| Environment | Trait | QTL | Chr. | Gene | Start | End | Distance (cM) |
| 2020 CIM QTL with LOD Scores > 2.5 | |||||||
| Carbondale, IL | Daidzein | qDAID01 | 12 | Glyma.12G067000 | 4,909,073 | 4,911,905 | - |
| Glyma.12G067100 | 4,919,960 | 4,922,998 | - | ||||
| Carbondale, IL | Daidzein | qDAID02 | 20 | Glyma.20G027800 | 3,179,955 | 3,183,453 | 0.62 cM |
| Carbondale, IL | Genistein | qGEN01 | 10 | Glyma.10G058200 | 5,328,963 | 5,333,501 | 0.6 cM |
| Carbondale, IL | Genistein | qGEN02 | 12 | Glyma.12G067000 | 4,909,073 | 4,911,905 | - |
| Glyma.12G067100 | 4,919,960 | 4,922,998 | - | ||||
| Carbondale, IL | Genistein | qGEN03 | 20 | Glyma.20G027800 | 3,179,955 | 3,183,453 | 2.44 cM |
| 2020 CIM QTL with LOD Scores 2.0 < LOD < 2.5 | |||||||
| Carbondale, IL | Daidzein | qDAID03 | 2 | Glyma.02G067900 | 5,986,285 | 5,987,684 | 3.70 cM |
| Glyma.02G130400 | 13,399,253 | 13,401,493 | 11.11 cM | ||||
| Carbondale, IL | Daidzein | qDAID04 | 10 | Glyma.10G058200 | 5,328,963 | 5,333,501 | 0.6 cM |
| Carbondale, IL | Daidzein | qDAID05 | 12 | Glyma.12G067000 | 4,909,073 | 4,911,905 | - |
| Glyma.12G067100 | 4,919,960 | 4,922,998 | - | ||||
| Carbondale, IL | Daidzein | qDAID06 | 20 | Glyma.20G027800 | 3,179,955 | 3,183,453 | - |
| Carbondale, IL | Genistein | qGEN04 | 4 | - | - | - | - |
| Carbondale, IL | Genistein | qGEN05 | 10 | Glyma.10G058200 | 5,328,963 | 5,333,501 | 0.6 cM |
| Carbondale, IL | Genistein | qGEN06 | 12 | Glyma.12G067000 | 4,909,073 | 4,911,905 | 3.48 cM |
| Glyma.12G067100 | 4,919,960 | 4,922,998 | 3.49 cM | ||||
| Carbondale, IL | Genistein | qGEN07 | 20 | Glyma.20G027800 | 3,179,955 | 3,183,453 | 2.44 cM |
| Carbondale, IL | Glycitein | qGLY01 | 15 | Glyma.15G001700 | 190,985 | 194,451 | 0.56 cM |
| Glyma.15G156300 | 13,098,492 | 13,100,036 | 11.02 cM | ||||
| Glyma.15G156100 | 13,076,997 | 13,079,333 | 11 cM | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knizia, D.; Yuan, J.; Bellaloui, N.; Vuong, T.; Usovsky, M.; Song, Q.; Betts, F.; Register, T.; Williams, E.; Lakhssassi, N.; et al. The Soybean High Density ‘Forrest’ by ‘Williams 82’ SNP-Based Genetic Linkage Map Identifies QTL and Candidate Genes for Seed Isoflavone Content. Plants 2021, 10, 2029. https://doi.org/10.3390/plants10102029
Knizia D, Yuan J, Bellaloui N, Vuong T, Usovsky M, Song Q, Betts F, Register T, Williams E, Lakhssassi N, et al. The Soybean High Density ‘Forrest’ by ‘Williams 82’ SNP-Based Genetic Linkage Map Identifies QTL and Candidate Genes for Seed Isoflavone Content. Plants. 2021; 10(10):2029. https://doi.org/10.3390/plants10102029
Chicago/Turabian StyleKnizia, Dounya, Jiazheng Yuan, Nacer Bellaloui, Tri Vuong, Mariola Usovsky, Qijian Song, Frances Betts, Teresa Register, Earl Williams, Naoufal Lakhssassi, and et al. 2021. "The Soybean High Density ‘Forrest’ by ‘Williams 82’ SNP-Based Genetic Linkage Map Identifies QTL and Candidate Genes for Seed Isoflavone Content" Plants 10, no. 10: 2029. https://doi.org/10.3390/plants10102029
APA StyleKnizia, D., Yuan, J., Bellaloui, N., Vuong, T., Usovsky, M., Song, Q., Betts, F., Register, T., Williams, E., Lakhssassi, N., Mazouz, H., Nguyen, H. T., Meksem, K., Mengistu, A., & Kassem, M. A. (2021). The Soybean High Density ‘Forrest’ by ‘Williams 82’ SNP-Based Genetic Linkage Map Identifies QTL and Candidate Genes for Seed Isoflavone Content. Plants, 10(10), 2029. https://doi.org/10.3390/plants10102029










