DNA Barcode Authentication of Devil’s Claw Herbal Dietary Supplements
Abstract
:1. Introduction
2. Results
2.1. Reference Sequences
| Species | Voucher Specimen | Locality | Sample Type | GenBank Accession | |||
|---|---|---|---|---|---|---|---|
| rbcL | matK | nrITS2 | psbA-trnH | ||||
| Dicerocaryum zanguebarium | Loeb and Koch 339 (NY) | Namibia: Oshikango | reference and validation | — | — | — | KT717163 |
| Harpagophytum procumbens | Allen 308 (M0) | Botswana: Orapa | reference and validation | — | KT717103 | KT717127 | KT717148 |
| Harpagophytum procumbens | Davidse and Loxton 6296 (MO) | Namibia: Keetmanshoop | reference and validation | KT717178 | KT717109 | KT717133 | KT717153 |
| Harpagophytum procumbens | de Koning 8142 (MO) | Mozambique: Chigubo | reference | — | KT717110 | — | — |
| Harpagophytum procumbens | Dinter 396 (MO) | Namibia: Okahandja | reference and validation | — | — | — | KT717150 |
| Harpagophytum procumbens | Grignon 239 (MO) | Botswana: Ghanzi | reference and validation | KT717174 | KT717104 | KT717128 | KT717149 |
| Harpagophytum procumbens | Hardy 6575 (MO) | Namibia: Aranos | reference and validation | KT717168 | KT717095 | KT717124 | KT717154 |
| Harpagophytum procumbens | Herman 1264 (MO) | South Africa: Blouberg Privaatnatuurreserwe | reference and validation | KT717176 | KT717107 | KT717131 | KT717151 |
| Harpagophytum procumbens | Lavranos and Bleck 22701 (MO) | Namibia: Otjiwarongo | reference and validation | KT717177 | KT717108 | KT717132 | KT717152 |
| Harpagophytum procumbens | Lavranos and Bleck 22703 (MO) | Namibia: Khorixas | reference and validation | KT717173 | KT717102 | KT717126 | KT717147 |
| Harpagophytum procumbens | Leach 10682 (MO) | Zimbabwe: Beit Bridge | reference | — | KT717099 | — | — |
| Harpagophytum procumbens | Long and Rae 44 (MO) | Botswana: Jwaneng | reference and validation | KT717171 | KT717101 | KT717120 | KT717145 |
| Harpagophytum procumbens | Ngoni 257 (MO) | Botswana: Mosu | reference | — | KT717105 | KT717129 | KY706349 |
| Harpagophytum procumbens | Owens 19 (MO) | Botswana: Deception Valley | reference and validation | KT717172 | KT717096 | KT717125 | KT717146 |
| Harpagophytum procumbens | Rodin 3539 (NY) | South Africa: Vryburg | reference | — | — | — | KY706351 |
| Harpagophytum procumbens | Rogers s.n. (MO) | South Africa: Bellville | reference | — | KT717097 | — | KY706348 |
| Harpagophytum procumbens | Sidey 305 (MO) | South Africa: Fauresmith | reference and validation | KT717169 | KT717098 | KT717119 | KT717143 |
| Harpagophytum procumbens | Skarpe S-319 (MO) | Botswana: Hukuntsi | reference and validation | KT717170 | KT717100 | KT717123 | KT717144 |
| Harpagophytum procumbens | Smuts and Gillelt 2130 (MO) | South Africa: Rooikop | validation | — | — | — | — |
| Harpagophytum procumbens | Venter 9637 (MO, NY) | South Africa: Glen Agricultural College | reference | KT717175 | KT717106 | KT717130 | KY706350 |
| Harpagophytum zeyheri | Germishuizen 00733 (MO) | South Africa: Bamboeskloof | reference and validation | — | KT717114 | KT717122 | KT717159 |
| Harpagophytum zeyheri | Germishuizen 990 (MO) | South Africa: Vaalwater | reference and validation | KT717183 | — | KT717138 | KT717160 |
| Harpagophytum zeyheri | Luwiika et al. 335 (MO) | Zambia: Lukona Basic School | reference | — | KT717116 | KT717137 | — |
| Harpagophytum zeyheri | Mashasha 111 (MO) | Zimbabwe: Victoria Falls | reference and validation | KT717179 | KT717111 | KT717134 | KT717155 |
| Harpagophytum zeyheri | Mogg 37171 (MO) | South Africa: Sandsloot | reference and validation | KT717182 | KT717113 | KT717136 | KT717157 |
| Harpagophytum zeyheri | Moyo 7 (MO) | Zimbabwe: Victoria Falls | reference | — | — | KT717118 | — |
| Harpagophytum zeyheri | Norlindh and Weimarck 5234 (NY) | South Africa: Pietersburg | reference | — | — | — | KY706353 |
| Harpagophytum zeyheri | Rodin 9140 (MO) | Namibia: Rundu | reference and validation | KT717184 | KT717115 | KT717121 | KT717158 |
| Harpagophytum zeyheri | Rushworth 110 (MO) | Zimbabwe: Dina Pan | reference and validation | KT717180 | KT717094 | KT717135 | KT717156 |
| Harpagophytum zeyheri | Yalala 300 (MO) | Botswana: Mahalapye | reference | KT717181 | KT717112 | KT717117 | KY706352 |
| Josephinia euginiae | Michell and Boyce 3144 (MO) | Australia: Nitmiluk National Park | reference and validation | — | — | — | KT717162 |
| Pedaliodiscus macrocarpus | Luke et al. TPR 73 (MO) | Kenya: Tana River National Primate Reserve | reference and validation | — | — | — | KT717139 |
| Pedalium murex | Comanor 608 (NY) | Sri Lanka: Potuvil—Panama Road | reference and validation | — | — | — | KT717140 |
| Pterodiscus auranthacus | Seydel 4135 (NY) | Namibia: Windhoek | reference and validation | — | — | — | KT717141 |
| Pterodiscus speciosus | Zietsman 4079 (NY) | South Africa: Hoopstad | reference and validation | — | — | — | KT717142 |
| Rogeria adenophylla | Seydel 4368 (NY) | Namibia: Windhoek | reference and validation | — | — | — | KT717167 |
| Sesamum indicum | Donmez 9932 (NY) | Turkey: Kula | reference and validation | — | — | — | KT717164 |
| Sesamum indicum | Nesbitt 1939 (RNG) | — | reference | — | — | — | EU531713 |
| Sesamum radiatum | Thomas 10563 (NY) | Brazil: Ilhéus | reference and validation | — | — | — | KT717165 |
| Sesamum triphyllum | Zietsman and Peyper 4061 (NY) | South Africa: Petrusburg | reference and validation | — | — | — | KT717161 |
| Uncarina grandidieri | Falk 97001 (NY) | cultivated | reference and validation | — | — | — | KT717166 |
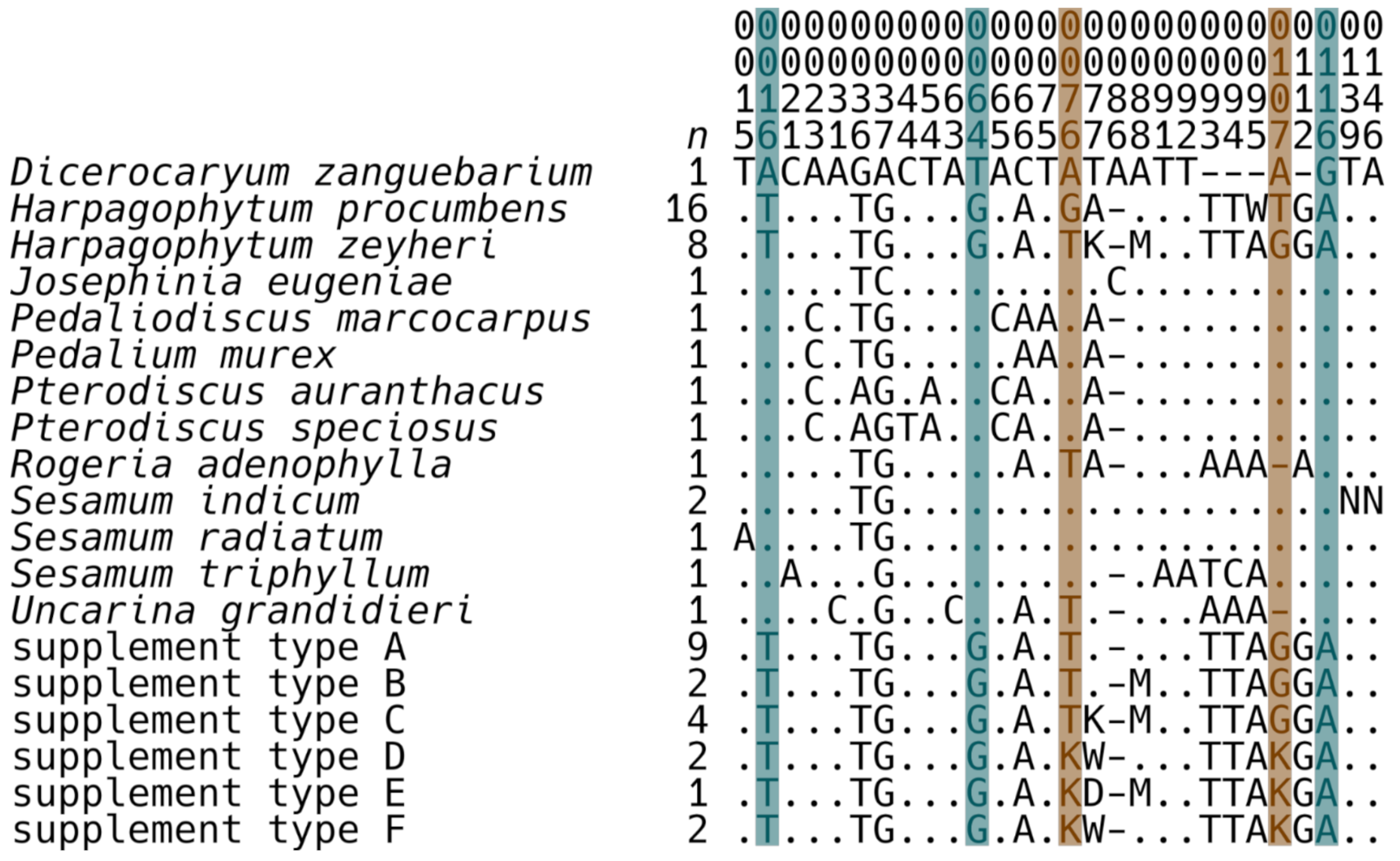
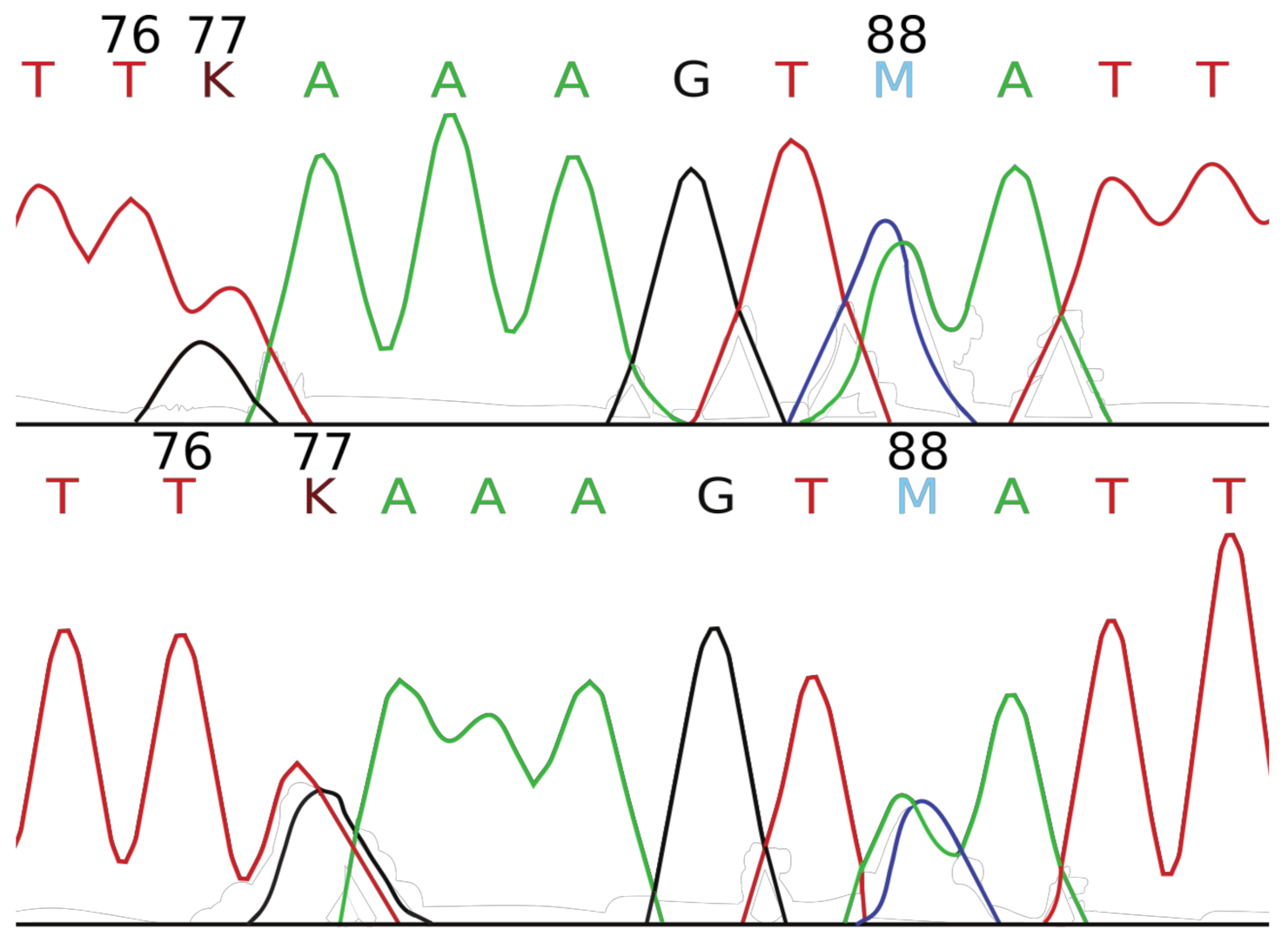
2.2. Mini-Barcode Validation
2.3. An Analysis of Herbal Supplements
| Supplement Sequence Type | Label Species | Devil’s Claw Material Type | Contains H. procumbens | Contains H. zeyheri |
|---|---|---|---|---|
| A | Harpagophytum procumbens, Curcuma longa, Crataegus oxyacantha, Arctium lappa, Smilax febrifuga, Yucca schidigera, Zingiber officinale, and Vaccinium myrtillus | root extract | no | yes |
| A | Harpagophytum procumbens | root | no | yes |
| A | Harpagophytum procumbens | root | no | yes |
| A | Boswellia serrata, Curcuma longa, and Harpagophytum procumbens | root extract | no | yes |
| A | Boswellia serrata, Uncaria tomentosa, Harpagophytum procumbens, Yucca schidigera, Gymnema sylvestre, Curcuma longa, Camellia sinensis, and Oryza sativa | root | no | yes |
| A | Harpagophytum procumbens and Oryza sativa | root extract | no | yes |
| A | Harpagophytum procumbens | root extract | no | yes |
| A | Harpagophytum procumbens | root | no | yes |
| A | Harpagophytum procumbens, Boswellia serrata, Curcuma longa, and Tanacetum parthenium | root extract | no | yes |
| B | Harpagophytum procumbens | root | no | yes |
| B | Harpagophytum procumbens | root extract | no | yes |
| C | Harpagophytum procumbens | root | no | yes |
| C | Harpagophytum procumbens and Oryza sativa | root extract | no | yes |
| C | Harpagophytum procumbens | root | no | yes |
| C | Harpagophytum procumbens | root | no | yes |
| D | Harpagophytum procumbens | root | yes | yes |
| D | Harpagophytum procumbens and/or Harpagophytum zeyheri | root extract | yes | yes |
| E | Harpagophytum procumbens | root and root extract | yes | yes |
| F | Harpagophytum procumbens | root | yes | yes |
| F | Harpagophytum procumbens and/or Harpagophytum zeyheri | root extract | yes | yes |
| — | Harpagophytum procumbens | root | unknown | unknown |
| — | Polygonum cuspidatum, Curcuma longa, Zingiber officinale, Camellia sinensis, Harpagophytum procumbens, and Salix alba | root extract | unknown | unknown |
| — | Harpagophytum procumbens | root extract | unknown | unknown |
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Stewart, K.M.; Cole, D. The commercial harvest of Devil’s Claw (Harpagophytum spp.) in Southern Africa: The Devil’s in the details. J. Ethnopharmacol. 2005, 100, 225–236. [Google Scholar] [CrossRef]
- Decaisne, J. Revue du groupe des pedalinees. Ann. Sci. Nat. Bot. 1865, 5, 321–336. [Google Scholar]
- Ihlenfeldt, H.-D.; Hartmann, H. Die gattung Harpagophytum (Burch.) DC. Ex Meissn. (Monographie Der Afrikanischen Pedaliaceae II). Mitt. Aus Dem Staatsinst. Fur Allg. Bot. Hambg. 1970, 13, 15–69. [Google Scholar]
- Muzila, M.; Setshogo, M.P.; Mpoloka, S.W. Multivariate Analysis of Harpagophytum DC. Ex Meisn (Pedaliaceae) based on fruit characters. Int. J. Biodivers. Conserv. 2011, 3, 101–109. [Google Scholar]
- Mncwangi, N.P.; Viljoen, A.M.; Zhao, J.; Vermaak, I.; Chen, W.; Khan, I. What the devil is in your phytomedicine? Exploring species substitution in Harpagophytum through chemometric modeling of 1H-NMR and UHPLC-MS datasets. Phytochemistry 2014, 106, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Ihlenfeldt, H.-D. Pedaliaceae. In Flora Zambesiaca; Royal Botanic Gardens Kew: Richmond, Surrey, UK, 1988; Volume 8, pp. 86–113. [Google Scholar]
- Muzila, M.; Werlemark, G.; Ortiz, R.; Sehic, J.; Fatih, M.; Setshogo, M.; Mpoloka, W.; Nybom, H. Assessment of diversity in Harpagophytum with RAPD and ISSR markers provides evidence of introgression. Hereditas 2014, 151, 91–101. [Google Scholar] [CrossRef]
- Pimental, R.A. A comparative study of data and ordination techniques based on a hybrid swarm of sand verbenas (Abronia Juss.). Syst. Zool. 1981, 30, 250–267. [Google Scholar] [CrossRef]
- Wilson, P. On inferring hybridity from morphological intermediacy. Taxon 1992, 41, 11–23. [Google Scholar] [CrossRef]
- McDade, L.A. Hybrids and phylogenetic systematics III. Comparison with distance methods. Syst. Bot. 1997, 22, 669–683. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Arbuthnott, J. An argument for divine providence, taken from the constant regularity observ’d in the births of both sexes. Philos. Trans. 1710, 27, 186–190. [Google Scholar]
- Scheffé, H. A method for judging all contrasts in the analysis of variance. Biometrika 1953, 40, 87–104. [Google Scholar]
- Marshall, N.T. Searching for a Cure: Conservation of Medicinal Wildlife Resources in East and Southern Africa; Traffic International: Cambridge, UK, 1998. [Google Scholar]
- McGuffin, M.; Kartesz, J.T.; Leung, A.Y.; Tucker, A.O. Herbs of Commerce, 2nd ed.American Herbal Products Association: Silver Spring, MD, USA, 2000. [Google Scholar]
- Grote, K. The Increased Harvest and Trade of Devil’s Claw (Harpagophytum procumbens) and Its Impacts on the Peoples and Environment of Namibia, Botswana and South Africa; Global Facilitation Unit for Underutilized Species: Maccarese, Italy, 2003. [Google Scholar]
- Raimondo, D.; Donaldson, J. The Trade, Management and Biological Status of Harpagophytum Spp. in Southern African Range States; Convention on International Trade in Endangered Species of Wild Fauna and Flora: Geneva, Switzerland, 2002. [Google Scholar]
- Chantre, P.; Cappelaere, A.; Leblan, D.; Guedon, D.; Vandermander, J.; Fournie, B. Efficacy and tolerance of Harpagophytum procumbens versus diacerhein in treatment of osteoarthritis. Phytomedicine 2000, 7, 177–183. [Google Scholar] [CrossRef]
- Chrubasik, S.; Thanner, J.; Künzel, O.; Conradt, C.; Black, A.; Pollak, S. Comparison of outcome measures during treatment with the proprietary Harpagophytum extract Doloteffin® in patients with pain in the lower back, knee or hip. Phytomedicine 2002, 9, 181–194. [Google Scholar] [CrossRef]
- Frerick, H.; Schmidt, U. Stufenschema bei coxarthrose. Der Kassenarzt 2001, 5, 34–41. [Google Scholar]
- Wegener, T.; Lüpke, N. Treatment of patients with arthrosis of hip or knee with an aqueous extract of Devil’s Claw (Harpagophytum procumbens DC). Phytother. Res. 2003, 17, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.L.; Santos, E.H.R.; Maria de Lourdes, V.S.; da Silva, A.A.B.; Tufik, S. Evaluation of acute and chronic treatments with Harpagophytum procumbens on Freund’s adjuvant–induced arthritis in rats. J. Ethnopharmacol. 2004, 91, 325–330. [Google Scholar] [CrossRef]
- Baghdikian, B.; Lanhers, M.C.; Fleurentin, J.; Ollivier, E.; Maillard, C.; Balansard, G.; Mortier, F. An analytical study and anti–inflammatory and analgesic effects of Harpagophytum procumbens and Harpagophytum zeyheri. Planta Med. 2007, 63, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.K.; Mossanda, K.S.; Na, H.-K.; Surh, Y.-J. Inhibitory effects of the extracts of Sutherlandia frutescens (L.) R. Br. and Harpagophytum procumbens DC. on phorbol ester–Induced COX-2 expression in mouse skin: AP-1 and CREB as potential upstream targets. Cancer Lett. 2005, 218, 21–31. [Google Scholar] [CrossRef]
- McLeod, D.W.; Revell, P.; Robinson, B.V. Investigations of Harpagophytum procumbens (Devil’s Claw) in the treatment of experimental inflammation and arthritis in the rat. Br. J. Pharmacol. 1979, 66, 140P–141P. [Google Scholar]
- Whitehouse, L.W.; Znamirowska, M.; Paul, C.J. Devil’s Claw (Harpagophytum procumbens): No evidence for anti–inflammatory activity in the treatment of arthritic disease. Can. Med Assoc. J. 1983, 129, 249–251. [Google Scholar]
- Abdelouahab, N.; Heard, C. Effect of the major glycosides of Harpagophytum procumbens (Devil’s Claw) on epidermal cyclooxygenase-2 (COX-2). Vitro. J. Nat. Prod. 2008, 71, 746–749. [Google Scholar] [CrossRef] [PubMed]
- Gyurkovska, V.; Alipieva, K.; Maciuk, A.; Dimitrova, P.; Ivanovska, N.; Haas, C.; Bley, T.; Georgiev, M. Anti–inflammatory activity of Devil’s Claw in vitro systems and their active constituents. Food Chem. 2011, 125, 171–178. [Google Scholar] [CrossRef]
- Jang, M.-H.; Lim, S.; Han, S.-M.; Park, H.-J.; Shin, I.; Kim, J.-W.; Kim, N.-J.; Lee, J.-S.; Kim, K.-A.; Kim, C.-J. Harpagophytum procumbens suppresses lipopolysaccharide–stimulated expressions of cyclooxygenase-2 and inducible nitric oxide synthase in fibroblast cell line L929. J. Pharmacol. Sci. 2003, 93, 367–371. [Google Scholar] [CrossRef] [Green Version]
- Fiebich, B.L.; Heinrich, M.; Hiller, K.O.; Kammerer, N. Inhibition of TNF-α synthesis in LPS–stimulated primary human monocytes by Harpagophytum extract SteiHap 69. Phytomedicine 2001, 8, 28–30. [Google Scholar] [CrossRef] [PubMed]
- Inaba, K.; Murata, K.; Naruto, S.; Matsuda, H. Inhibitory effects of Devil’s Claw (secondary root of Harpagophytum procumbens) extract and harpagoside on cytokine production in mouse macrophages. J. Nat. Med. 2010, 64, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, M.I.; Ivanovska, N.; Alipieva, K.; Dimitrova, P.; Verpoorte, R. Harpagoside: From Kalahari Desert to pharmacy shelf. Phytochemistry 2013, 92, 8–15. [Google Scholar] [CrossRef]
- Ichim, M.C.; Häser, A.; Nick, P. Microscopic authentication of commercial herbal products in the globalized market: Potential and limitations. Front. Pharmacol. 2020, 11, 876. [Google Scholar] [CrossRef] [PubMed]
- Grazina, L.; Amaral, J.S.; Mafra, I. Botanical origin authentication of dietary supplements by DNA–based approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1080–1109. [Google Scholar] [CrossRef] [Green Version]
- Raclariu, A.C.; Heinrich, M.; Ichim, M.C.; Boer, H. Benefits and limitations of DNA barcoding and metabarcoding in herbal product authentication. Phytochem. Anal. 2018, 29, 123–128. [Google Scholar] [CrossRef]
- Anantha Narayana, D.B.; Johnson, S.T. DNA Barcoding in authentication of herbal raw materials, extracts and dietary supplements: A perspective. Plant Biotechnol. Rep. 2019, 13, 201–210. [Google Scholar] [CrossRef]
- Grant, L.; McBean, D.E.; Fyfe, L.; Warnock, A.M. A review of the biological and potential therapeutic actions of Harpagophytum procumbens. Phytother. Res. 2007, 21, 199–209. [Google Scholar] [CrossRef]
- Little, D.P. A Unified index of sequence quality and contig overlap for DNA barcoding. Bioinformatics 2010, 26, 2780–2781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanner, R. Proposed Standards for BARCODE Records in INSDC (BRIs); Database Working Group, Consortium for the Barcode of Life: Washington, DC, USA, 2009. [Google Scholar]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 25 August 2021).
- Kool, A.; de Boer, H.J.; Krüger, Å.; Rydberg, A.; Abbad, A.; Björk, L.; Martin, G. Molecular identification of commercialized medicinal plants in Southern Morocco. PLoS ONE 2012, 7, e39459. [Google Scholar] [CrossRef]
- Little, D.P. DNA Barcode sequence identification incorporating taxonomic hierarchy and within taxon variability. PLoS ONE 2011, 6, e20552. [Google Scholar] [CrossRef]
- Thorner, R.M.; Remein, Q.R. Principals and Procedures in the Evaluation of Screening for Disease; United States Public Health Service: Washington, DC, USA, 1961; p. 24.
- McNemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Nixon, K.C.; Wheeler, Q.D. An amplification of the phylogenetic species concept. Cladistics 1990, 6, 211–223. [Google Scholar] [CrossRef]
- Baker, D.A.; Stevenson, D.W.; Little, D.P. DNA barcode identification of black cohosh herbal dietary supplements. J. AOAC Int. 2012, 95, 1023–1034. [Google Scholar] [CrossRef]
- Little, D.P. Authentication of Ginkgo biloba herbal dietary supplements using DNA barcoding. Genome 2014, 57, 513–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Little, D.P.; Jeanson, M.L. DNA barcode authentication of saw palmetto herbal dietary supplements. Sci. Rep. 2013, 3, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busconi, M.; Foroni, C.; Corradi, M.; Bongiorni, C.; Cattapan, F.; Fogher, C. DNA Extraction from olive oil and its use in the identification of the production cultivar. Food Chem. 2003, 83, 127–134. [Google Scholar] [CrossRef]
- Fernandes, T.J.R.; Oliveira, M.B.P.P.; Mafra, I. Tracing transgenic maize as affected by breadmaking process and raw material for the production of a traditional maize bread, broa. Food Chem. 2013, 138, 687–692. [Google Scholar] [CrossRef]
- Gryson, N.; Messens, K.; Dewettinck, K. PCR detection of soy ingredients in bread. Eur. Food Res. Technol. 2008, 227, 345–351. [Google Scholar] [CrossRef]
- Hellebrand, M.; Nagy, M.; Morsel, J.T. Determination of DNA traces in rapeseed oil. Z. Lebensm. Forsch. A 1998, 206, 237–242. [Google Scholar] [CrossRef]
- Meyer, R. Development and application of DNA analytical methods for the detection of GMOs in food. Food Control 1999, 10, 391–399. [Google Scholar] [CrossRef]
- Murray, S.R.; Butler, R.C.; Hardacre, A.K.; Timmerman–Vaughan, G.M. Use of quantitative real-time PCR to estimate maize endogenous DNA degradation after cooking and extrusion or in food products. J. Agric. Food Chem. 2007, 55, 2231–2239. [Google Scholar] [CrossRef]
- Oguchi, T.; Onishi, M.; Chikagawa, Y.; Kodama, T.; Suzuki, E.; Kasahara, M.; Akiyama, H.; Teshima, R.; Futo, S.; Hino, A.; et al. Investigation of residual DNAs in sugar from sugar beet (Beta vulgaris, L.). Food Hyg. Saf. Sci. 2009, 50, 41–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staats, M.; Cuenca, A.; Richardson, J.E.; Vrielink–van Ginkel, R.; Petersen, G.; Seberg, O.; Bakker, F.T. DNA damage in plant herbarium tissue. PLoS ONE 2011, 6, e28448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilley, M. PCR amplification of wheat sequences from DNA extracted during milling and baking. Cereal Chem. 2004, 81, 44–47. [Google Scholar] [CrossRef]
- Bryan, G.J.; Dixon, A.; Gale, M.D.; Wiseman, G. A PCR–based method for the detection of hexaploid bread wheat adulteration of durum wheat and pasta. J. Cereal Sci. 1998, 28, 135–145. [Google Scholar] [CrossRef]
- Hupfer, C.; Hotzel, H.; Sachse, K.; Engel, K.-H. Detection of the genetic modification in heat–treated products of Bt maize by polymerase chain reaction. Z. Lebensm. Forsch. A 1998, 206, 203–207. [Google Scholar] [CrossRef]
- Straub, J.A.; Hertel, C.; Hammes, W.P. Limits of a PCR–based detection method for genetically modified soya beans in wheat bread production. Z. Lebensm. Forsch. A 1999, 208, 77–82. [Google Scholar] [CrossRef]
- Bauer, T.; Weller, P.; Hammes, W.P.; Hertel, C. The effect of processing parameters on DNA degradation in food. Eur. Food Res. Technol. 2003, 217, 338–343. [Google Scholar] [CrossRef]
- Duggan, P.S.; Chambers, P.A.; Heritage, J.; Forbes, J.M. Fate of genetically modified maize DNA in the oral cavity and rumen of sheep. Br. J. Nutr. 2003, 89, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, M.; Lundberg, L.; Ferm, M.; Malmheden Yman, I. Real time PCR for the detection and discrimination of cereal contamination in gluten free foods. Eur. Food Res. Technol. 2003, 217, 344–349. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Ge, Y.; Xu, B. Degradation of endogenous and exogenous genes of roundup–ready soybean during food processing. J. Agric. Food Chem. 2005, 53, 10239–10243. [Google Scholar] [CrossRef] [PubMed]
- Bergerová, E.; Godalova, Z.; Siekel, P. Combined effects of temperature, pressure and low pH on the amplification of DNA of plant derived foods. Czech J. Food Sci. 2011, 29, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Bergerová, E.; Hrncirova, Z.; Stankovska, M.; Lopasovska, M.; Siekel, P. Effect of thermal treatment on the amplification and quantification of transgenic and non–transgenic soybean and maize DNA. Food Anal. Methods 2010, 3, 211–218. [Google Scholar] [CrossRef]
- Costa, J.; Mafra, I.; Amaral, J.S.; Oliveira, M.B.P.P. Monitoring genetically modified soybean along the industrial soybean oil extraction and refining processes by polymerase chain reaction techniques. Food Res. Int. 2010, 43, 301–306. [Google Scholar] [CrossRef]
- Särkinen, T.; Staats, M.; Richardson, J.E.; Cowan, R.S.; Bakker, F.T. How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS ONE 2012, 7, e43808. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Rubinsky, M.; Babajanian, S.; Zhang, Y.; Chang, P.; Swanson, G. Visualization of DNA in highly processed botanical materials. Food Chem. 2018, 245, 1042–1051. [Google Scholar] [CrossRef] [PubMed]
- Boiteux, L.S.; Fonseca, M.E.N.; Simon, P.W. Effects of plant tissue and DNA purification method on randomly amplified polymorphic DNA-based genetic fingerprinting analysis in carrot. J. Am. Soc. Hortic. Sci. 1999, 124, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Bowman, M.J.; Simon, P.W. Quantification of the relative abundance of plastome to nuclear genome in leaf and root tissues of carrot (Daucus carota, L.) using quantitative PCR. Plant. Mol. Biol. Rep. 2013, 31, 1040–1047. [Google Scholar] [CrossRef]
- Schmidt, A.H. Validation of a fast–HPLC method for the separation of iridoid glycosides to distinguish between the Harpagophytum species. J. Liq. Chromatogr. Relat. Technol. 2005, 28, 2339–2347. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Ihlenfeldt, H.-D. Pedaliaceae. In The Families and Genera of Vascular Plants VII: Flowering Plants; Dicotyledons: Lamiales (except Acanthaceae including Avicenniaceae); Kadereit, J.W., Ed.; Springer: Berlin, Germany, 2004; pp. 307–322. [Google Scholar]
- Gormley, I.C.; Bedigian, D.; Olmstead, R.G. Phylogeny of Pedaliaceae and Martyniaceae and the placement of Trapella in Plantaginaceae s. l. Syst. Bot. 2015, 40, 259–268. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef] [Green Version]
- Lledo, M.D.; Crespo, M.B.; Cameron, K.M.; Fay, M.F.; Chase, M.W. Systematics of Plumbaginaceae based upon cladistic analysis of rbcL sequence data. Syst. Bot. 1998, 23, 21–29. [Google Scholar] [CrossRef]
- Levin, R.A.; Wagner, W.L.; Hoch, P.C.; Nepokroeff, M.; Pires, J.C.; Zimmer, E.A.; Sytsma, K.J. Family–level relationships of Onagraceae based on chloroplast rbcL and ndhF data. Am. J. Bot. 2003, 90, 107–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Marker | Primer Name | Sequence (5′–3′) | Source |
|---|---|---|---|
| matK | 1R | ACCCAGTCCATCTGGAAATCTTGGTTC | K.J. Kim (pers. com.) |
| matK | 3F | CGTACAGTACTTTTGTGTTTACGAG | K.J. Kim (pers. com.) |
| nrITS2 | S2F | ATGCGATACTTGGTGTGAAT | [77] |
| nrITS2 | S3R | GACGCTTCTCCAGACTACAAT | [77] |
| psbA-trnH | psbAF | GTTATGCATGAACGTAATGCTC | [78] |
| psbA-trnH | trnHR | CGCGCATGGTGGATTCACAAATC | [78] |
| psbA-trnH mini-barcode | F | GAAGATAAATGAAATGATTGAAATGC | novel |
| psbA-trnH mini-barcode | R | TGGATTCACAAATCCACTGC | novel |
| rbcL | 32F | TTGGATTCAAAGCTGGTGTT | [79] |
| rbcL | a_F | ATGTCACCACAAACAGAGACTAAAGC | [80] |
| rbcL | ajf634R | GAAACGGTCTCTCCAACGCAT | [81] |
| Marker | Primers | Cycling |
|---|---|---|
| matK | 1R & 3F | 10 × {30 s, 95 °C; 30 s, 56 °C; 30 s, 72 °C}; 25 × {30 s, 88 °C; 30 s, 56 °C; 30 s, 72 °C} |
| nrITS2 | S2F & S3R | 35 × {30 s, 95 °C; 30 s, 56 °C; 30 s, 72 °C} |
| psbA-trnH | psbAF & trnHR | 10 × {30 s, 95 °C; 120 s, 55 °C}; 23 × {45 s, 90 °C; 120 s, 55 °C} |
| psbA-trnH mini-barcode | F & R | 35 × {30 s, 95 °C; 120 s, 58 °C} |
| rbcL | 32F & ajf634R | 35 × {30 s, 95 °C; 30 s, 58 °C; 30 s, 72 °C} |
| rbcL | a_F & ajf634R | 35 × {30 s, 95 °C; 30 s, 58 °C; 30 s, 72 °C} |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diaz-Silveira, G.L.; Deutsch, J.; Little, D.P. DNA Barcode Authentication of Devil’s Claw Herbal Dietary Supplements. Plants 2021, 10, 2005. https://doi.org/10.3390/plants10102005
Diaz-Silveira GL, Deutsch J, Little DP. DNA Barcode Authentication of Devil’s Claw Herbal Dietary Supplements. Plants. 2021; 10(10):2005. https://doi.org/10.3390/plants10102005
Chicago/Turabian StyleDiaz-Silveira, Genelle L., Joan Deutsch, and Damon P. Little. 2021. "DNA Barcode Authentication of Devil’s Claw Herbal Dietary Supplements" Plants 10, no. 10: 2005. https://doi.org/10.3390/plants10102005
APA StyleDiaz-Silveira, G. L., Deutsch, J., & Little, D. P. (2021). DNA Barcode Authentication of Devil’s Claw Herbal Dietary Supplements. Plants, 10(10), 2005. https://doi.org/10.3390/plants10102005





