Distinct Activities of Gli1 and Gli2 in the Absence of Ift88 and the Primary Cilia
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Immunofluorescence Assay on Cryosections
2.3. Immunoblot Analyses
2.4. RNA In Situ Hybridization on Cryosections
3. Results
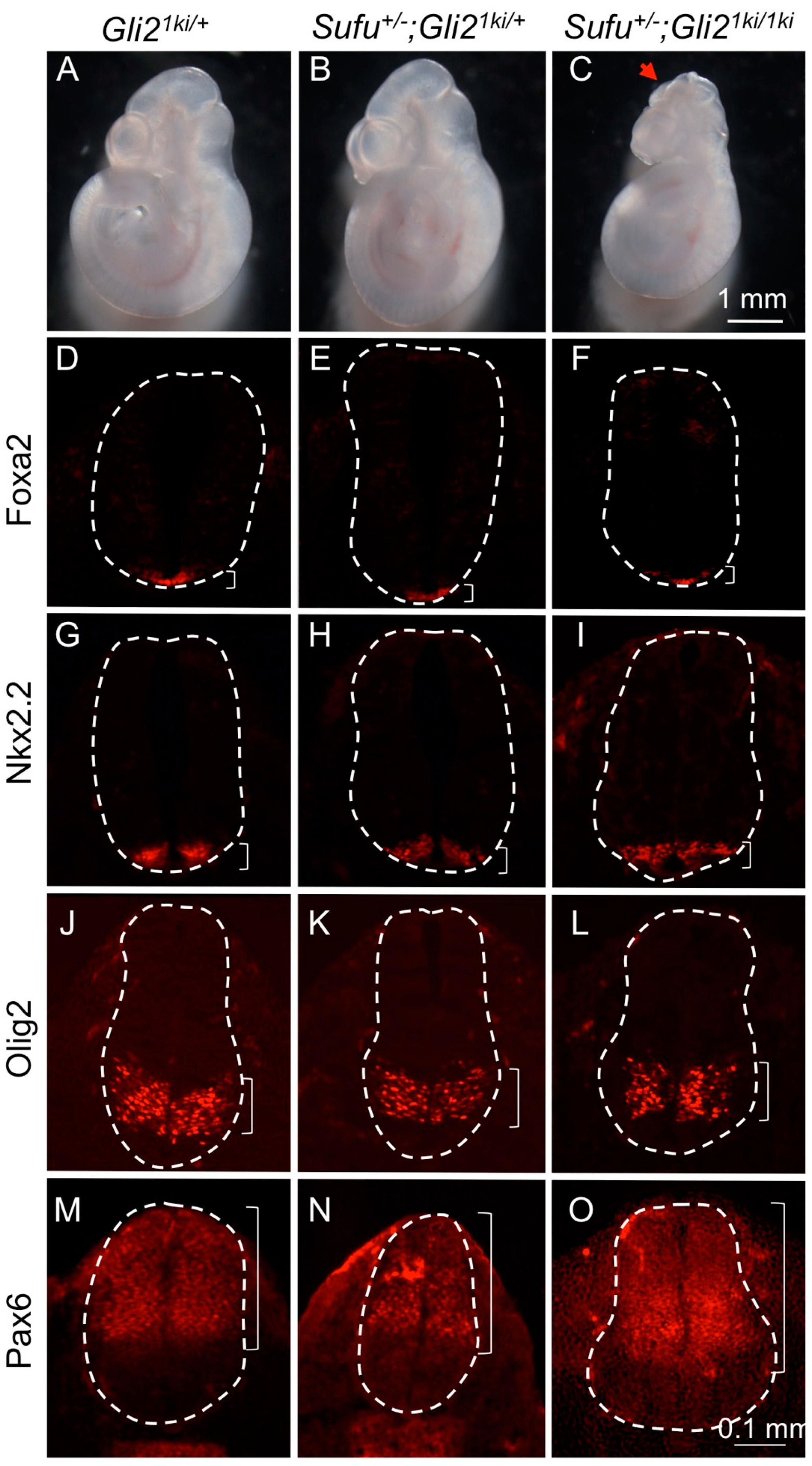
3.1. Gli1 Expression Was Uncoupled from Hh Signaling in Gli21ki Embryos
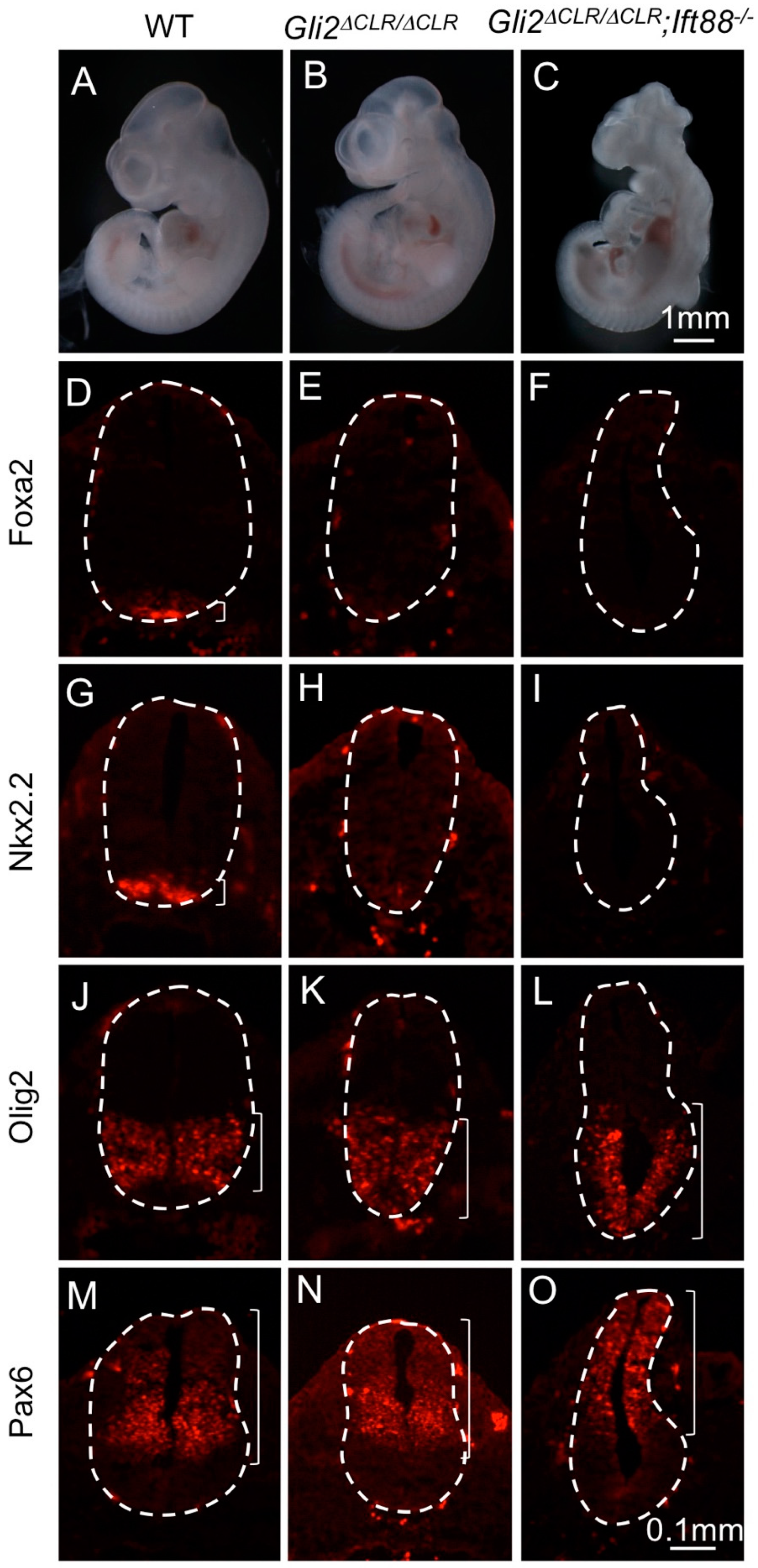
3.2. The Cilia Are Essential for Maximal Activation of Gli1
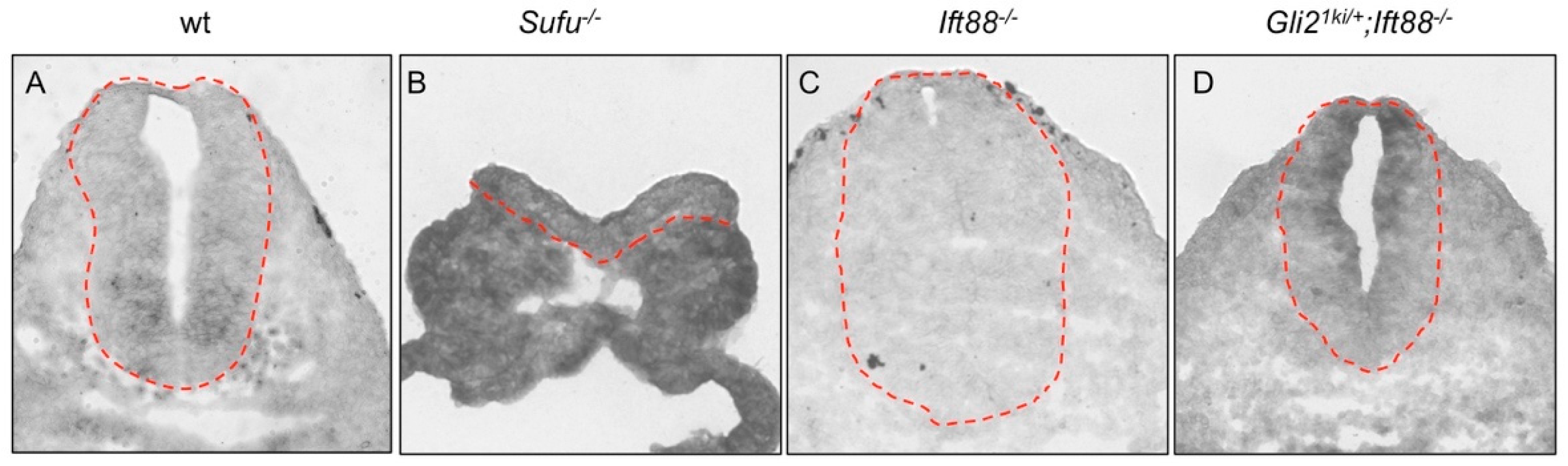
3.3. Gli1 Was Partially Activated in the Absence of Cilia
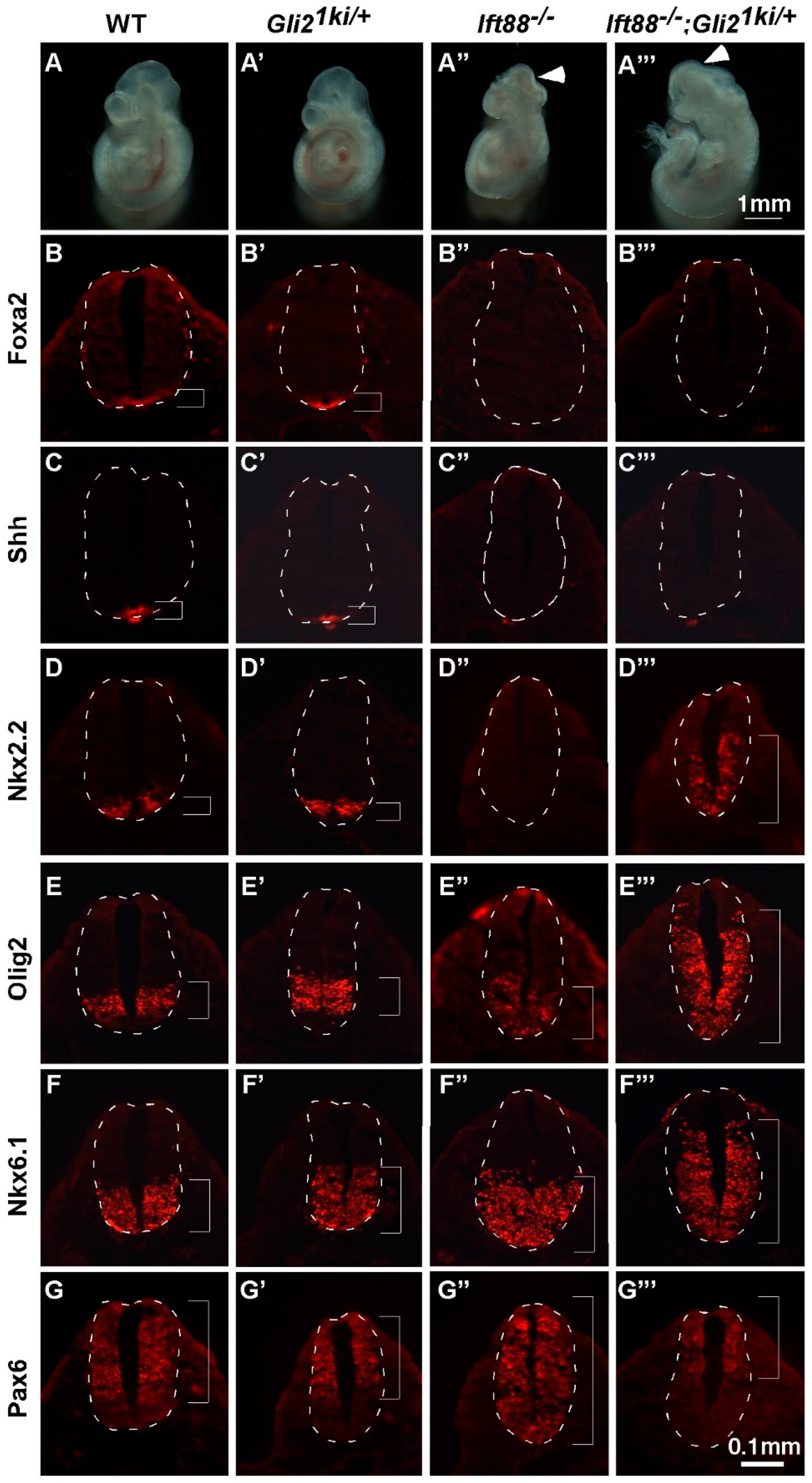
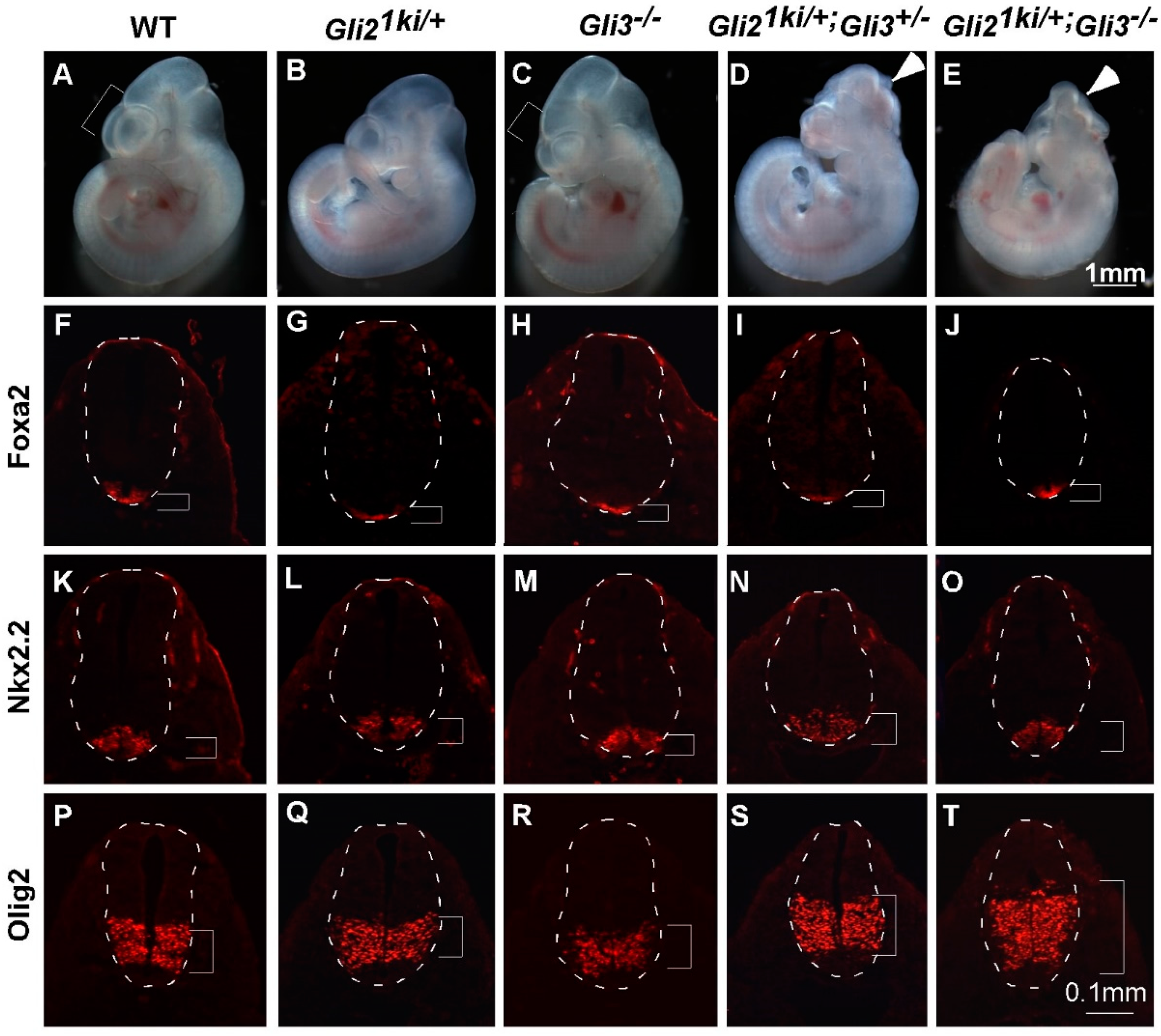
3.4. Ectopic Gli1 Partially Ventralized the Neural Tube in the Presence of Reduced Gli3
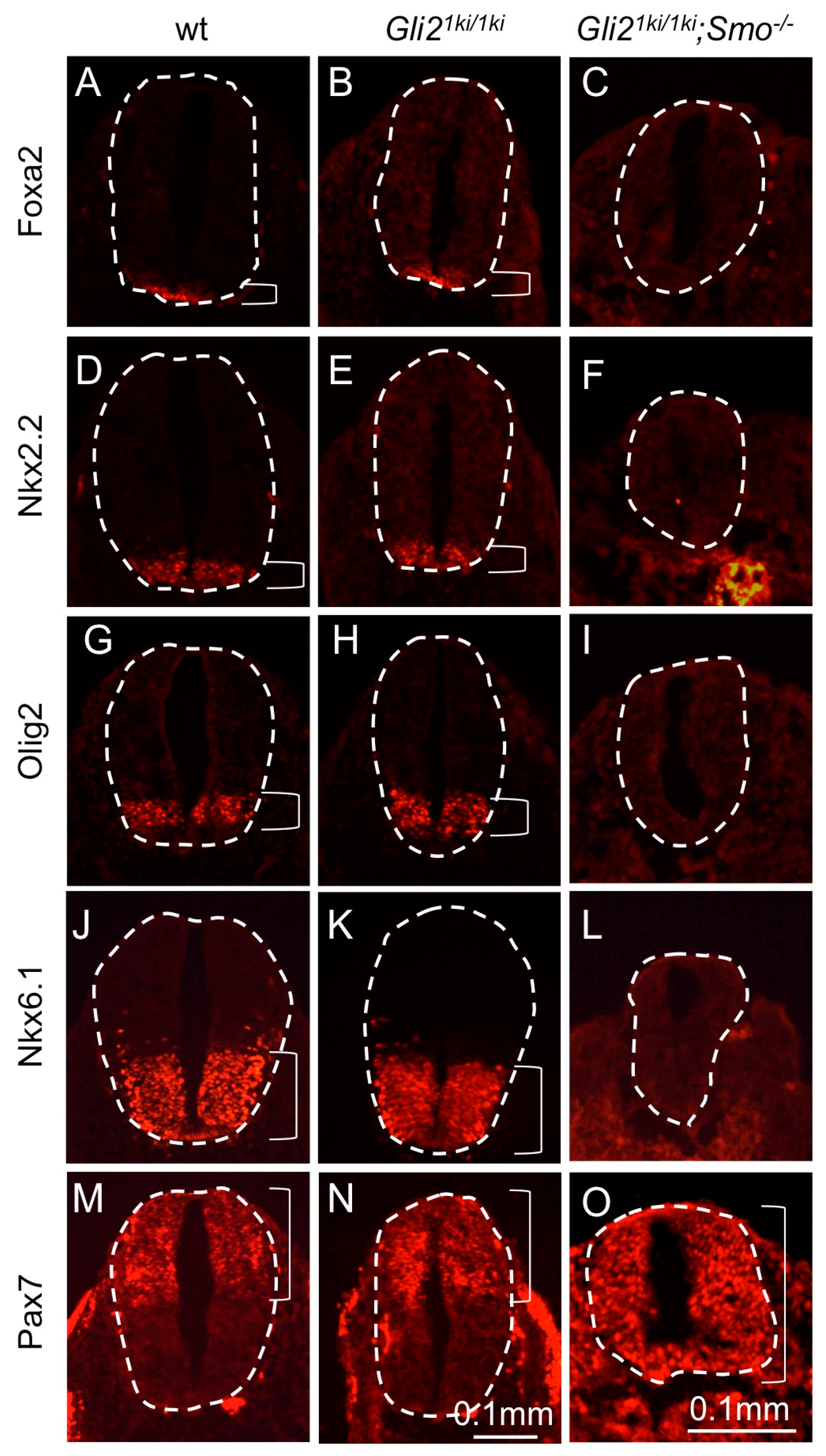
3.5. Gli1 Expression from the Gli2 Locus Failed to Alter the Smo Mutant Phenotype
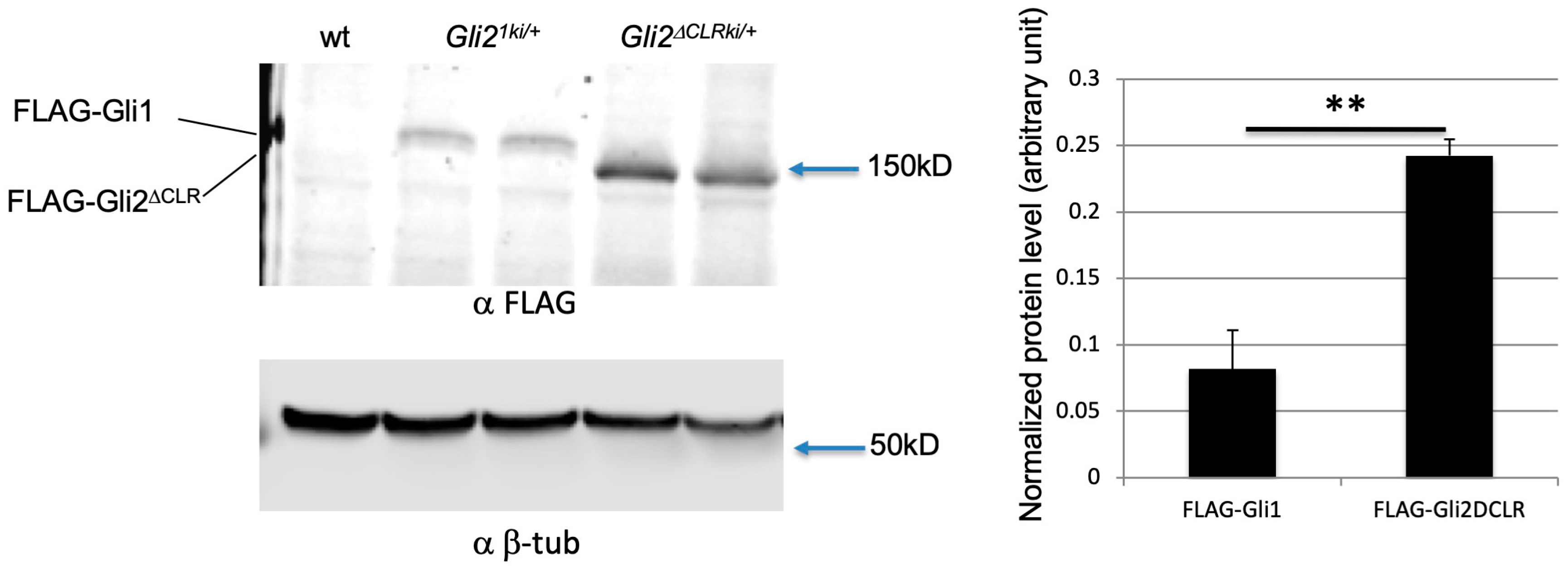
3.6. Increased Protein Level Was Not the Major Reason for the Cilia-Independent Gli1 Activation
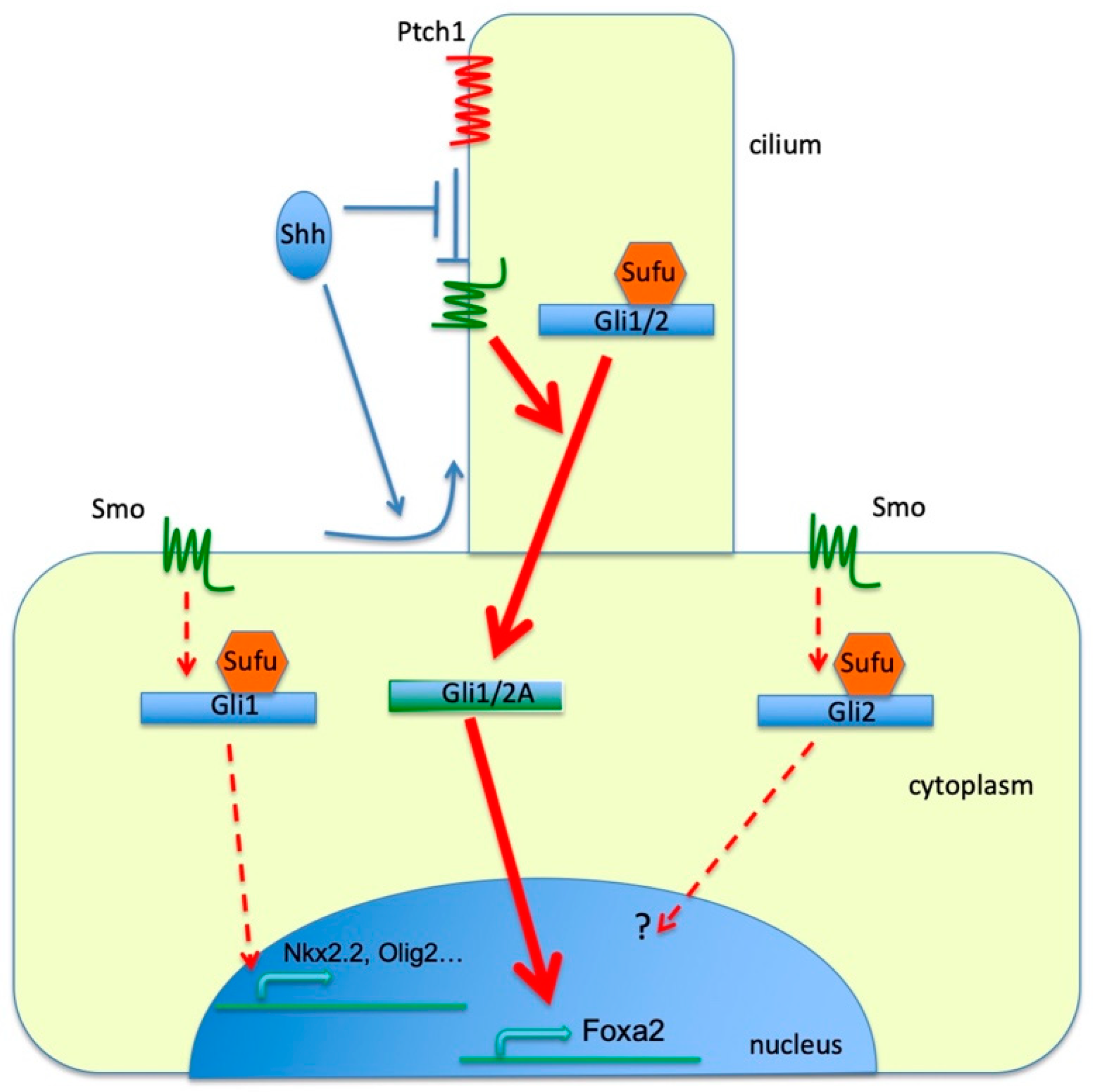
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Briscoe, J.; Therond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Re. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, Y.I.; Lin, C.; Chuang, P.T. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr. Opin. Genet. Dev. 2013, 23, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Matise, M.P.; Joyner, A.L. Gli genes in development and cancer. Oncogene 1999, 18, 7852–7859. [Google Scholar] [CrossRef]
- Haycraft, C.J.; Banizs, B.; Aydin-Son, Y.; Zhang, Q.; Michaud, E.J.; Yoder, B.K. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005, 1, e53. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Litingtung, Y.; Lee, E.; Young, K.E.; Corden, J.L.; Westphal, H.; Beachy, P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 1996, 383, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Motoyama, J.; Gasca, S.; Mo, R.; Sasaki, H.; Rossant, J.; Hui, C.C. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 1998, 125, 2533–2543. [Google Scholar] [PubMed]
- Matise, M.P.; Epstein, D.J.; Park, H.L.; Platt, K.A.; Joyner, A.L. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development 1998, 125, 2759–2770. [Google Scholar] [PubMed]
- Litingtung, Y.; Chiang, C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat. Neurosci. 2000, 3, 979–985. [Google Scholar] [CrossRef]
- Park, H.L.; Bai, C.; Platt, K.A.; Matise, M.P.; Beeghly, A.; Hui, C.C.; Nakashima, M.; Joyner, A.L. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 2000, 127, 1593–1605. [Google Scholar]
- Bai, C.B.; Auerbach, W.; Lee, J.S.; Stephen, D.; Joyner, A.L. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 2002, 129, 4753–4761. [Google Scholar]
- Bai, C.B.; Stephen, D.; Joyner, A.L. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 2004, 6, 103–115. [Google Scholar] [CrossRef]
- Mastrangelo, E.; Milani, M. Role and inhibition of GLI1 protein in cancer. Lung Cancer (Auckl) 2018, 9, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Kasai, K. GLI1, a master regulator of the hallmark of pancreatic cancer. Pathol. Int. 2016, 66, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wu, M.; Zhao, F.; Fu, W.; Li, W.; Li, X.; Liu, T. Prognostic and clinicopathological value of Gli-1 expression in gastric cancer: A meta-analysis. Oncotarget 2016, 7, 69087–69096. [Google Scholar] [CrossRef] [PubMed]
- Palle, K.; Mani, C.; Tripathi, K.; Athar, M. Aberrant GLI1 Activation in DNA Damage Response, Carcinogenesis and Chemoresistance. Cancers 2015, 7, 2330–2351. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.; Liu, A.; Rakeman, A.S.; Murcia, N.S.; Niswander, L.; Anderson, K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 2003, 426, 83–87. [Google Scholar] [CrossRef]
- Huangfu, D.; Anderson, K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA 2005, 102, 11325–11330. [Google Scholar] [CrossRef]
- Liu, A.; Wang, B.; Niswander, L.A. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development 2005, 132, 3103–3111. [Google Scholar] [CrossRef]
- May, S.R.; Ashique, A.M.; Karlen, M.; Wang, B.; Shen, Y.; Zarbalis, K.; Reiter, J.; Ericson, J.; Peterson, A.S. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol. 2005, 287, 378–389. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, H.; Liu, A. The loss of Hh responsiveness by a non-ciliary Gli2 variant. Development 2015, 142, 1651–1660. [Google Scholar] [CrossRef]
- Cooper, A.F.; Yu, K.P.; Brueckner, M.; Brailey, L.L.; Johnson, L.; McGrath, J.M.; Bale, A.E. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development 2005, 132, 4407–4417. [Google Scholar] [CrossRef] [PubMed]
- Svard, J.; Heby-Henricson, K.; Persson-Lek, M.; Rozell, B.; Lauth, M.; Bergstrom, A.; Ericson, J.; Toftgard, R.; Teglund, S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell 2006, 10, 187–197. [Google Scholar] [CrossRef]
- Liu, J.; Heydeck, W.; Zeng, H.; Liu, A. Dual function of suppressor of fused in Hh pathway activation and mouse spinal cord patterning. Dev. Biol. 2012, 362, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Kogerman, P.; Grimm, T.; Kogerman, L.; Krause, D.; Unden, A.B.; Sandstedt, B.; Toftgard, R.; Zaphiropoulos, P.G. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1999, 1, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Dunaeva, M.; Michelson, P.; Kogerman, P.; Toftgard, R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 2003, 278, 5116–5122. [Google Scholar] [CrossRef] [PubMed]
- Paces-Fessy, M.; Boucher, D.; Petit, E.; Paute-Briand, S.; Blanchet-Tournier, M.F. The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem. J. 2004, 378, 353–362. [Google Scholar] [CrossRef]
- Lin, C.; Yao, E.; Wang, K.; Nozawa, Y.; Shimizu, H.; Johnson, J.R.; Chen, J.N.; Krogan, N.J.; Chuang, P.T. Regulation of Sufu activity by p66beta and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes Dev. 2014, 28, 2547–2563. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Wilson, C.W.; Li, Y.J.; Law, K.K.; Lu, C.S.; Gacayan, R.; Zhang, X.; Hui, C.C.; Chuang, P.T. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009, 23, 1910–1928. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Kolterud, A.; Zeng, H.; Hoover, A.; Teglund, S.; Toftgard, R.; Liu, A. Suppressor of Fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev. Biol. 2009, 330, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Humke, E.W.; Dorn, K.V.; Milenkovic, L.; Scott, M.P.; Rohatgi, R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010, 24, 670–682. [Google Scholar] [CrossRef]
- Tukachinsky, H.; Lopez, L.V.; Salic, A. A mechanism for vertebrate Hedgehog signaling: Recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 2010, 191, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.B.; Joyner, A.L. Gli1 can rescue the in vivo function of Gli2. Development 2001, 128, 5161–5172. [Google Scholar] [PubMed]
- Murcia, N.S.; Richards, W.G.; Yoder, B.K.; Mucenski, M.L.; Dunlap, J.R.; Woychik, R.P. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development 2000, 127, 2347–2355. [Google Scholar] [PubMed]
- Buscher, D.; Grotewold, L.; Ruther, U. The XtJ allele generates a Gli3 fusion transcript. Mamm Genome 1998, 9, 676–678. [Google Scholar] [PubMed]
- Lakso, M.; Pichel, J.G.; Gorman, J.R.; Sauer, B.; Okamoto, Y.; Lee, E.; Alt, F.W.; Westphal, H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 1996, 93, 5860–5865. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Jia, J.; Liu, A. Coordinated translocation of mammalian Gli proteins and suppressor of fused to the primary cilium. PLoS ONE 2010, 5, e15900. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, A. Immunohistochemistry and RNA in situ hybridization in mouse brain development. Methods Mol. Biol. 2014, 1082, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Walterhouse, D.; Ahmed, M.; Slusarski, D.; Kalamaras, J.; Boucher, D.; Holmgren, R.; Iannaccone, P. Gli, a zinc finger transcription factor and oncogene, is expressed during normal mouse development. Dev. Dyn. 1993, 196, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.C.; Slusarski, D.; Platt, K.A.; Holmgren, R.; Joyner, A.L. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev. Biol. 1994, 162, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, L.V.; Milenkovic, L.; Higgins, K.M.; Scott, M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 1997, 277, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Fallon, J.F.; Beachy, P.A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 2000, 100, 423–434. [Google Scholar] [CrossRef]
- Pan, Y.; Bai, C.B.; Joyner, A.L.; Wang, B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell Biol. 2006, 26, 3365–3377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, L.; Wang, B.; Ou, C.Y.; Chien, C.T.; Jiang, J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell 2006, 10, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, Q.; Chen, Y.; Yue, T.; Li, S.; Wang, B.; Jiang, J. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2009, 106, 21191–21196. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.G.; Kim, H.J.; Dlugosz, A.A.; Ellison, D.W.; Gilbertson, R.J.; Alvarez-Buylla, A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 2009, 15, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Seol, A.D.; So, P.L.; Ermilov, A.N.; Bichakjian, C.K.; Epstein, E.H., Jr.; Dlugosz, A.A.; Reiter, J.F. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat. Med. 2009, 15, 1055–1061. [Google Scholar] [CrossRef]
- Moore, E.R.; Yang, Y.; Jacobs, C.R. Primary cilia are necessary for Prx1-expressing cells to contribute to postnatal skeletogenesis. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Finetti, F.; Paccani, S.R.; Riparbelli, M.G.; Giacomello, E.; Perinetti, G.; Pazour, G.J.; Rosenbaum, J.L.; Baldari, C.T. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat. Cell Biol. 2009, 11, 1332–1339. [Google Scholar] [CrossRef]
- Boehlke, C.; Janusch, H.; Hamann, C.; Powelske, C.; Mergen, M.; Herbst, H.; Kotsis, F.; Nitschke, R.; Kuehn, E.W. A Cilia Independent Role of Ift88/Polaris during Cell Migration. PLoS ONE 2015, 10, e0140378. [Google Scholar] [CrossRef]
- Delaval, B.; Bright, A.; Lawson, N.D.; Doxsey, S. The cilia protein IFT88 is required for spindle orientation in mitosis. Nat. Cell Biol. 2011, 13, 461–468. [Google Scholar] [CrossRef]
- Borovina, A.; Ciruna, B. IFT88 plays a cilia- and PCP-independent role in controlling oriented cell divisions during vertebrate embryonic development. Cell Rep. 2013, 5, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.H.; Arastoo, M.; Georgiou, I.; Manning, D.R.; Riobo-Del Galdo, N.A. Activation of the Gi protein-RHOA axis by non-canonical Hedgehog signaling is independent of primary cilia. PLoS ONE 2018, 13, e0203170. [Google Scholar] [CrossRef]
- Yuan, X.; Cao, J.; He, X.; Serra, R.; Qu, J.; Cao, X.; Yang, S. Ciliary IFT80 balances canonical versus non-canonical hedgehog signalling for osteoblast differentiation. Nat. Commun. 2016, 7, 11024. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, M.F.; Damhofer, H.; Roelink, H. Hedgehog-stimulated chemotaxis is mediated by smoothened located outside the primary cilium. Sci. Signal. 2012, 5, ra60. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zeng, H.; Liu, A. Distinct Activities of Gli1 and Gli2 in the Absence of Ift88 and the Primary Cilia. J. Dev. Biol. 2019, 7, 5. https://doi.org/10.3390/jdb7010005
Wang Y, Zeng H, Liu A. Distinct Activities of Gli1 and Gli2 in the Absence of Ift88 and the Primary Cilia. Journal of Developmental Biology. 2019; 7(1):5. https://doi.org/10.3390/jdb7010005
Chicago/Turabian StyleWang, Yuan, Huiqing Zeng, and Aimin Liu. 2019. "Distinct Activities of Gli1 and Gli2 in the Absence of Ift88 and the Primary Cilia" Journal of Developmental Biology 7, no. 1: 5. https://doi.org/10.3390/jdb7010005
APA StyleWang, Y., Zeng, H., & Liu, A. (2019). Distinct Activities of Gli1 and Gli2 in the Absence of Ift88 and the Primary Cilia. Journal of Developmental Biology, 7(1), 5. https://doi.org/10.3390/jdb7010005





