Sonic Hedgehog Is a Member of the Hh/DD-Peptidase Family That Spans the Eukaryotic and Bacterial Domains of Life
Abstract
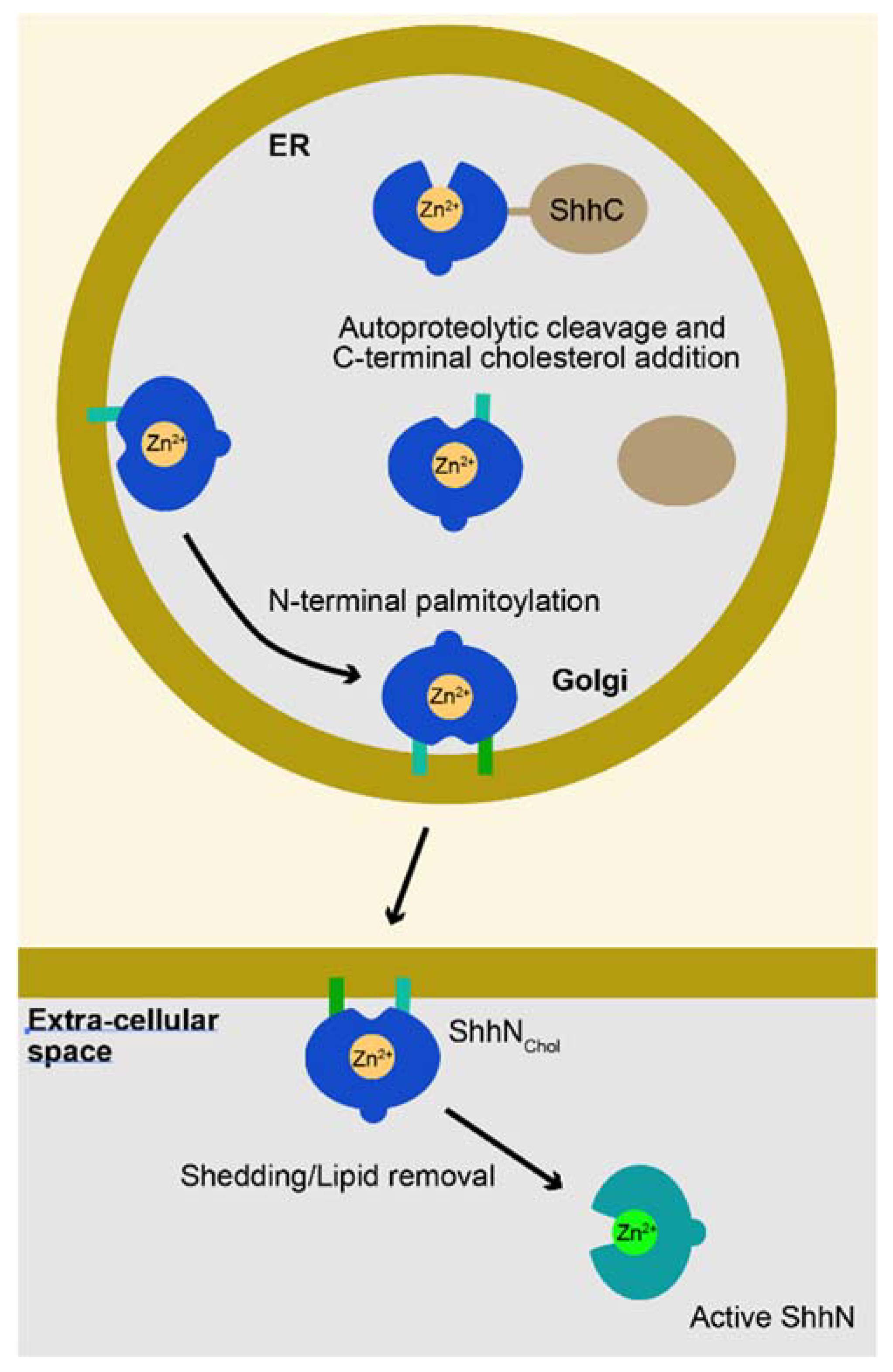
1. The Sonic Hedgehog Pro-Protein Gives Rise to the Mature Ligand after an Autoproteolytic Cleavage Event
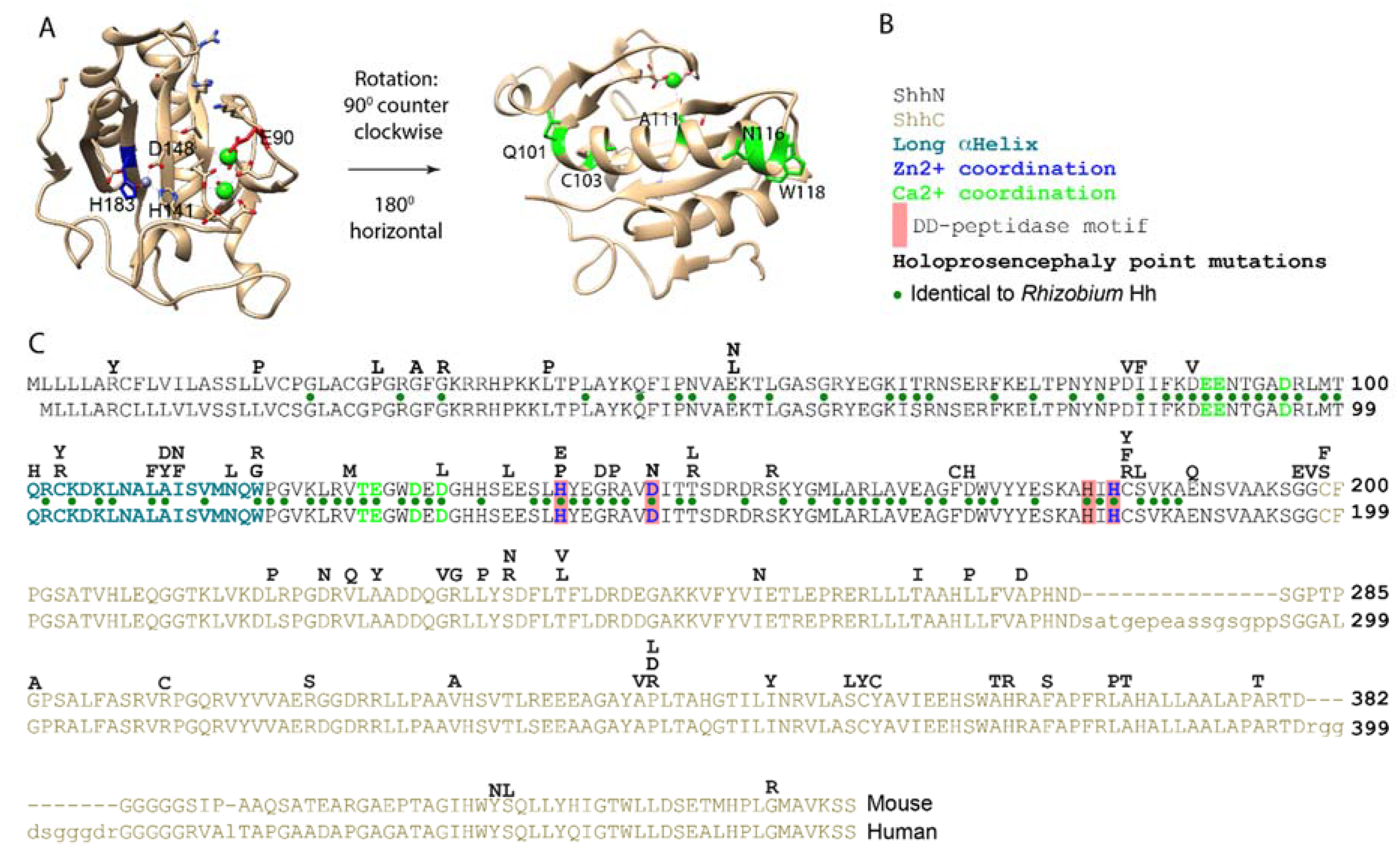
2. Both the N- and C-Terminal Domains of Shh Are the Targets for Point Mutations Found in Holoprosencephaly
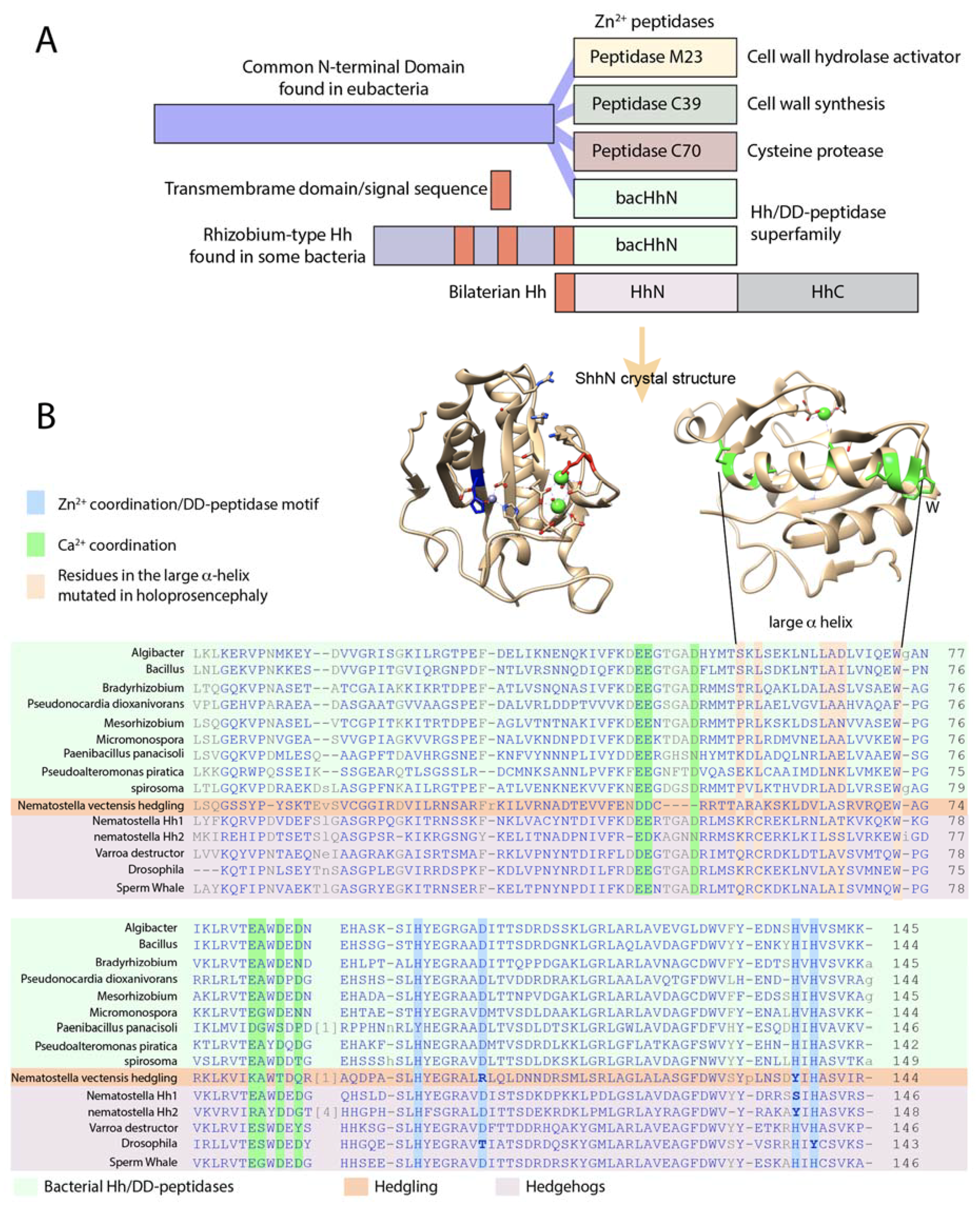
3. Shh Has All the Hallmarks of a DD-Peptidase
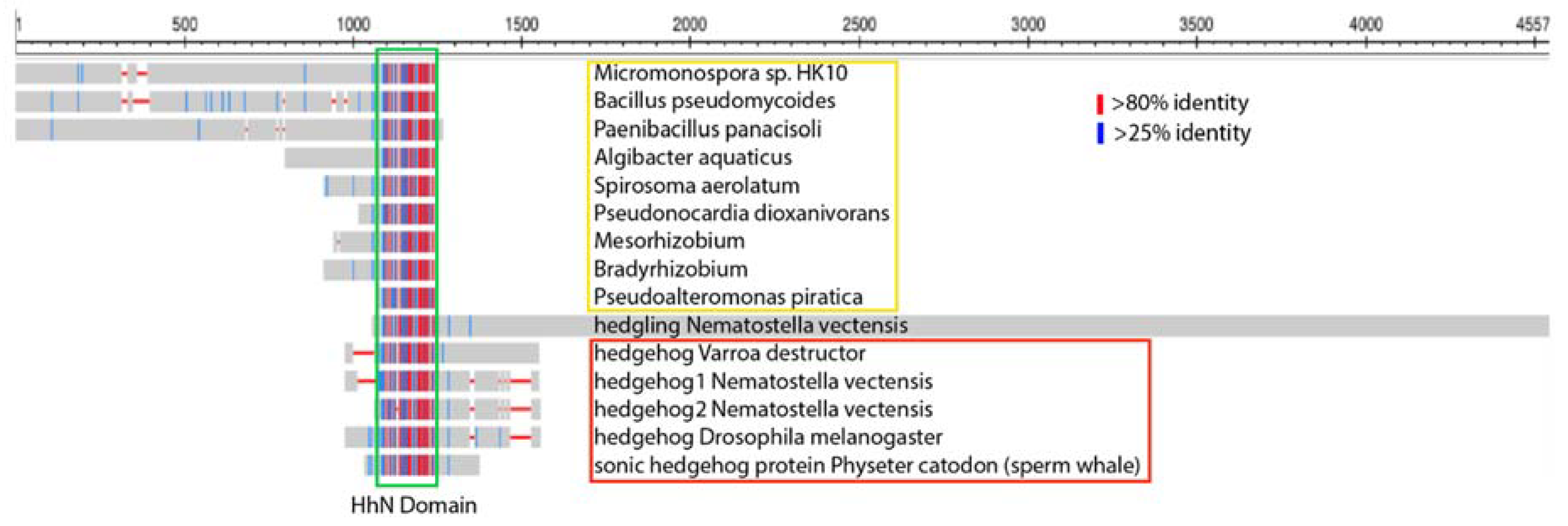
4. Several Bacterial Species Have Highly Conserved HhN Domains
5. What Are the Possible Substrates for the Shh Peptidase Activity?
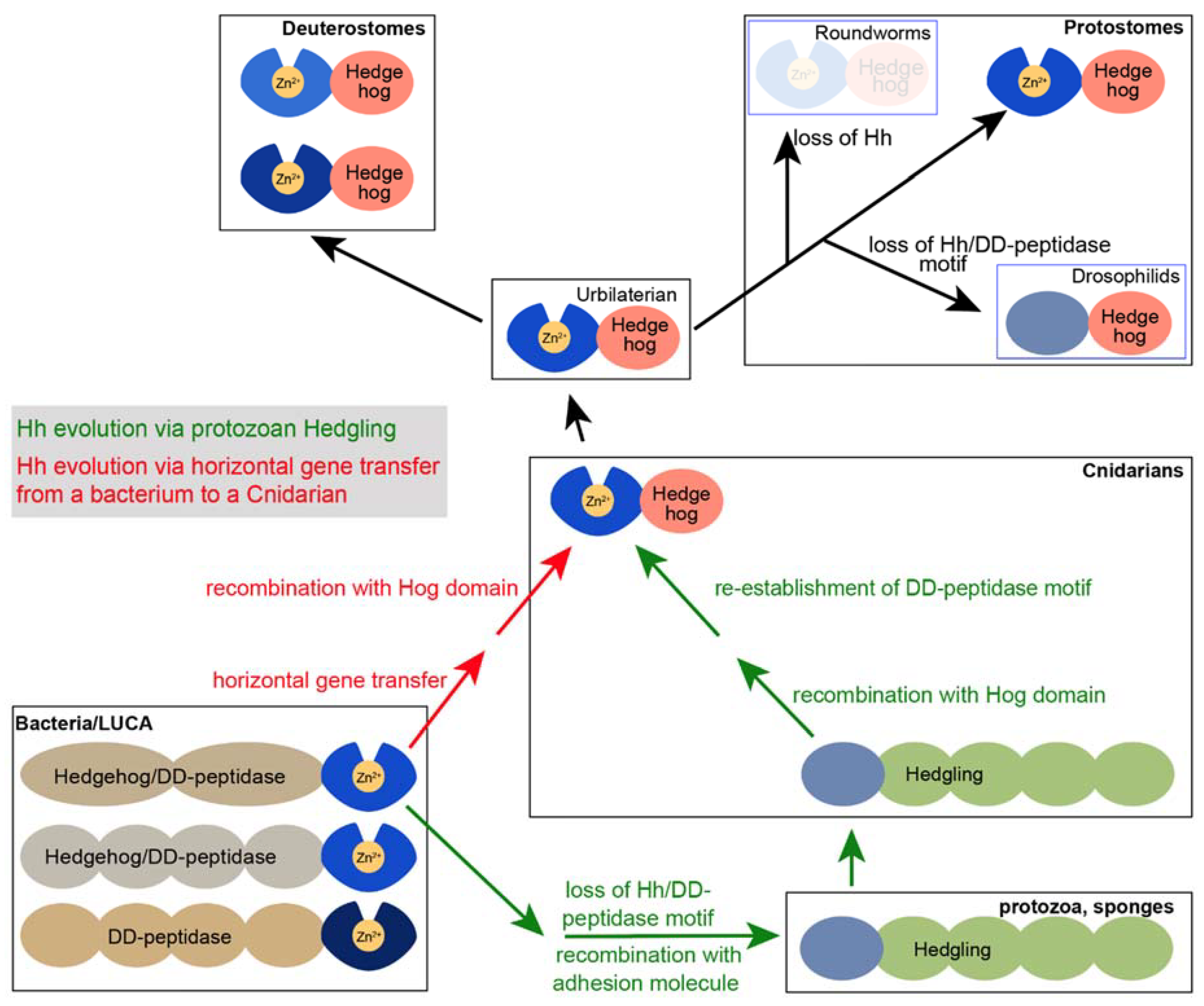
6. Drosophila Hh Is Not a Member of the Hh/DD-Peptidase Family
7. The Presence of bacHhs Suggests an Alternative Hypothesis Regarding the Evolution of Modern Hh
8. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Nüsslein-Volhard, C.; Wieshaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Echelard, Y.; Epstein, D.J.; St-Jacques, B.; Shen, L.; Mohler, J.; McMahon, J.A.; McMahon, A.P. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 1993, 75, 1417–1430. [Google Scholar] [CrossRef]
- Hall, T.M.; Porter, J.A.; Beachy, P.A.; Leahy, D.J. A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature 1995, 378, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Rebollido-Rios, R.; Bandari, S.; Wilms, C.; Jakuschev, S.; Vortkamp, A.; Grobe, K.; Hoffmann, D. Signaling domain of Sonic Hedgehog as cannibalistic calcium-regulated zinc-peptidase. PLoS Comput. Biol. 2014, 10, e1003707. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.A.; Ekker, S.C.; Park, W.J.; von Kessler, D.P.; Young, K.E.; Chen, C.H.; Ma, Y.; Woods, A.S.; Cotter, R.J.; Koonin, E.V.; et al. Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 1996, 86, 21–34. [Google Scholar] [CrossRef]
- Porter, J.A.; Young, K.E.; Beachy, P.A. Cholesterol modification of hedgehog signaling proteins in animal development. Science 1996, 274, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Taylor, F.R.; Wen, D.; Garber, E.A.; Carmillo, A.N.; Baker, D.P.; Arduini, R.M.; Williams, K.P.; Weinreb, P.H.; Rayhorn, P.; Hronowski, X.; et al. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry 2001, 40, 4359–4371. [Google Scholar] [CrossRef] [PubMed]
- Pepinsky, R.B.; Zeng, C.; Wen, D.; Rayhorn, P.; Baker, D.P.; Williams, K.P.; Bixler, S.A.; Ambrose, C.M.; Garber, E.A.; Miatkowski, K.; et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 1998, 273, 14037–14045. [Google Scholar] [CrossRef] [PubMed]
- Chamoun, Z.; Mann, R.K.; Nellen, D.; von Kessler, D.P.; Bellotto, M.; Beachy, P.A.; Basler, K. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 2001, 293, 2080–2084. [Google Scholar] [CrossRef] [PubMed]
- Tukachinsky, H.; Kuzmickas, R.P.; Jao, C.Y.; Liu, J.; Salic, A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2012, 2, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Roelink, H.; Porter, J.A.; Chiang, C.; Tanabe, Y.; Chang, D.T.; Beachy, P.A.; Jessell, T.M. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 1995, 81, 445–455. [Google Scholar] [CrossRef]
- Ohlig, S.; Farshi, P.; Pickhinke, U.; van den Boom, J.; Höing, S.; Jakuschev, S.; Hoffmann, D.; Dreier, R.; Schöler, H.R.; Dierker, T.; et al. Sonic hedgehog shedding results in functional activation of the solubilized protein. Dev. Cell 2011, 20, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.M.; Porter, J.A.; Young, K.E.; Koonin, E.V.; Beachy, P.A.; Leahy, D.J. Crystal structure of a Hedgehog autoprocessing domain: Homology between Hedgehog and self-splicing proteins. Cell 1997, 91, 85–97. [Google Scholar] [CrossRef]
- Tokhunts, R.; Singh, S.; Chu, T.; D’Angelo, G.; Baubet, V.; Goetz, J.A.; Huang, Z.; Yuan, Z.; Ascano, M.; Zavros, Y.; et al. The full-length unprocessed hedgehog protein is an active signaling molecule. J. Biol. Chem. 2010, 285, 2562–2568. [Google Scholar] [CrossRef] [PubMed]
- Traiffort, E.; Dubourg, C.; Faure, H.; Rognan, D.; Odent, S.; Durou, M.-R.; David, V.; Ruat, M. Functional characterization of sonic hedgehog mutations associated with holoprosencephaly. J. Biol. Chem. 2004, 279, 42889–42897. [Google Scholar] [CrossRef] [PubMed]
- Inouye, K.; Kusano, M.; Hashida, Y.; Minoda, M.; Yasukawa, K. Engineering, expression, purification, and production of recombinant thermolysin. Biotechnol. Annu. Rev. 2007, 13, 43–64. [Google Scholar] [PubMed]
- Zeng, X.; Goetz, J.A.; Suber, L.M.; Scott, W.J.J.; Schreiner, C.M.; Robbins, D.J. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature 2001, 411, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Fuse, N.; Maiti, T.; Wang, B.; Porter, J.A.; Hall, T.M.; Leahy, D.J.; Beachy, P.A. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc. Natl. Acad. Sci. USA 1999, 96, 10992–10999. [Google Scholar] [CrossRef] [PubMed]
- Bosanac, I.; Maun, H.R.; Scales, S.J.; Wen, X.; Lingel, A.; Bazan, J.F.; de Sauvage, F.J.; Hymowitz, S.G.; Lazarus, R.A. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nat. Struct. Mol. Biol. 2009, 16, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Maun, H.R.; Wen, X.; Lingel, A.; de Sauvage, F.J.; Lazarus, R.A.; Scales, S.J.; Hymowitz, S.G. Hedgehog pathway antagonist 5E1 binds hedgehog at the pseudo-active site. J. Biol. Chem. 2010, 285, 26570–26580. [Google Scholar] [CrossRef] [PubMed]
- Odent, S.; Atti Bitach, T.; Blayau, M.; Mathieu, M.; Aug, J.; Delezo de, A.L.; Gall, J.Y.; Le Marec, B.; Munnich, A.; David, V.; et al. Expression of the Sonic hedgehog (SHH) gene during early human development and phenotypic expression of new mutations causing holoprosencephaly. Hum. Mol. Genet. 1999, 8, 1683–1689. [Google Scholar] [CrossRef] [PubMed]
- Hehr, U.; Pineda-Alvarez, D.E.; Uyanik, G.; Hu, P.; Zhou, N.; Hehr, A.; Schell-Apacik, C.; Altus, C.; Daumer-Haas, C.; Meiner, A.; et al. Heterozygous mutations in SIX3 and SHH are associated with schizencephaly and further expand the clinical spectrum of holoprosencephaly. Hum. Genet. 2010, 127, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Roessler, E.; El-Jaick, K.B.; Dubourg, C.; Vélez, J.I.; Solomon, B.D.; Pineda-Alvarez, D.E.; Lacbawan, F.; Zhou, N.; Ouspenskaia, M.; Paulussen, A.; et al. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum. Mutat. 2009, 30, E921–E935. [Google Scholar] [CrossRef] [PubMed]
- Day, E.S.; Wen, D.; Garber, E.A.; Hong, J.; Avedissian, L.S.; Rayhorn, P.; Shen, W.; Zeng, C.; Bailey, V.R.; Reilly, J.O.; et al. Zinc-dependent structural stability of human Sonic hedgehog. Biochemistry 1999, 38, 14868–14880. [Google Scholar] [CrossRef] [PubMed]
- Hitzenberger, M.; Schuster, D.; Hofer, T.S. The Binding Mode of the Sonic Hedgehog Inhibitor Robotnikinin, a Combined Docking and QM/MM MD Study. Front. Chem. 2017, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Stanton, B.Z.; Peng, L.F.; Maloof, N.; Nakai, K.; Wang, X.; Duffner, J.L.; Taveras, K.M.; Hyman, J.M.; Lee, S.W.; Koehler, A.N.; et al. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat. Chem. Biol. 2009, 5, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Himmelstein, D.S.; Cajigas, I.; Bi, C.; Clark, B.S.; Van Der Voort, G.; Kohtz, J.D. SHH E176/E177-Zn(2+) conformation is required for signaling at endogenous sites. Dev. Biol. 2017, 424, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Van Heijenoort, J. Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev. 2011, 75, 636–663. [Google Scholar] [CrossRef] [PubMed]
- Bochtler, M.; Odintsov, S.G.; Marcyjaniak, M.; Sabala, I. Similar active sites in lysostaphins and D-Ala-D-Ala metallopeptidases. Protein Sci. 2004, 13, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, H.; Olivares, G.H.; Faunes, F.; Oliva, C.; Larraín, J. Heparan sulfate proteoglycans exert positive and negative effects in Shh activity. J. Cell. Biochem. 2005, 96, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Capurro, M.I.; Xu, P.; Shi, W.; Li, F.; Jia, A.; Filmus, J. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev. Cell 2008, 14, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Witt, R.M.; Hecht, M.-L.; Pazyra-Murphy, M.F.; Cohen, S.M.; Noti, C.; van Kuppevelt, T.H.; Fuller, M.; Chan, J.A.; Hopwood, J.J.; Seeberger, P.H.; et al. Heparan sulfate proteoglycans containing a glypican 5 core and 2-O-sulfo-iduronic acid function as Sonic Hedgehog co-receptors to promote proliferation. J. Biol. Chem. 2013, 288, 26275–26288. [Google Scholar] [CrossRef] [PubMed]
- Siekmann, A.F.; Brand, M. Distinct tissue-specificity of three zebrafish ext1 genes encoding proteoglycan modifying enzymes and their relationship to somitic Sonic hedgehog signaling. Dev. Dyn. 2005, 232, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Bellaiche, Y.; The, I.; Perrimon, N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 1998, 394, 85–88. [Google Scholar] [PubMed]
- Krauss, S.; Concordet, J.P.; Ingham, P.W. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 1993, 75, 1431–1444. [Google Scholar] [CrossRef]
- Villarreal, C.M.; Darakananda, K.; Wang, V.R.; Jayaprakash, P.M.; Suzuki, Y. Hedgehog signaling regulates imaginal cell differentiation in a basally branching holometabolous insect. Dev. Biol. 2015, 404, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Adamska, M.; Matus, D.Q.; Adamski, M.; Green, K.; Rokhsar, D.S.; Martindale, M.Q.; Degnan, B.M. The evolutionary origin of hedgehog proteins. Curr. Biol. 2007, 17, R836–R837. [Google Scholar] [CrossRef] [PubMed]
- Fairclough, S.R.; Chen, Z.; Kramer, E.; Zeng, Q.; Young, S.; Robertson, H.M.; Begovic, E.; Richter, D.J.; Russ, C.; Westbrook, M.J.; et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013, 14, R15. [Google Scholar] [CrossRef] [PubMed]
- Pettigrew, C.A.; Asp, E.; Emerson, C.P. A new role for Hedgehogs in juxtacrine signaling. Mech. Dev. 2014, 131, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Ochi, H.; Pearson, B.J.; Chuang, P.-T.; Hammerschmidt, M.; Westerfield, M. Hhip regulates zebrafish muscle development by both sequestering Hedgehog and modulating localization of Smoothened. Dev. Biol. 2006, 297, 127–140. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roelink, H. Sonic Hedgehog Is a Member of the Hh/DD-Peptidase Family That Spans the Eukaryotic and Bacterial Domains of Life. J. Dev. Biol. 2018, 6, 12. https://doi.org/10.3390/jdb6020012
Roelink H. Sonic Hedgehog Is a Member of the Hh/DD-Peptidase Family That Spans the Eukaryotic and Bacterial Domains of Life. Journal of Developmental Biology. 2018; 6(2):12. https://doi.org/10.3390/jdb6020012
Chicago/Turabian StyleRoelink, Henk. 2018. "Sonic Hedgehog Is a Member of the Hh/DD-Peptidase Family That Spans the Eukaryotic and Bacterial Domains of Life" Journal of Developmental Biology 6, no. 2: 12. https://doi.org/10.3390/jdb6020012
APA StyleRoelink, H. (2018). Sonic Hedgehog Is a Member of the Hh/DD-Peptidase Family That Spans the Eukaryotic and Bacterial Domains of Life. Journal of Developmental Biology, 6(2), 12. https://doi.org/10.3390/jdb6020012




