An Overview of Hox Genes in Lophotrochozoa: Evolution and Functionality
Abstract
:1. Introduction
2. Hox Presence in Lophotrochozoa
3. Expression Patterns
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gehring, W.J.; Affolter, M.; Burglin, T. Homeodomain proteins. Annu. Rev. Biochem. 1994, 63, 487–526. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, W.; Krumlauf, R. Homeobox genes and axial patterning. Cell 1992, 68, 283–302. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Dessain, S.; Gross, C.T.; Kuziora, M.A.; McGinnis, W. Antp-type homeodomains have distinct DNA binding specificities that correlate with their different regulatory functions in embryos. EMBO J. 1992, 11, 991–1002. [Google Scholar] [PubMed]
- Meyer, A.; van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). BioEssays 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Duboule, D. Temporal colinearity and phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 1994, 135–142. [Google Scholar]
- Ikuta, T.; Yoshida, N.; Satoh, N.; Saiga, H. Ciona intestinalis Hox gene cluster: Its dispersed structure and residual collinear expression in development. Proc. Natl. Acad. Sci. USA 2004, 101, 15118–15123. [Google Scholar] [CrossRef] [PubMed]
- Fröbius, A.C.; Matus, D.Q.; Seaver, E.C. Genomic organization and expression demonstrate spatial and temporal Hox gene colinearity in the lophotrochozoan Capitella sp I. PLoS ONE 2008, 3, e4004. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E.; Permanyer, J.; Martinez, P. The origin of patterning systems in bilateria—Insights from the Hox and ParaHox genes in Acoelomorpha. Genom. Proteom. Bioinf. 2011, 9, 65–76. [Google Scholar] [CrossRef]
- Garcia-Fernàndez, J. The genesis and evolution of Homeobox gene clusters. Nat. Rev. Genet. 2005, 6, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Kourakis, M.J.; Martindale, M.K. Hox gene duplication and deployment in the annelid leech Helobdella. Evol. Dev. 2001, 3, 145–153. [Google Scholar] [PubMed]
- Arthur, W. The Origin of Animal Body Plans: A Study in evolutionary Developmental Biology, 1st ed.; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar]
- Garcia-Fernàndez, J. Hox, ParaHox, ProtoHox: Facts and guesses. Heredity 2005, 94, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Holland, P.W. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E.; Nadal, M.; Baguñà, J.; Martínez, P. Tracking the origins of the bilaterian Hox patterning system: Insights from the acoel flatworm Symsagittifera roscoffensis. Evol. Dev. 2009, 11, 574–581. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, R.; Grenier, J.K.; Andreeva, T.; Cook, C.E.; Adoutte, A.; Akam, M.; Carroll, S.B.; Balavoine, G. Hox genes in brachiopods and priapulids and protostome evolution. Nature 1999, 399, 772–776. [Google Scholar] [PubMed]
- Balavoine, G.; de Rosa, R.; Adoutte, A. Hox clusters and bilaterian phylogeny. Mol. Phylogenet. Evol. 2002, 24, 366–373. [Google Scholar] [CrossRef]
- Dunn, C.W.; Hejnol, A.; Matus, D.Q.; Pang, K.; Browne, W.E.; Smith, S.A.; Seaver, E.; Rouse, G.W.; Obst, M.; Edgecombe, G.D.; et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 2008, 452, 745–749. [Google Scholar] [CrossRef] [PubMed]
- Paps, J.; Baguñà, J.; Riutort, M. Lophotrochozoa internal phylogeny: New insights from an up-to-date analysis of nuclear ribosomal genes. Proc. Biol. Sci. 2009, 276, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Simakov, O.; Marletaz, F.; Cho, S.-J.; Edsinger-Gonzales, E.; Havlak, P.; Hellsten, U.; Kuo, D.H.; Larsson, T.; Lv, J.; Arendt, D.; et al. Insights into bilaterian evolution from three spiralian genomes. Nature 2013, 493, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Laumer, C.E.; Bekkouche, N.; Kerbl, A.; Goetz, F.; Neves, R.C.; Sørensen, M.V.; Kristensen, R.M.; Hejnol, A.; Dunn, C.W.; Giribet, G.; et al. Spiralian phylogeny informs the evolution of microscopic lineages. Curr. Biol. 2015, 25, 2000–2006. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.J.; Takeuchi, T.; Koyanagi, R.; Yamada, L.; Kanda, M.; Khalturina, M.; Fujie, M.; Yamasaki, S.; Endo, K.; Satoh, N. The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Flot, J.F.; Hespeels, B.; Li, X.; Noel, B.; Arkhipova, I.; Danchin, E.G.; Hejnol, A.; Henrissat, B.; Koszul, R.; Aury, J.M.; et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 2013, 500, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Koyanagi, R.; Gyoja, F.; Kanda, M.; Hisata, K.; Fujie, M.; Goto, H.; Yamasaki, S.; Nagai, K.; Morino, Y.; et al. Bivalve-specific gene expansion in the pearl oyster genome: Implications of adaptation to a sessile lifestyle. Zool. Lett. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Albertin, C.B.; Simakov, O.; Mitros, T.; Wang, Z.Y.; Pungor, J.R.; Edsinger-Gonzales, E.; Brenner, S.; Ragsdale, C.W.; Rokhsar, D.S. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 2015, 524, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, W.; Zhang, L.; Zhang, Z.; Li, J.; Lu, G.; Zhu, Y.; Wang, Y.; Huang, Y.; Liu, J.; et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nat. Genet. 2013, 45, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Koziol, U.; Lalanne, A.I.; Castillo, E. Hox genes in the parasitic Platyhelminthes Mesocestoides corti, Echinococcus multilocularis, and Schistosoma mansoni: Evidence for a reduced Hox complement. Biochem. Genet. 2009, 47, 100–116. [Google Scholar] [CrossRef] [PubMed]
- Tsai, I.J.; Zarowiecki, M.; Holroyd, N.; Garciarrubio, A.; Sánchez-Flores, A.; Brooks, K.L.; Tracey, A.; Bobes, R.J.; Fragoso, G.; Sciutto, E.; et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 2013, 496, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Jex, A.R.; Li, B.; Liu, S.; Yang, L.; Xiong, Z.; Li, Y.; Cantacessi, C.; Hall, R.S.; Xu, X.; et al. Whole-genome sequence of Schistosoma haematobium. Nat. Genet. 2012, 44, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.L.; Chen, S.X.; Dou, T.H.; Xu, M.J.; Xu, J.X.; Zhang, L.; Hu, W.; Wang, S.J.; Zhou, Y. Hox genes from the parasitic flatworm Schistosoma japonicum. Genomics 2012, 99, 59–65. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hahn, C.; Fromm, B.; Bachmann, L. Comparative genomics of flatworms (Platyhelminthes) reveals shared genomic features of ecto- and endoparastic Neodermata. Genome Biol. Evol. 2015, 6, 1105–1117. [Google Scholar] [CrossRef] [PubMed]
- Wasik, K.; Gurtowski, J.; Zhou, X.; Ramos, O.M.; Delás, M.J.; Battistoni, G.; El Demerdash, O.; Falciatori, I.; Vizoso, D.B.; Smith, A.D.; et al. Genome and transcriptome of the regeneration-competent flatworm, Macrostomum lignano. Proc. Natl. Acad. Sci. USA 2015, 112, 12462–12467. [Google Scholar] [CrossRef] [PubMed]
- Passamaneck, Y.J.; Halanych, K.M. Evidence from Hox genes that bryozoans are lophotrochozoans. Evol. Dev. 2004, 6, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Kmita-Cunisse, M.; Loosli, F.; Bièrne, J.; Gehring, W.J. Homeobox genes in the ribbonworm Lineus sanguineus: Evolutionary implications. Proc. Natl. Acad. Sci. USA 1998, 95, 3030–3035. [Google Scholar] [CrossRef] [PubMed]
- Hiebert, L.S.; Maslakova, S.A. Hox genes pattern the anterior-posterior axis of the juvenile but not the larva in a maximally indirect developing invertebrate, Micrura alaskensis (Nemertea). BMC Biol. 2015, 13. [Google Scholar] [CrossRef] [PubMed]
- Hiebert, L.S.; Maslakova, S.A. Expression of Hox, Cdx, and Six3/6 genes in the hoplonemertean Pantinonemertes californiensis offers insight into the evolution of maximally indirect development in the phylum Nemertea. Evodevo 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Paps, J.; Xu, F.; Zhang, G.; Holland, P.W. Reinforcing the egg-timer: Recruitment of novel lophotrochozoa homeobox genes to early and late development in the pacific oyster. Genome Biol. Evol. 2015, 7, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Barucca, M.; Olmo, E.; Canapa, A. Hox and ParaHox genes in bivalve molluscs. Gene 2003, 317, 97–102. [Google Scholar] [CrossRef]
- Iijima, M.; Akiba, N.; Sarashina, I.; Kuratani, S.; Endo, K. Evolution of Hox genes in molluscs: A comparison among seven morphologically diverse classes. J. Molluscan Stud. 2006, 72, 259–266. [Google Scholar] [CrossRef]
- Mesías-Gansbiller, C.; Sánchez, J.L.; Pazos, A.J.; Lozano, V.; Martínez-Escauriaza, R.; Luz Pérez-Parallé, M. Conservation of Gbx genes from EHG homeobox in bivalve molluscs. Mol. Phylogenet. Evol. 2012, 63, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Peréz-Parallé, M.L.; Carpintero, P.; Pazos, A.J.; Abad, M.; Sánchez, J.L. The Hox cluster in the bivalve mollusc Mytilus galloprovincialis. Biochem. Genet. 2005, 43, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Canapa, A.; Biscotti, M.A.; Olmo, E.; Barucca, M. Isolation of Hox and ParaHox genes in the bivalve Pecten maximus. Gene 2005, 348, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Carpintero, P.; Pazos, A.J.; Abad, M.; Sánchez, J.L.; Pérez-Parallé, M.L. Presence Proboscipedia and Caudal gene homologues in a bivalve mollusc. J. Biochem. Mol. Biol. 2004, 37, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Morino, Y.; Okada, K.; Niikura, M.; Honda, M.; Satoh, N.; Wada, H. A genome-wide survey of genes encoding transcription factors in the Japanese pearl oyster, Pinctada fucata: I. Homeobox genes. Zool. Sci. 2013, 30, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Gates, R.D.; Jacobs, D.K. Gene fishing: The use of a simple protocol to isolate multiple homeodomain classes from diverse invertebrate taxa. J. Mol. Evol. 2003, 56, 509–516. [Google Scholar] [PubMed]
- Samadi, L.; Steiner, G. Involvement of Hox genes in shell morphogenesis in the encapsulated development of a top shell gastropod (Gibbula varia L.). Dev. Genes Evol. 2009, 219, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Samadi, L.; Steiner, G. Expression of Hox genes during the larval development of the snail, Gibbula varia [L]-further evidence of non-colinearity in molluscs. Dev. Genes Evol. 2010, 220, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Giusti, A.F.; Hinman, V.F.; Degnam, S.M.; Degnan, B.M.; Morse, D.E. Expression of a Scr/Hox5 gene in the larval central nervous system of the gastropod Haliotis, a non-segmented spiralian lophotrochozoan. Evol. Dev. 2000, 2, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Hinman, V.F.; Degnan, B.M. Mox homeobox expression in muscle lineage of the gastropod Haliotis asinina: Evidence for a conserved role in bilaterian myogenesis. Dev. Genes Evol. 2002, 212, 141–144. [Google Scholar] [PubMed]
- Hinman, V.F.; O’Brien, E.K.; Richards, G.S.; Degnan, B.M. Expression of anterior Hox genes during larval development of the gastropod Haliotis asinina. Evol. Dev. 2003, 5, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Degnan, B.M.; Morse, D.E. Identification of eight homeobox-containing transcripts expressed during larval development and at metamorphosis in the gastropod mollusc Haliotis rufescens. Mol. Mar. Biol. Biotechnol. 1993, 2, 1–9. [Google Scholar] [PubMed]
- Lambert, J.D.; Chan, X.Y.; Spiecker, B.; Sweet, H.C. Characterizing the embryonic transcriptome of the snail Ilyanassa. Integr. Comp. Biol. 2010, 50, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Kenny, N.J.; Namigai, E.K.; Marlétaz, F.; Hui, J.H.; Shimeld, S.M. Draft genome assemblies and predicted microRNA complements of the intertidal lophotrochozoans Patella vulgata (Mollusca, Patellogastropoda) and Spirobranchus (Pomatoceros) lamarcki (Annelida, Serpulida). Mar. Genom. 2015, 24, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Callaerts, P.; Lee, P.N.; Hartmann, B.; Farfan, C.; Choy, D.W.; Ikeo, K.; Fischbach, K.-F.; Gehring, W.J.; Gert de Couet, H. Hox genes in the sepiolid squid Euprymna scolopes: Implications for the evolution of complex body plans. Proc. Natl. Acad. Sci. USA 2002, 99, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- Pernice, M.; Deutsch, J.S.; Andouche, A.; Boucher-Rodoni, R.; Bonnaud, L. Unexpected variation of Hox genes’ homeodomains in cephalopods. Mol. Phylogenet. Evol. 2006, 40, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, M.; Wollesen, T.; de Oliveira, A.L.; Wanninger, A. Unexpected co-linearity of Hox gene expression in an aculiferan mollusk. BMC Evol. Biol. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Canapa, A.; Olmo, E.; Barucca, M. Hox genes in the Antarctic polyplacophoran Nuttallochiton mirandus. J. Exp. Zool. Part B 2007, 308, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Dick, M.H.; Buss, L.W. A PCR-based Survey of Homeobox Genes in Ctenodrilus serratus (Annelida: Polychaeta). Mol. Phylogenet. Evol. 1994, 3, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Cho, P.Y.; Cho, S.J.; Lee, M.S.; Lee, J.A.; Tak, E.S.; Shin, C.; Choo, J.K.; Park, S.C.; Lee, K.-S.; Park, H.-Y.; et al. Note: A PCR-Based Analysis of Hox Genes in an Earthworm, Eisenia andrei (Annelida: Oligochaeta). Biochem. Genet. 2004, 42, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Zwarycz, A.S.; Nossa, C.W.; Putnam, N.H.; Ryan, J. Timing and scope of genomic expansion within Annelida: Evidence from homeoboxes in the genome of the earthworm Eisenia fetida. Genome Biol. Evol. 2015. [Google Scholar] [CrossRef]
- Kourakis, M.J.; Master, V.A.; Lokhorst, D.K.; Nardelli-Haefliger, D.; Wedeen, C.J.; Martindale, M.Q.; Shankland, M. Conserved anterior boundaries of Hox gene expression in the central nervous system of the leech Helobdella. Dev. Biol. 1997, 190, 284–300. [Google Scholar] [CrossRef] [PubMed]
- Aisemberg, G.O.; Macagno, E.R. Lox1, an Antennapedia-class homeobox gene, is expressed during leech gangliogenesis in both transient and stable central neurons. Dev. Biol. 1994, 161, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.Y.; Aisemberg, G.O.; Gan, W.B.; Macagno, E.R. The leech homeobox gene Lox4 may determine segmental differentiation of identified neurons. J. Neurosci. 1995, 15, 5551–5559. [Google Scholar] [PubMed]
- Wong, V.Y.; Macagno, E.R. Lox6, a leech Dfd ortholog, is expressed in the central nervous system and in peripheral sensory structures. Dev. Genes Evol. 1998, 208, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Wysocka-Diller, J.W.; Aisemberg, G.O.; Baumgarten, M.; Levine, M.; Macagno, E.R. Characterization of a homologue of bithorax-complex genes in the leech Hirudo medicinalis. Nature 1989, 341, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Vallès, Y.; Kim, K.M.; Ji, S.C.; Han, S.J.; Park, S.C. Additional duplicated Hox genes in the earthworm: Perionyx excavatus Hox genes consist of eleven paralog groups. Gene 2012, 493, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Snow, P.; Buss, L.W. HOM/Hox-type homeoboxes from Stylaria lacustris (Annelida: Oligochaeta). Mol. Phylogenet. Evol. 1994, 3, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, T.F.; Cook, C.; Korchagina, N.M.; Akam, M.; Dondua, A.K. Cloning and analysis of structural organization of Hox genes in the polychaete Nereis virens. Ontogenez 2001, 32, 225–233. [Google Scholar] [PubMed]
- Kulakova, M.; Bakalenko, N.; Novikova, E.; Cook, C.E.; Eliseeva, E.; Steinmetz, P.R.; Kostyuchenko, R.P.; Dondua, A.; Arendt, D.; Akam, M.; et al. Hox gene expression in larval development of the polychaetes Nereis virens and Platynereis dumerilii (Annelida, Lophotrochozoa). Dev. Genes Evol. 2007, 217, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Bleidorn, C.; Lanterbecq, D.; Eeckhaut, I.; Tiedemann, R. A PCR survey of Hox genes in the myzostomid Myzostoma cirriferum. Dev. Genes Evol. 2009, 219, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Lee, D.H.; Kwon, H.J.; Ahn, C.H.; Park, S.C.; Shin, K.S. Hox genes in the echiuroid Urechis unicinctus. Dev. Genes Evol. 2006, 216, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Irvine, S.Q.; Martindale, M.Q. Expression patterns of anterior Hox genes in the polychaete Chaetopterus: Correlation with morphological boundaries. Dev. Biol. 2000, 217, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Oliver, G.; Vispo, M.; Mailhos, A.; Martinez, C.; Sosa-Pineda, B.; Fielitz, W.; Ehrlich, R. Homeoboxes in flatworms. Gene 1992, 121, 337–342. [Google Scholar] [CrossRef]
- Olson, P.D. Hox genes and the parasitic flatworms: New opportunities, challenges and lessons from the free-living. Parasitol. Int. 2008, 57, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, Y.S.; Jeon, H.K.; Park, J.K.; Kim, C.B.; Eom, K.S. Hox genes from the tapeworm Taenia asiatica (Platyhelminthes: Cestoda). Biochem. Genet. 2007, 45, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Bartels, J.L.; Murtha, M.T.; Ruddle, F.H. Multiple Hox/HOM-class homeoboxes in Platyhelminthes. Mol. Phylogenet. Evol. 1993, 2, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Webster, P.J.; Mansour, T.E. Conserved classes of homeodomains in Schistosoma mansoni, an early bilateral metazoan. Mech. Dev. 1992, 38, 25–32. [Google Scholar] [CrossRef]
- Pierce, R.; Wu, W.; Hirai, H.; Ivens, A.; Murphy, L.D.; Noël, C.; Johnston, D.A.; Artiguenave, F.; Adams, M.; Cornette, J.; et al. Evidence for a dispersed Hox gene cluster in the platyhelminth parasite Schistosoma mansoni. Mol. Biol. Evol. 2005, 22, 2491–2503. [Google Scholar] [CrossRef] [PubMed]
- Badets, M.; Verneau, O. Hox genes from Polystomatidae (Platyhelmintes, Monogenea). Int. J. Parassitol. 2009, 39, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Balavoine, G.; Telford, M.J. Identification of planarian homeobox sequences indicates the antiquity of most Hox/homeotic gene subclasses. Proc. Natl. Acad. Sci. USA. 1995, 92, 7227–7231. [Google Scholar] [CrossRef] [PubMed]
- Orii, H.; Kato, K.; Umesono, Y.; Sakurai, T.; Agata, K.; Watanabe, K. The planarian HOM/HOX homeobox genes (Plox) expressed along the anteroposterior axis. Dev. Biol. 1999, 210, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Nogi, T.; Watanabe, K. Position-specific and non-colinear expression of the planarian posterior (Abdominal-B-like) gene. Dev. Growth Differ. 2001, 43, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Saló, E.; Tauler, J.; Jmenéz, E.; Bayascas, J.R.; González-Linares, J.; Garcia-Fernàndez, J.; Baguñà, J. Hox and ParaHox Genes in Flatworms. Characterization and Expression. Am. Zool. 2001, 41, 652–663. [Google Scholar]
- Tarabykin, V.S.; Lukyanov, K.A.; Potapov, V.K.; Lukyanov, S.A. Detection of planarian Antennapedialike, homeobox genes expressed during regeneration. Gene 1995, 158, 197–202. [Google Scholar] [CrossRef]
- Bayascas, J.R.; Castillo, E.; Muñoz-Mármol, A.M.; Saló, E. Planarian Hox genes: Novel patterns of expression during re generation. Development 1997, 124, 141–148. [Google Scholar] [PubMed]
- Bayascas, J.R.; Castillo, E.; Saló, A.M. Platyhelminthes have a Hox code differentially activated during regeneration, with genes closely related to those of spiralian protostomes. Dev. Genes Evol. 1998, 208, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Balavoine, G. Identification of members of several homeobox genes in a planarian using a ligation-mediated polymerase chain reaction technique. Nucleic Acids. Res. 1996, 8, 1547–1553. [Google Scholar] [CrossRef]
- Iglesias, M.; Gomez-Skarmeta, J.L.; Saló, E.; Adell, T. Silencing of Smed-catenin1 generates radial-like hypercephalized planarians. Development 2008, 135, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Martín-Durán, J.M.; Amaya, E.; Romero, R. Germ layer specification and axial patterning in the embryonic development of the freshwater planarian Schmidtea polychroa. Dev. Biol. 2010, 340, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Welch, M.J.L.; Welch, M.D.B.; Center for Comparative Molecular Biology and Evolution, Woods Hole, MA, USA. Unpublished work. 2008.
- Smith, S.A.; Wilson, N.G.; Goetz, F.E.; Feehery, C.; Andrade, S.C.; Rouse, G.W.; Giribet, G.; Dunn, C.W. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 2011, 480, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Wilson, N.G.; Goetz, F.E.; Feehery, C.; Andrade, S.C.; Rouse, G.W.; Giribet, G.; Dunn, C.W. Corrigendum: Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 2013, 493, 708. [Google Scholar] [CrossRef]
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Barucca, M. Hox and ParaHox genes: A review on molluscs. Genesis 2014, 52, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Struck, T.H.; Paul, C.; Hill, N.; Hartmann, S.; Hösel, C.; Kube, M.; Lieb, B.; Meyer, A.; Tiedemann, R.; Purschke, G.; et al. Phylogenomic analyses unravel annelid evolution. Nature 2011, 471, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Kristof, A.; Wollesen, T.; Wanninger, A. Segmental mode of neural patterning in sipuncula. Curr. Biol. 2008, 18, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Hessling, R. Metameric organisation of the nervous system in developmental stages of Urechis caupo (Echiura) and its phylogenetic implications. Zoomorphology 2002, 121, 221–234. [Google Scholar] [CrossRef]
- Struck, T.H.; Wey-Fabrizius, A.R.; Golombek, A.; Hering, L.; Weigert, A.; Bleidorn, C.; Klebow, S.; Iakovenko, N.; Hausdorf, B.; Petersen, M.; et al. Platyzoan paraphyly based on phylogenomic data supports a noncoelomate ancestry of spiralia. Mol. Biol. Evol. 2014, 31, 1833–1849. [Google Scholar] [CrossRef] [PubMed]
- Sperling, E.A.; Pisani, D.; Peterson, K.J. Molecular paleobiological insights into the origin of the Brachiopoda. Evol. Dev. 2011, 13, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Riutort, M.; Alvarez-Presas, M.; Lazaro, E.; Solà, E.; Paps, J. Evolutionary history of the Tricladida and the Platyhelminthes: An up-to-date phylogenetic and systematic account. Int. J. Dev. Biol. 2012, 56, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Ferrier, D.E.K.; Holland, P.W.H. Ciona intestinalis ParaHox genes: Evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol. Phylogenet. Evol. 2002, 24, 412–417. [Google Scholar] [CrossRef]
- Focareta, L.; Sesso, S.; Cole, A.G. Characterization of homeobox genes reveals sophisticated regionalization of the central nervous system in the European cuttlefish Sepia officinalis. PLoS ONE 2014, 9, e109627. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, P.R.H.; Kostyuchenko, R.P.; Fischer, A.; Arendt, D. The segmental pattern of otx, gbx, and Hox genes in the annelid Platynereis dumerilii. Evol. Dev. 2011, 13, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Badets, M.; Mitta, G.; Galinier, R.; Verneau, O. Expression patterns of Abd-A/Lox4 in a monogenean parasite with alternative developmental paths. Mol. Biochem. Parasit. 2010, 173, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Bayascas, J.R.; Castillo, E.; Muñoz-Mármol, A.M.; Baguñà, J.; Saló, E. Synchronous and early activation of planarian Hox genes and re-specification of body axes during regeneration. Hidrobiologia 1998, 383, 125–130. [Google Scholar] [CrossRef]
- Baguñà, J.; Saló, E.; Romero, R.; Garcia-Fernàndez, J.; Bueno, D.; Muñoz-Mármol, A.M.; Bayascas-Ramírez, J.R.; Casali, A. Regeneration and pattern formation in planarians: Cells, molecules and genes . Zool. Sci. 1994, 11, 781–795. [Google Scholar]
- Saló, E.; Baguñà, J. Regeneration in planarians and other worms: New findings, new tools, and new perspectives. J. Exp. Zool. 2002, 292, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Saló, E.; Abril, J.F.; Adell, T.; Cebrià, F.; Eckelt, K.; Fernandez-Taboada, E.; Handberg-Thorsager, M.; Iglesias, M.; Molina, D.; Rodriguez-Esteban, G. Planarian regeneration: Achievements and future directions after 20 years of research. Int. J. Dev. Biol. 2009, 53, 1317–1327. [Google Scholar] [CrossRef] [PubMed]

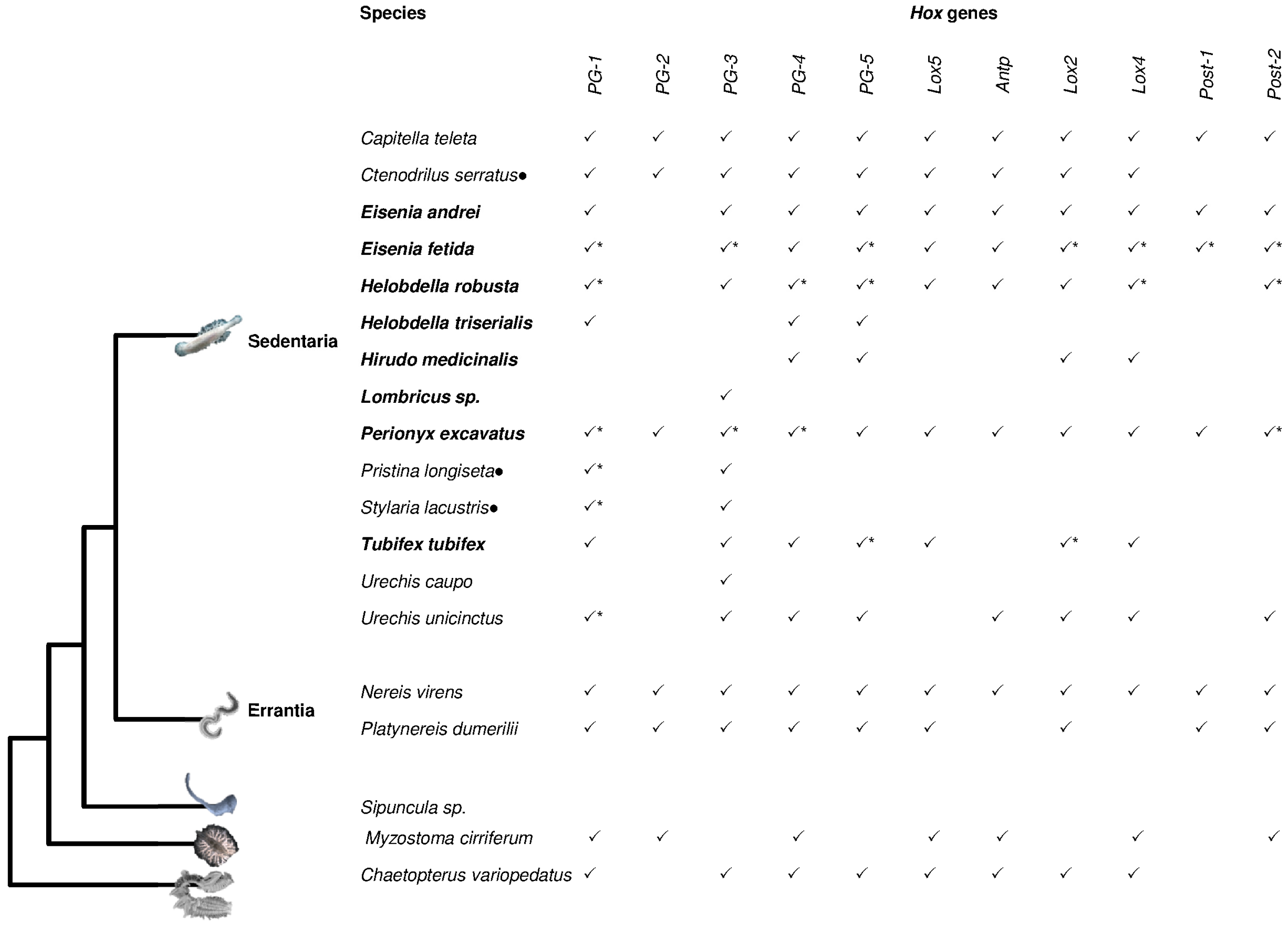
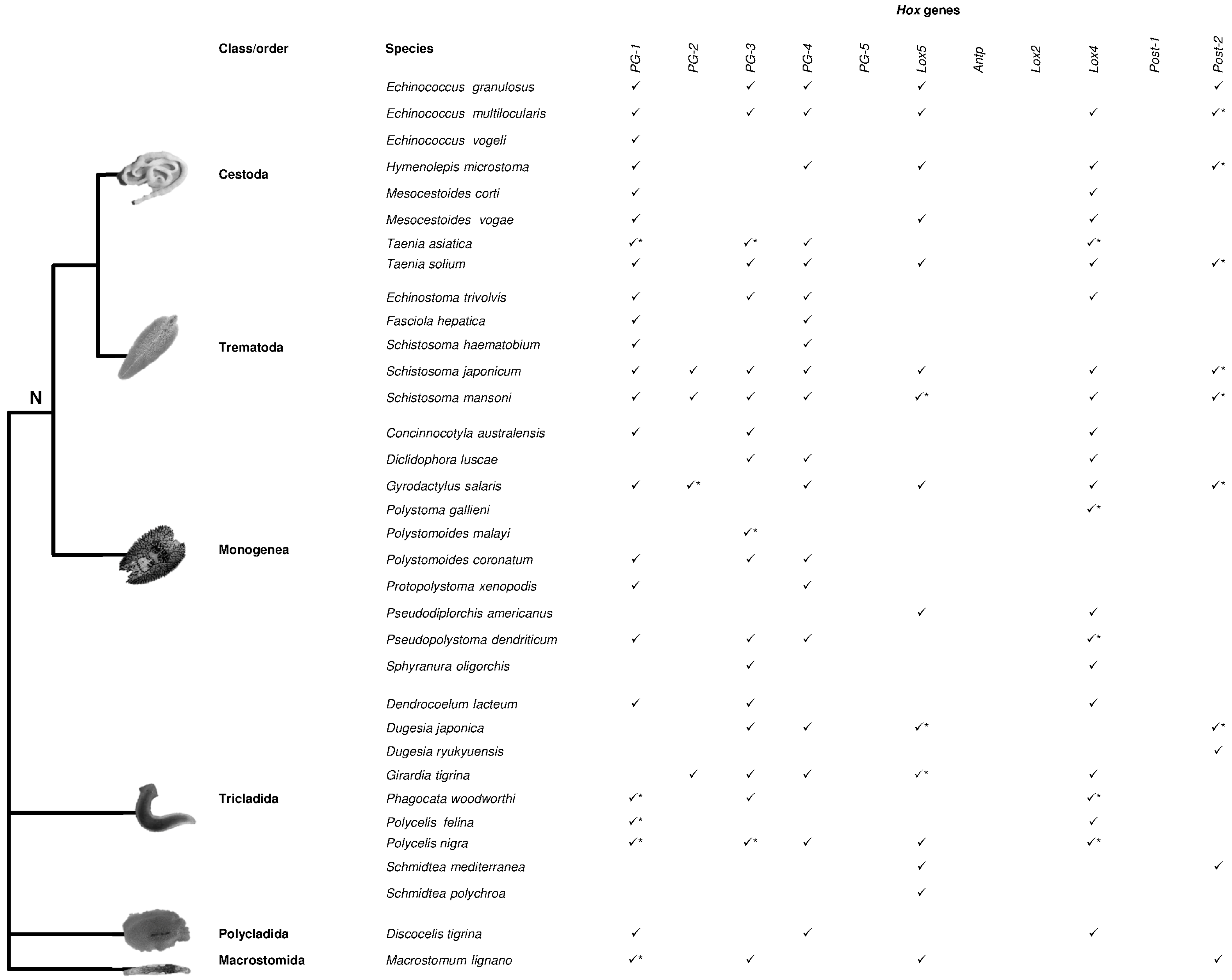
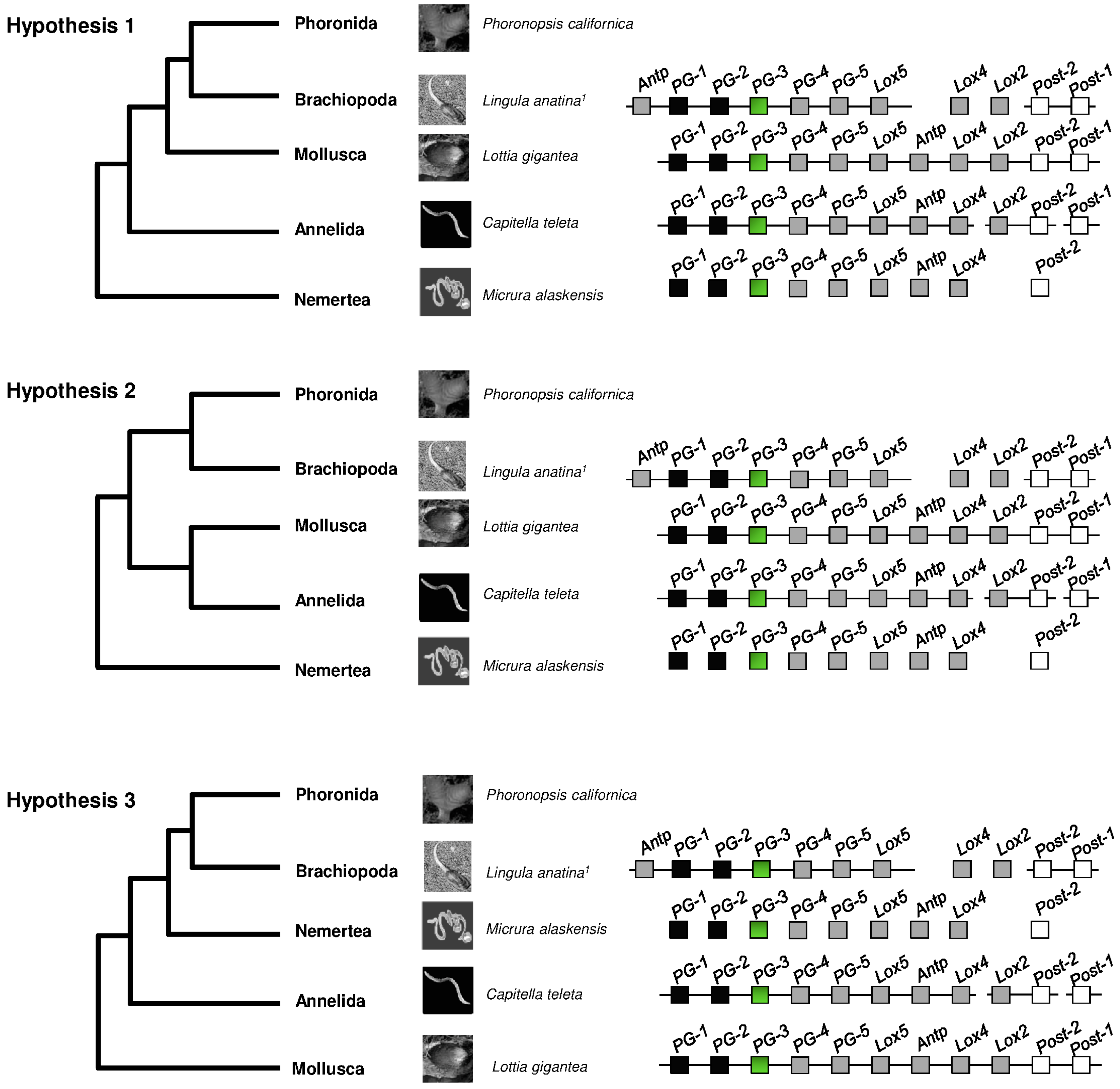
| Hox Genes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Species | PG-1 | PG-2 | PG-3 | PG-4 | PG-5 | Lox5 | Antp | Lox2 | Lox4 | Post-1 | Post-2 | References |
| Brachiopoda | Lingula anatina | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | [16,22] |
| Bryozoa | Bugula turrita | √ | √ | √ * | √ | √ | [34] | ||||||
| Rotifera | Adineta vaga | √ * | √ | √ * | √ * | √ * | √ * | √ * | [23] | ||||
| Philodina roseola | √ * | √ * | [91] | ||||||||||
| Nemertea | Lineus sanguineus | √ | √ | √ | √ | √ | √ | [35] | |||||
| Micrura alaskensis | √ | √ | √ | √ | √ | √ | √ | √ | √ | [36] | |||
| Pantinonemertes californiensis | √ | √ | √ | √ | √ | √ | [37] | ||||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barucca, M.; Canapa, A.; Biscotti, M.A. An Overview of Hox Genes in Lophotrochozoa: Evolution and Functionality. J. Dev. Biol. 2016, 4, 12. https://doi.org/10.3390/jdb4010012
Barucca M, Canapa A, Biscotti MA. An Overview of Hox Genes in Lophotrochozoa: Evolution and Functionality. Journal of Developmental Biology. 2016; 4(1):12. https://doi.org/10.3390/jdb4010012
Chicago/Turabian StyleBarucca, Marco, Adriana Canapa, and Maria Assunta Biscotti. 2016. "An Overview of Hox Genes in Lophotrochozoa: Evolution and Functionality" Journal of Developmental Biology 4, no. 1: 12. https://doi.org/10.3390/jdb4010012
APA StyleBarucca, M., Canapa, A., & Biscotti, M. A. (2016). An Overview of Hox Genes in Lophotrochozoa: Evolution and Functionality. Journal of Developmental Biology, 4(1), 12. https://doi.org/10.3390/jdb4010012








