The Presence of Two MyoD Genes in a Subset of Acanthopterygii Fish Is Associated with a Polyserine Insert in MyoD1
Abstract
1. Introduction
2. Methods
2.1. Sequence Analysis and Phylogeny of MyoD Genes in Teleosts
2.2. Cloning and Expression Analysis
2.3. Protein Expression
2.4. Western Blotting
2.5. Immunofluorescence
2.6. qPCR
3. Results
3.1. Phylogenetic Analysis Reveals Relationship among MyoD Genes in Teleost Fish
3.2. Expression Patterns of MyoD1 and MyoD2 in Developing O. alcalica Embryos
3.3. O. alcalica and D. rerio MyoD1 Proteins Show Nuclear Localisation
3.4. O. alcalica MyoD1 Protein Persists Longer Than D. rerio MyoD1
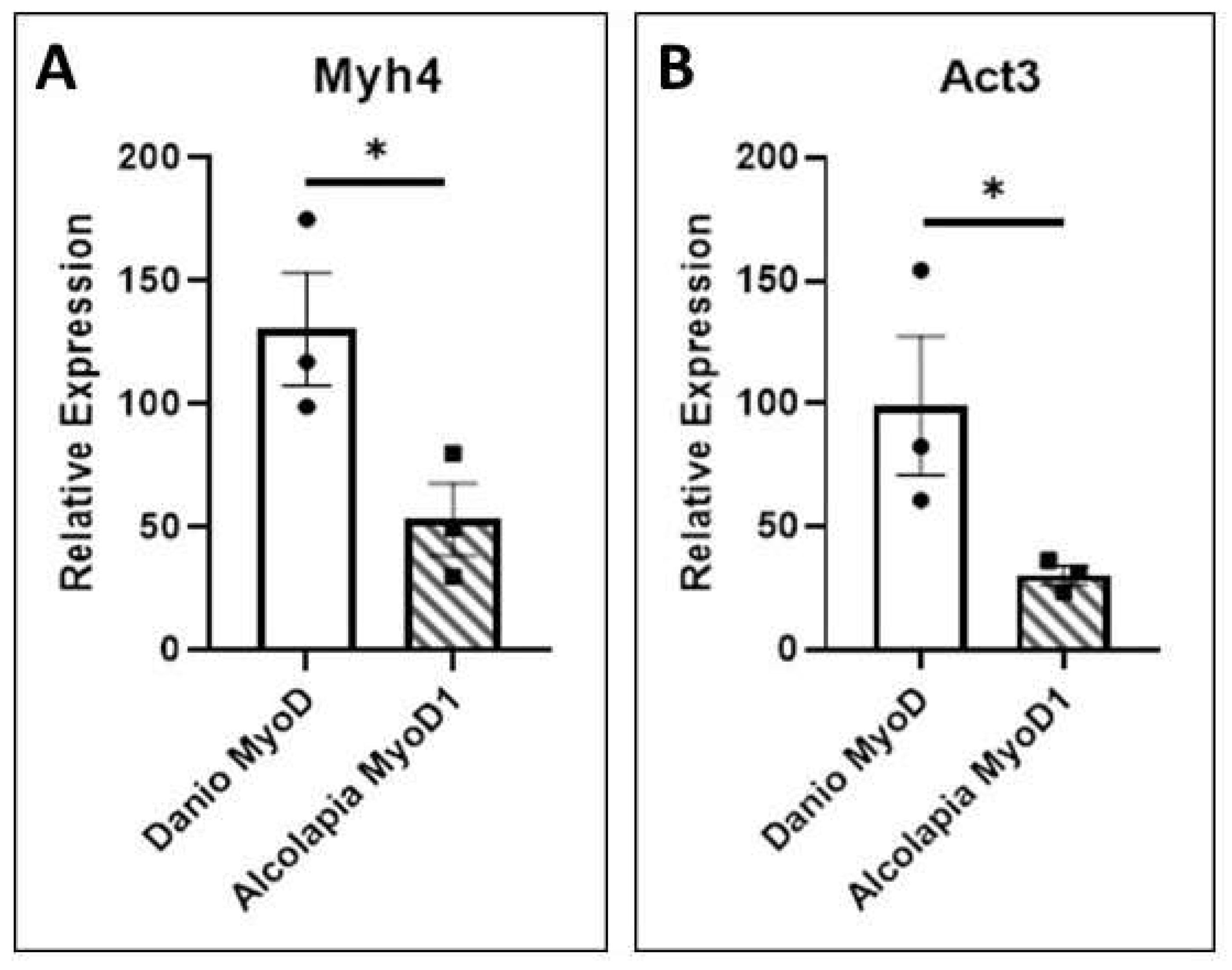
3.5. Transcriptional Activity
4. Discussion
4.1. Phylogenetic Analysis Reveals a Single Evolutionary Event for Inclusion of the Poly-Serine Domain in MyoD1
4.2. Preliminary Functional Analysis of MyoD1 Polyserine Region and Future Directions
4.3. Differences in the Expression Patterns of MyoD1 and MyoD2 in O. alcalica
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pownall, M.E.; Gustafsson, M.K.; Emerson, C.P., Jr. Myogenic Regulatory Factors and the Specification of Muscle Progenitors in Vertebrate Embryos. Annu. Rev. Cell Dev. Biol. 2002, 18, 747–783. [Google Scholar] [CrossRef] [PubMed]
- Zammit, P.S. Function of the Myogenic Regulatory Factors Myf5, MyoD, Myogenin and MRF4 in Skeletal Muscle, Satellite Cells and Regenerative Myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The Myogenic Regulatory Factors, Determinants of Muscle Development, Cell Identity and Regeneration. Semin. Cell Dev. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.V.; Hughes, S.M. Mef2 and the Skeletal Muscle Differentiation Program. Semin. Cell Dev. Biol. 2017, 72, 33–44. [Google Scholar] [CrossRef]
- Baylies, M.K.; Michelson, A.M. Invertebrate Myogenesis: Looking back to the Future of Muscle Development. Curr. Opin. Genet. Dev. 2001, 11, 431–439. [Google Scholar] [CrossRef]
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome Evolution in the Allotetraploid Frog Xenopus Laevis. Nature 2016, 538, 336–343. [Google Scholar] [CrossRef]
- Fisher, M.E.; Peck, W.; Branney, P.A.; Pownall, M.E. Cloning and Characterisation of Myf5 and MyoD Orthologues in Xenopus Tropicalis. Biol. Cell 2003, 95, 555–561. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. An Update on MyoD Evolution in Teleosts and a Proposed Consensus Nomenclature to Accommodate the Tetraploidization of Different Vertebrate Genomes. PLoS ONE 2008, 3, e1567. [Google Scholar] [CrossRef]
- Aase-Remedios, M.E.; Coll-Lladó, C.; Ferrier, D.E.K. More Than One-to-Four via 2R: Evidence of an Independent Amphioxus Expansion and Two-Gene Ancestral Vertebrate State for MyoD-Related Myogenic Regulatory Factors (MRFs). Mol. Biol. Evol. 2020, 37, 2966–2982. [Google Scholar] [CrossRef]
- Postlethwait, J.H.; Yan, Y.L.; Gates, M.A.; Horne, S.; Amores, A.; Brownlie, A.; Donovan, A.; Egan, E.S.; Force, A.; Gong, Z.; et al. Vertebrate Genome Evolution and the Zebrafish Gene Map. Nat. Genet. 1998, 18, 345–349. [Google Scholar] [CrossRef]
- Tan, X.; Du, S.J. Differential Expression of Two MyoD Genes in Fast and Slow Muscles of Gilthead Seabream (Sparus aurata). Dev. Genes Evol. 2002, 212, 207–217. [Google Scholar] [CrossRef]
- Andersen, Ø.; Dahle, S.W.; van Nes, S.; Bardal, T.; Tooming-Klunderud, A.; Kjørsvik, E.; Galloway, T.F. Differential Spatio-Temporal Expression and Functional Diversification of the Myogenic Regulatory Factors MyoD1 and MyoD2 in Atlantic Halibut (Hippoglossus hippoglossus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 154, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Kavembe, G.D.; Kautt, A.F.; Machado-Schiaffino, G.; Meyer, A. Eco-Morphological Differentiation in Lake Magadi Tilapia, an Extremophile Cichlid Fish Living in Hot, Alkaline and Hypersaline Lakes in East Africa. Mol. Ecol. 2016, 25, 1610–1625. [Google Scholar] [CrossRef] [PubMed]
- Ford, A.G.P.; Dasmahapatra, K.K.; Rüber, L.; Gharbi, K.; Cezard, T.; Day, J.J. High Levels of Interspecific Gene Flow in an Endemic Cichlid Fish Adaptive Radiation from an Extreme Lake Environment. Mol. Ecol. 2015, 24, 3421–3440. [Google Scholar] [CrossRef] [PubMed]
- Sutton, G.; White, L.J.; Ford, A.G.P.; Shechonge, A.; Day, J.J.; Dasmahapatra, K.K.; Pownall, M.E. Exploring the Expression of Cardiac Regulators in a Vertebrate Extremophile: The Cichlid Fish Oreochromis (Alcolapia) Alcalica. J. Dev. Biol. 2020, 8, 22. [Google Scholar] [CrossRef]
- Hardwick, L.J.A.; Davies, J.D.; Philpott, A. MyoD Phosphorylation on Multiple C Terminal Sites Regulates Myogenic Conversion Activity. Biochem. Biophys. Res. Commun. 2016, 481, 97–103. [Google Scholar] [CrossRef]
- Sun, L.; Trausch-Azar, J.S.; Ciechanover, A.; Schwartz, A.L. Ubiquitin-Proteasome-Mediated Degradation, Intracellular Localization, and Protein Synthesis of MyoD and Id1 during Muscle Differentiation. J. Biol. Chem. 2005, 280, 26448–26456. [Google Scholar] [CrossRef]
- Jo, C.; Cho, S.-J.; Jo, S.A. Mitogen-Activated Protein Kinase Kinase 1 (MEK1) Stabilizes MyoD through Direct Phosphorylation at Tyrosine 156 during Myogenic Differentiation. J. Biol. Chem. 2011, 286, 18903–18913. [Google Scholar] [CrossRef]
- Basu, S.; Mackowiak, S.D.; Niskanen, H.; Knezevic, D.; Asimi, V.; Grosswendt, S.; Geertsema, H.; Ali, S.; Jerković, I.; Ewers, H.; et al. Unblending of Transcriptional Condensates in Human Repeat Expansion Disease. Cell 2020, 181, 1062–1079. [Google Scholar] [CrossRef]
- Hughes, L.C.; Ortí, G.; Huang, Y.; Sun, Y.; Baldwin, C.C.; Thompson, A.W.; Arcila, D.; Betancur-R, R.; Li, C.; Becker, L.; et al. Comprehensive Phylogeny of Ray-Finned Fishes (Actinopterygii) Based on Transcriptomic and Genomic Data. Proc. Natl. Acad. Sci. USA 2018, 115, 6249–6254. [Google Scholar] [CrossRef]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Pennell, M.; Eastman, J.; Slater, G.; Brown, J.; Uyeda, J.; Fitzjohn, R.; Alfaro, M.; Harmon, L. geiger v2.0: An expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 2014, 30, 2216–2218. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Christen, B.; Slack, J.M. Spatial response to fibroblast growth factor signalling in Xenopus embryos. Development 1999, 126, 119–125. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.M.O.; Kinghorn, J.R.; Johnston, I.A. Differential Regulation of Multiple Alternatively Spliced Transcripts of MyoD. Gene 2007, 391, 178–185. [Google Scholar] [CrossRef]
- Hall, T.E.; Cole, N.J.; Johnston, I.A. Temperature and the Expression of Seven Muscle-Specific Protein Genes during Embryogenesis in the Atlantic Cod Gadus morhua L. J. Exp. Biol. 2003, 206, 3187–3200. [Google Scholar] [CrossRef][Green Version]
- Bower, N.I.; Johnston, I.A. Paralogs of Atlantic Salmon Myoblast Determination Factor Genes Are Distinctly Regulated in Proliferating and Differentiating Myogenic Cells. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2010, 298, R1615–R1626. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. A Novel Salmonid myoD Gene Is Distinctly Regulated during Development and Probably Arose by Duplication after the Genome Tetraploidization. FEBS Lett. 2006, 580, 4996–5002. [Google Scholar] [CrossRef]
- Hinits, Y.; Osborn, D.P.S.; Carvajal, J.J.; Rigby, P.W.J.; Hughes, S.M. Mrf4 (myf6) Is Dynamically Expressed in Differentiated Zebrafish Skeletal Muscle. Gene Expr. Patterns 2007, 7, 738–745. [Google Scholar] [CrossRef]
- Song, A.; Wang, Q.; Goebl, M.G.; Harrington, M.A. Phosphorylation of Nuclear MyoD Is Required for Its Rapid Degradation. Mol. Cell Biol. 1998, 18, 4994–4999. [Google Scholar] [CrossRef]
- Sadeh, R.; Breitschopf, K.; Bercovich, B.; Zoabi, M.; Kravtsova-Ivantsiv, Y.; Kornitzer, D.; Schwartz, A.; Ciechanover, A. The N-Terminal Domain of MyoD Is Necessary and Sufficient for Its Nuclear Localization-Dependent Degradation by the Ubiquitin System. Proc. Natl. Acad. Sci. USA 2008, 105, 15690–15695. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, N.D.; Gurdon, J.B. Activation of Muscle Genes without Myogenesis by Ectopic Expression of MyoD in Frog Embryo Cells. Nature 1990, 347, 197–200. [Google Scholar] [CrossRef]
- Kashi, Y.; King, D.; Soller, M. Simple Sequence Repeats as a Source of Quantitative Genetic Variation. Trends Genet. 1997, 13, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Kashi, Y.; King, D.G. Simple Sequence Repeats as Advantageous Mutators in Evolution. Trends Genet. 2006, 22, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Radó-Trilla, N.; Arató, K.; Pegueroles, C.; Raya, A.; de la Luna, S.; Albà, M.M. Key Role of Amino Acid Repeat Expansions in the Functional Diversification of Duplicated Transcription Factors. Mol. Biol. Evol. 2015, 32, 2263–2272. [Google Scholar] [CrossRef] [PubMed]
- Gerber, H.P.; Seipel, K.; Georgiev, O.; Höfferer, M.; Hug, M.; Rusconi, S.; Schaffner, W. Transcriptional Activation Modulated by Homopolymeric Glutamine and Proline Stretches. Science 1994, 263, 808–811. [Google Scholar] [CrossRef]
- Strzyz, P. Concentrating on Intrinsic Disorder. Nat. Rev. Genet. 2018, 19, 534. [Google Scholar] [CrossRef] [PubMed]
- Davesne, D.; Friedman, M.; Schmitt, A.D.; Fernandez, V.; Carnevale, G.; Ahlberg, P.E.; Sanchez, S.; Benson, R.B.J. Fossilized Cell Structures Identify an Ancient Origin for the Teleost Whole-Genome Duplication. Proc. Natl. Acad. Sci. USA 2021, 118, e2101780118. [Google Scholar] [CrossRef]
- Robertson, F.M.; Gundappa, M.K.; Grammes, F.; Hvidsten, T.R.; Redmond, A.K.; Lien, S.; Martin, S.A.M.; Holland, P.W.H.; Sandve, S.R.; Macqueen, D.J. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol. 2017, 18, 111. [Google Scholar] [CrossRef] [PubMed]
- Parey, E.; Louis, A.; Montfort, J.; Guiguen, Y.; Crollius, H.R.; Berthelot, C. An atlas of fish genome evolution reveals delayed rediploidization following the teleost whole-genome duplication. Genome Res. 2022, 32, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Betancur-R, R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Ortí, G. Phylogenetic Classification of Bony Fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef] [PubMed]
- Levinson, G.; Gutman, G.A. Slipped-Strand Mispairing: A Major Mechanism for DNA Sequence Evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar]
- Pâques, F.; Leung, W.Y.; Haber, J.E. Expansions and Contractions in a Tandem Repeat Induced by Double-Strand Break Repair. Mol. Cell Biol. 1998, 18, 2045–2054. [Google Scholar] [CrossRef] [PubMed]
- Persi, E.; Wolf, Y.I.; Koonin, E.V. Positive and Strongly Relaxed Purifying Selection Drive the Evolution of Repeats in Proteins. Nat. Commun. 2016, 7, 13570. [Google Scholar] [CrossRef]
- Haerty, W.; Golding, G.B. Genome-Wide Evidence for Selection Acting on Single Amino Acid Repeats. Genome Res. 2010, 20, 755–760. [Google Scholar] [CrossRef]
- Siwach, P.; Pophaly, S.D.; Ganesh, S. Genomic and Evolutionary Insights into Genes Encoding Proteins with Single Amino Acid Repeats. Mol. Biol. Evol. 2006, 23, 1357–1369. [Google Scholar] [CrossRef]
- Kumar, A.S.; Sowpati, D.T.; Mishra, R.K. Single Amino Acid Repeats in the Proteome World: Structural, Functional, and Evolutionary Insights. PLoS ONE 2016, 11, e0166854. [Google Scholar] [CrossRef]
- Kitzmann, M.; Vandromme, M.; Schaeffer, V.; Carnac, G.; Labbé, J.C.; Lamb, N.; Fernandez, A. cdk1-and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: Role in modulating MyoD half-life and myogenic activity. Mol. Cell. Biol. 1999, 19, 3167–3176. [Google Scholar] [CrossRef]
- Tapscott, S.J.; Davis, R.L.; Thayer, M.J.; Cheng, P.F.; Weintraub, H.; Lassar, A.B. MyoD1: A Nuclear Phosphoprotein Requiring a Myc Homology Region to Convert Fibroblasts to Myoblasts. Science 1988, 242, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Bergstrom, D.A.; Penn, B.H.; Strand, A.; Perry, R.L.; Rudnicki, M.A.; Tapscott, S.J. Promoter-Specific Regulation of MyoD Binding and Signal Transduction Cooperate to Pattern Gene Expression. Mol. Cell 2002, 9, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Berkes, C.A.; Tapscott, S.J. MyoD and the Transcriptional Control of Myogenesis. Semin. Cell Dev. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Devoto, S.H.; Melançon, E.; Eisen, J.S.; Westerfield, M. Identification of Separate Slow and Fast Muscle Precursor Cells in vivo, prior to Somite Formation. Development 1996, 122, 3371–3380. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
White, L.J.; Russell, A.J.; Pizzey, A.R.; Dasmahapatra, K.K.; Pownall, M.E. The Presence of Two MyoD Genes in a Subset of Acanthopterygii Fish Is Associated with a Polyserine Insert in MyoD1. J. Dev. Biol. 2023, 11, 19. https://doi.org/10.3390/jdb11020019
White LJ, Russell AJ, Pizzey AR, Dasmahapatra KK, Pownall ME. The Presence of Two MyoD Genes in a Subset of Acanthopterygii Fish Is Associated with a Polyserine Insert in MyoD1. Journal of Developmental Biology. 2023; 11(2):19. https://doi.org/10.3390/jdb11020019
Chicago/Turabian StyleWhite, Lewis J., Alexander J. Russell, Alastair R. Pizzey, Kanchon K. Dasmahapatra, and Mary E. Pownall. 2023. "The Presence of Two MyoD Genes in a Subset of Acanthopterygii Fish Is Associated with a Polyserine Insert in MyoD1" Journal of Developmental Biology 11, no. 2: 19. https://doi.org/10.3390/jdb11020019
APA StyleWhite, L. J., Russell, A. J., Pizzey, A. R., Dasmahapatra, K. K., & Pownall, M. E. (2023). The Presence of Two MyoD Genes in a Subset of Acanthopterygii Fish Is Associated with a Polyserine Insert in MyoD1. Journal of Developmental Biology, 11(2), 19. https://doi.org/10.3390/jdb11020019





