Polarimetric Target Decompositions and Light Gradient Boosting Machine for Crop Classification: A Comparative Evaluation
Abstract
1. Introduction
- (1)
- The performances of incoherent polarimetric target decompositions (eigenvector-based versus model-based) for discriminating crops using multi-temporal C-band PolSAR data were compared.
- (2)
- For the first time, LightGBM was used for crop classification using the polarimetric features of multi-temporal C-band PolSAR data.
- (3)
- The performance of polarimetric target decomposed parameters for crop classification was compared with the original polarimetric features (linear backscatter coefficients, and T and C matrices).
2. Study Site and Dataset
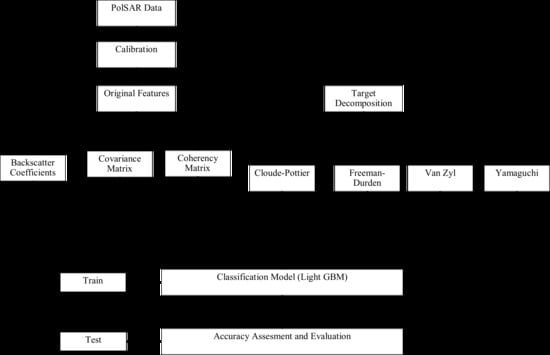
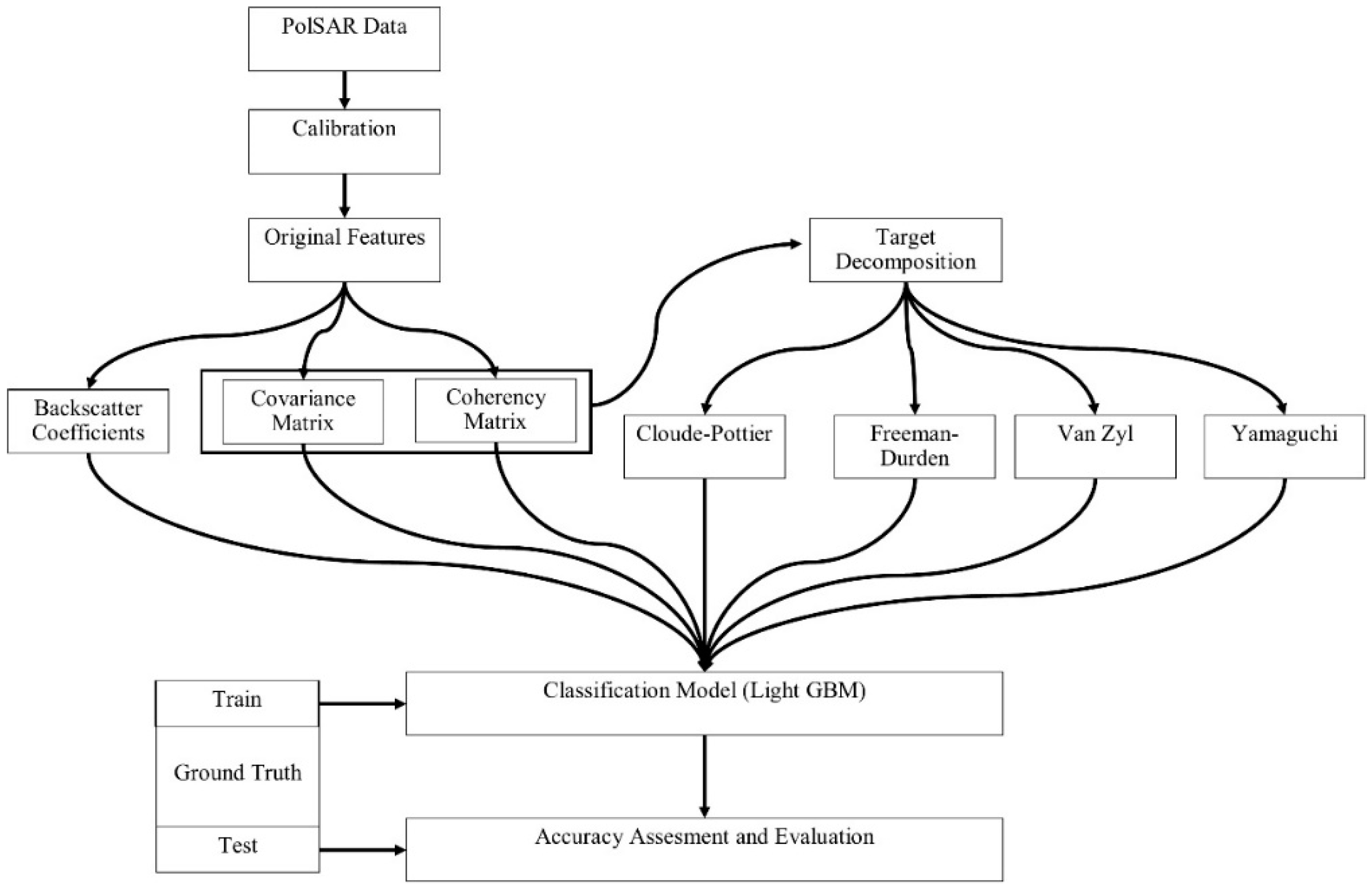
3. Methodology
3.1. Pre-Processing
3.2. Polarimetric Target Decomposition
3.3. Light Gradient Boosting Machine
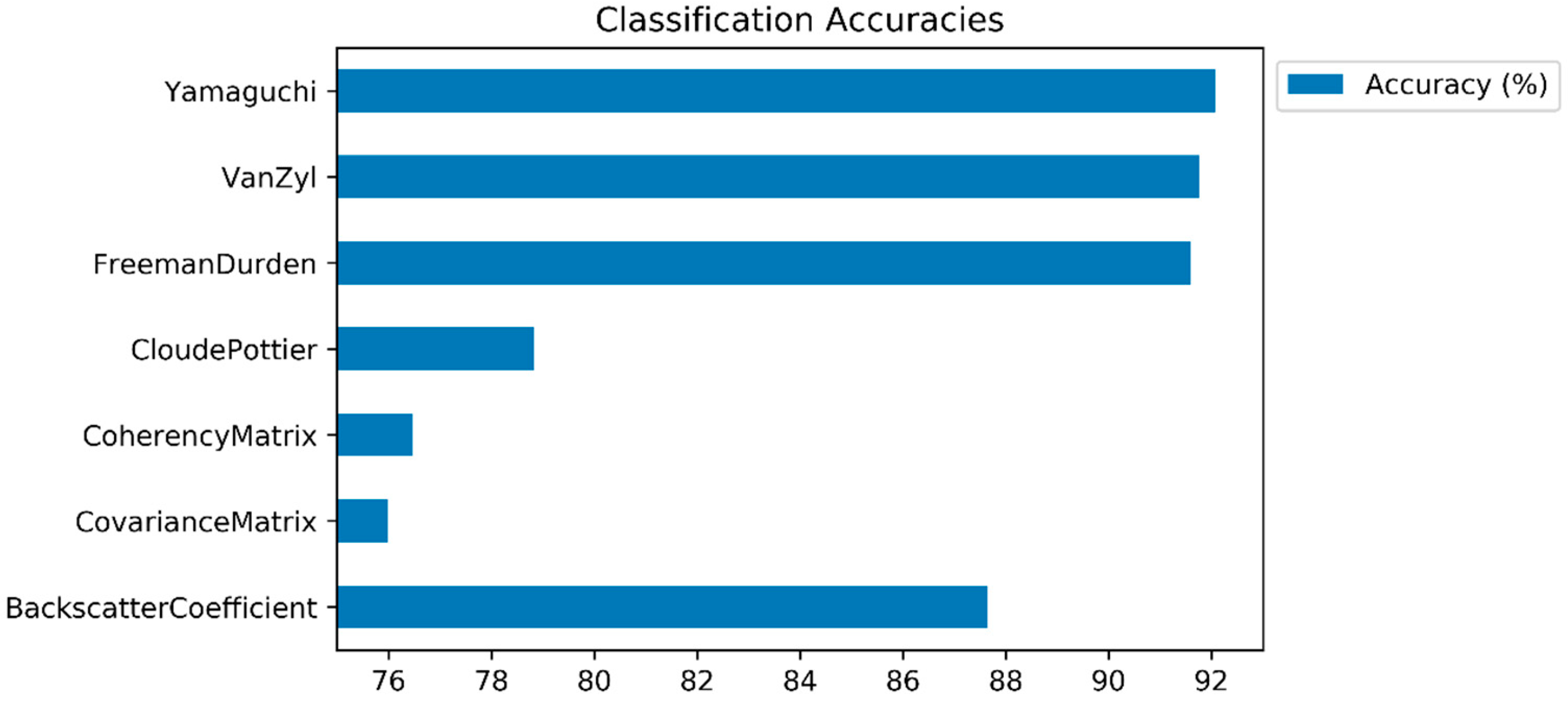
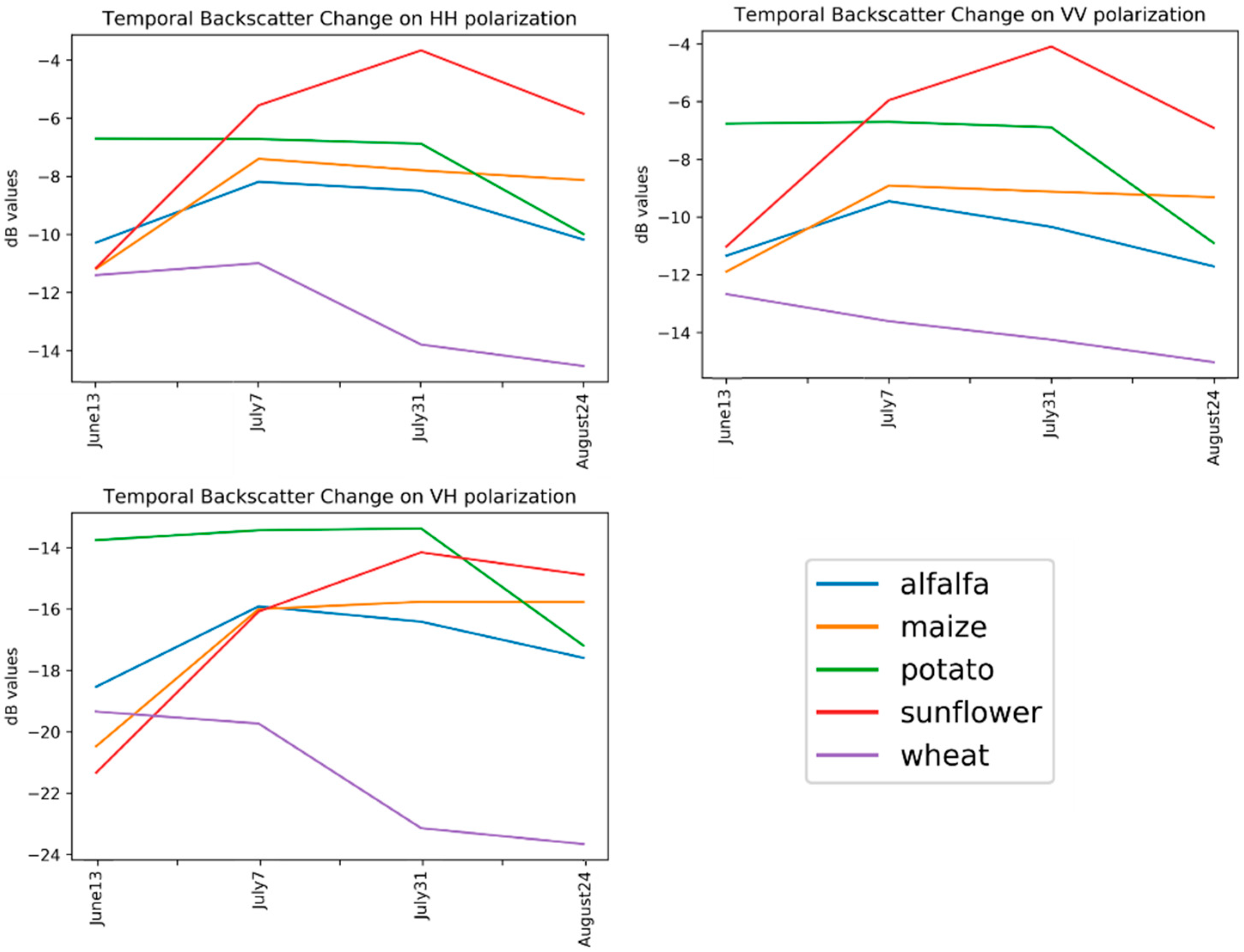
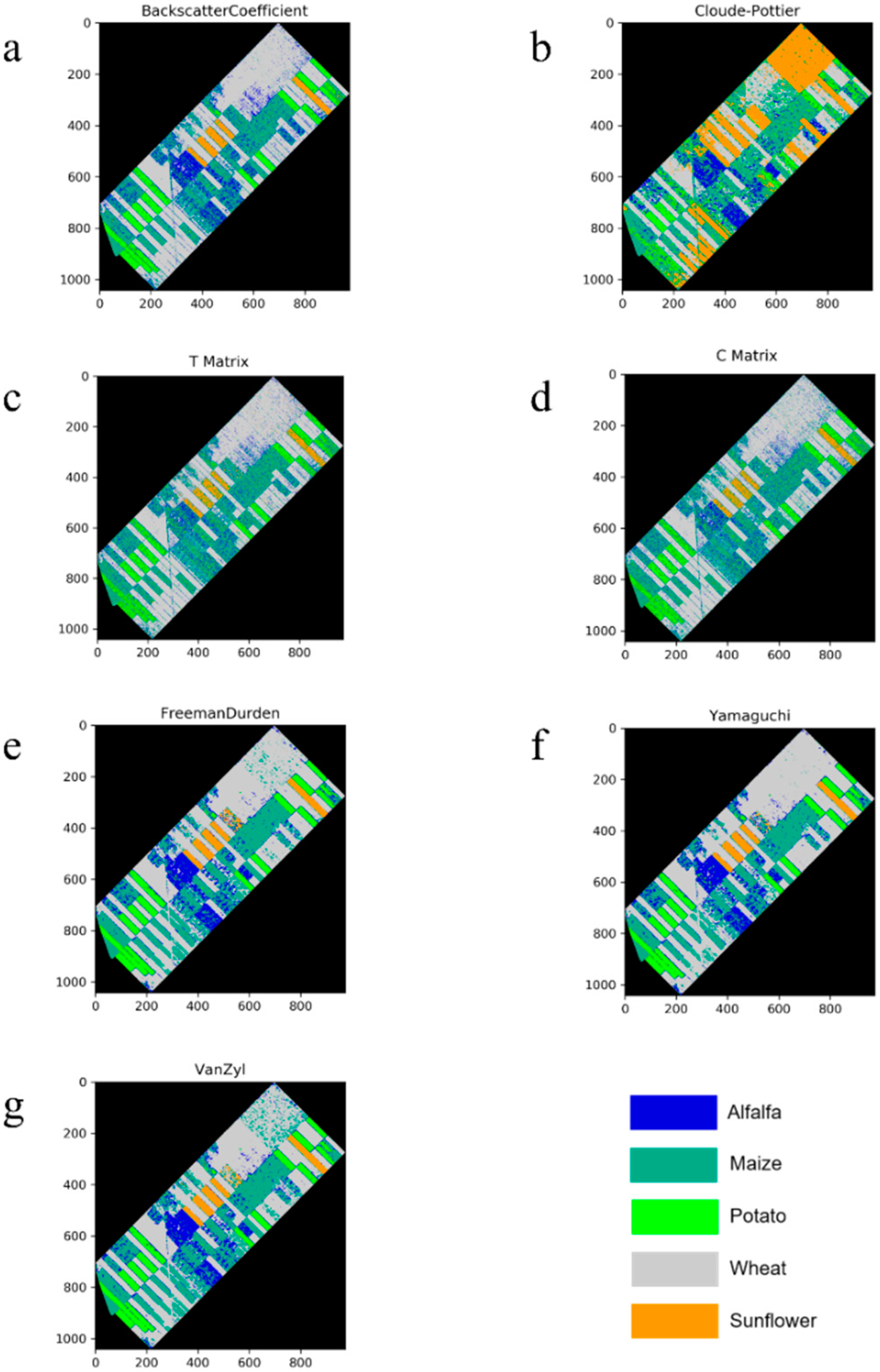
4. Experimental Results and Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- McNairn, H.; Shang, J. A Review of Multitemporal Synthetic Aperture Radar (SAR) for Crop Monitoring. In Multitemporal Remote Sensing: Methods and Applications; Ban, Y., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 317–340. [Google Scholar]
- Larrañaga, A.; Álvarez-Mozos, J. On the Added Value of Quad-Pol Data in a Multi-Temporal Crop Classification Framework Based on RADARSAT-2 Imagery. Remote Sens. 2016, 8, 335. [Google Scholar] [CrossRef]
- McNairn, H.; Brisco, B. The application of C-band polarimetric SAR for agriculture: A review. Can. J. Remote Sens. 2004, 30, 525–542. [Google Scholar] [CrossRef]
- Jiao, X.; Kovacs, J.M.; Shang, J.; McNairn, H.; Walters, D.; Ma, B.; Geng, X. Object-oriented crop mapping and monitoring using multi-temporal polarimetric RADARSAT-2 data. ISPRS J. Photogramm. Remote Sens. 2014, 96, 38–46. [Google Scholar] [CrossRef]
- Huang, X.; Wang, J.; Shang, J.; Liao, C.; Liu, J. Application of polarization signature to land cover scattering mechanism analysis and classification using multi-temporal C-band polarimetric RADARSAT-2 imagery. Remote Sens. Environ. 2017, 193, 11–28. [Google Scholar] [CrossRef]
- Skriver, H.; Mattia, F.; Satalino, G.; Balenzano, A.; Pauwels, V.R.N.; Verhoest, N.E.C.; Davidson, M. Crop Classification Using Short-Revisit Multitemporal SAR Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 4, 423–431. [Google Scholar] [CrossRef]
- Lee, J.-S.; Pottier, E. Polarimetric Radar Imaging: From Basics to Applications; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Schmullius, C.; Thiel, C.; Pathe, C.; Santoro, M. Radar Time Series for Land Cover and Forest Mapping. In Remote Sensing Time Series: Revealing Land Surface Dynamics; Kuenzer, C., Dech, S., Wagner, W., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 323–356. [Google Scholar]
- Tamiminia, H.; Homayouni, S.; McNairn, H.; Safari, A. A particle swarm optimized kernel-based clustering method for crop mapping from multi-temporal polarimetric L-band SAR observations. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 201–212. [Google Scholar] [CrossRef]
- Heine, I.; Jagdhuber, T.; Itzerott, S. Classification and Monitoring of Reed Belts Using Dual-Polarimetric TerraSAR-X Time Series. Remote Sens. 2016, 8, 552. [Google Scholar] [CrossRef]
- Cloude, S.R.; Pottier, E. A review of target decomposition theorems in radar polarimetry. IEEE Trans. Geosci. Remote Sens. 1996, 34, 498–518. [Google Scholar] [CrossRef]
- Freeman, A.; Durden, S.L. A three-component scattering model for polarimetric SAR data. IEEE Trans. Geosci. Remote Sens. 1998, 36, 963–973. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Moriyama, T.; Ishido, M.; Yamada, H. Four-component scattering model for polarimetric SAR image decomposition. IEEE Trans. Geosci. Remote Sens. 2005, 43, 1699–1706. [Google Scholar] [CrossRef]
- Marino, A.; Cloude, S.R.; Lopez-Sanchez, J.M. A New Polarimetric Change Detector in Radar Imagery. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2986–3000. [Google Scholar] [CrossRef]
- Qi, Z.; Yeh, A.G.-O.; Li, X.; Lin, Z. A novel algorithm for land use and land cover classification using RADARSAT-2 polarimetric SAR data. Remote Sens. Environ. 2012, 118, 21–39. [Google Scholar] [CrossRef]
- Samat, A.; Du, P.; Baig, M.H.A.; Chakravarty, S.; Cheng, L. Ensemble learning with multiple classifiers and polarimetric features for polarized SAR image classification. Photogramm. Eng. Remote Sens. 2014, 80, 239–251. [Google Scholar] [CrossRef]
- Du, P.; Samat, A.; Waske, B.; Liu, S.; Li, Z. Random Forest and Rotation Forest for fully polarized SAR image classification using polarimetric and spatial features. ISPRS J. Photogramm. Remote Sens. 2015, 105, 38–53. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, J.; Shang, J.; Wang, P. Assessment of multi-temporal RADARSAT-2 polarimetric SAR data for crop classification in an urban/rural fringe area. In Proceedings of the 2013 Second International Conference on Agro-Geoinformatics (Agro-Geoinformatics), Fairfax, VA, USA, 12–16 August 2013; pp. 314–319. [Google Scholar]
- Liu, C.; Shang, J.; Vachon, P.W.; McNairn, H. Multiyear Crop Monitoring Using Polarimetric RADARSAT-2 Data. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2227–2240. [Google Scholar] [CrossRef]
- Shuai, G.; Zhang, J.; Basso, B.; Pan, Y.; Zhu, X.; Zhu, S.; Liu, H. Multi-temporal RADARSAT-2 polarimetric SAR for maize mapping supported by segmentations from high-resolution optical image. Int. J. Appl. Earth Obs. Geoinf. 2019, 74, 1–15. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Colkesen, I. An assessment of the effectiveness of a rotation forest ensemble for land-use and land-cover mapping. Int. J. Remote Sens. 2013, 34, 4224–4241. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Rainforth, T.; Wood, F. Canonical correlation forests. arXiv, 2015; arXiv:1507.05444. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 3146–3154. [Google Scholar]
- Xia, J.; Yokoya, N.; Iwasaki, A. Hyperspectral Image Classification With Canonical Correlation Forests. IEEE Trans. Geosci. Remote Sens. 2017, 55, 421–431. [Google Scholar] [CrossRef]
- Colkesen, I.; Kavzoglu, T. Ensemble-based canonical correlation forest (CCF) for land use and land cover classification using sentinel-2 and Landsat OLI imagery. Remote Sens. Lett. 2017, 8, 1082–1091. [Google Scholar] [CrossRef]
- Dong, H.; Xu, X.; Wang, L.; Pu, F. Gaofen-3 PolSAR Image Classification via XGBoost and Polarimetric Spatial Information. Sensors 2018, 18, 611. [Google Scholar] [CrossRef] [PubMed]
- Georganos, S.; Grippa, T.; Vanhuysse, S.; Lennert, M.; Shimoni, M.; Wolff, E. Very High Resolution Object-Based Land Use–Land Cover Urban Classification Using Extreme Gradient Boosting. IEEE Geosci. Remote Sens. Lett. 2018, 15, 607–611. [Google Scholar] [CrossRef]
- Zheng, H.; Cui, Z.; Zhang, X. Identifying Modes of Driving Railway Trains from GPS Trajectory Data: An Ensemble Classifier-Based Approach. ISPRS Int. J. Geo-Inf. 2018, 7, 308. [Google Scholar] [CrossRef]
- Liu, L.; Ji, M.; Buchroithner, M. Combining Partial Least Squares and the Gradient-Boosting Method for Soil Property Retrieval Using Visible Near-Infrared Shortwave Infrared Spectra. Remote Sens. 2017, 9, 1299. [Google Scholar] [CrossRef]
- Meier, U. Growth Stages of Mono-and Dicotyledonous Plants; Federal Biological Research Centre for Agriculture and Forestry: Kleinmachnow, Germany, 2001; p. 158. [Google Scholar]
- ESA Sentinel Application Platform (SNAP) V.6.0. Available online: http://step.esa.int/main/toolboxes/snap/ (accessed on 4 December 2018).
- Moreira, A.; Prats-Iraola, P.; Younis, M.; Krieger, G.; Hajnsek, I.; Papathanassiou, K.P. A tutorial on synthetic aperture radar. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–43. [Google Scholar] [CrossRef]
- Chen, S.; Li, Y.; Wang, X.; Xiao, S.; Sato, M. Modeling and Interpretation of Scattering Mechanisms in Polarimetric Synthetic Aperture Radar: Advances and perspectives. IEEE Signal Process. Mag. 2014, 31, 79–89. [Google Scholar] [CrossRef]
- Lim, Y.X.; Burgin, M.S.; van Zyl, J.J. An Optimal Nonnegative Eigenvalue Decomposition for the Freeman and Durden Three-Component Scattering Model. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2167–2176. [Google Scholar] [CrossRef]
- Park, S.-E. The Effect of Topography on Target Decomposition of Polarimetric SAR Data. Remote Sens. 2015, 7, 4997–5011. [Google Scholar] [CrossRef]
- Van Zyl, J.J.; Arii, M.; Kim, Y. Model-Based Decomposition of Polarimetric SAR Covariance Matrices Constrained for Nonnegative Eigenvalues. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3452–3459. [Google Scholar] [CrossRef]
- Cloude, S.R.; Pottier, E. An entropy based classification scheme for land applications of polarimetric SAR. IEEE Trans. Geosci. Remote Sens. 1997, 35, 68–78. [Google Scholar] [CrossRef]
- Mahdianpari, M.; Salehi, B.; Mohammadimanesh, F.; Brisco, B.; Mahdavi, S.; Amani, M.; Granger, J.E. Fisher Linear Discriminant Analysis of coherency matrix for wetland classification using PolSAR imagery. Remote Sens. Environ. 2018, 206, 300–317. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Sato, A.; Boerner, W.; Sato, R.; Yamada, H. Four-Component Scattering Power Decomposition with Rotation of Coherency Matrix. IEEE Trans. Geosci. Remote Sens. 2011, 49, 2251–2258. [Google Scholar] [CrossRef]
- Waske, B.; Braun, M. Classifier ensembles for land cover mapping using multitemporal SAR imagery. ISPRS J. Photogramm. Remote Sens. 2009, 64, 450–457. [Google Scholar] [CrossRef]
- Machine Learning Challenge Winning Solutions. Available online: https://github.com/Microsoft/LightGBM/blob/master/examples/README.md#machine-learning-challenge-winning-solutions (accessed on 4 December 2018).
- LightGBM Python Package. Available online: https://pypi.org/project/lightgbm/ (accessed on 4 December 2018).
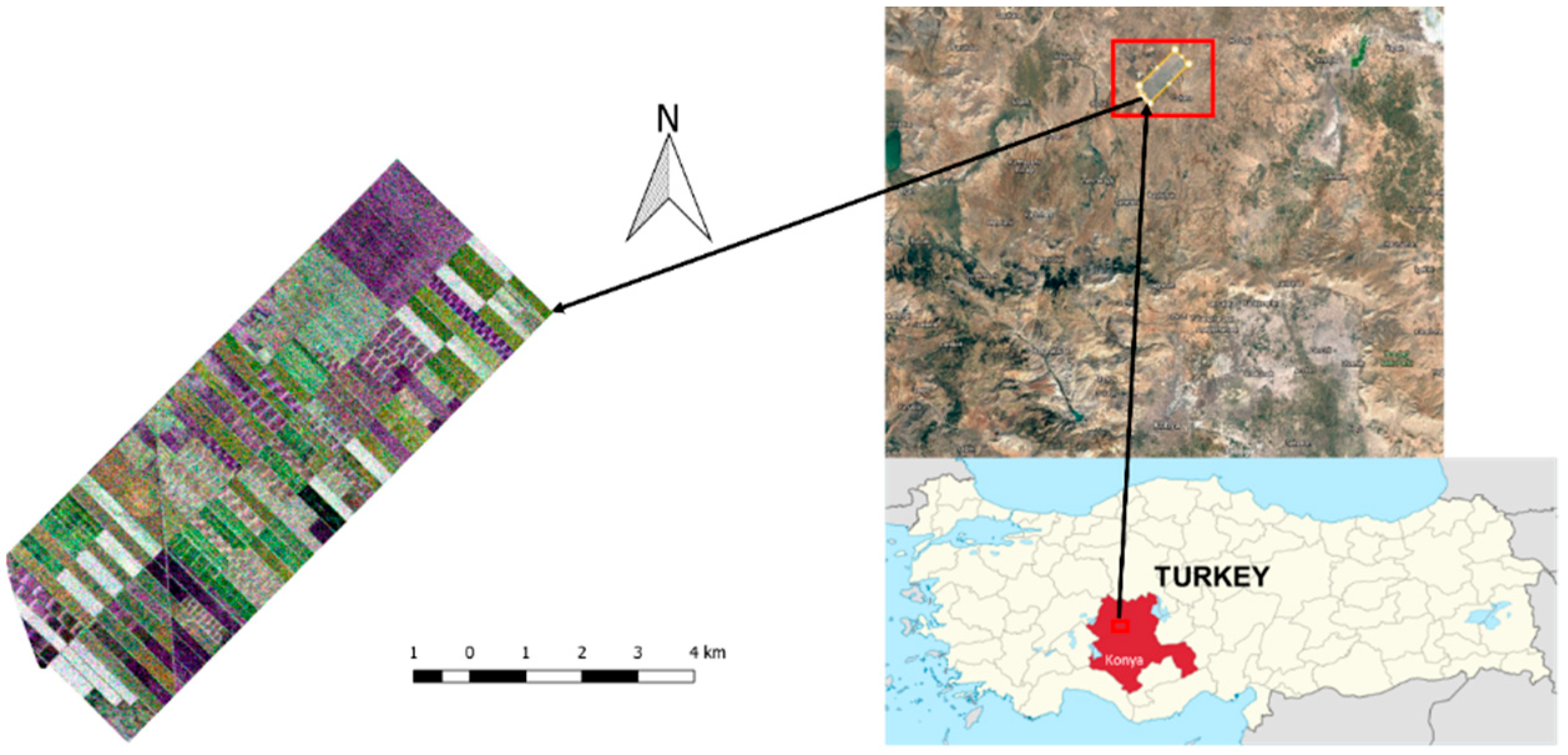

| Sensor Type | RADARSAT-2 |
|---|---|
| Wavelength | C band—5.6 cm |
| Resolution | 4.7 m × 5.1 m (range × azimuth) |
| Incidence angle | 400 |
| Pass direction | Descending |
| Acquisition type | Fine quad pol |
| Polarization | Full polarimetric |
| Nominal Scene Size | 25 × 25 km |
| Product type | Single look complex |
| Class | Training Set (in Pixel) | Testing Set (in Pixel) |
|---|---|---|
| Alfalfa | 1918 | 3542 |
| Maize | 5581 | 14,217 |
| Potato | 2274 | 10,604 |
| Sunflower | 3524 | 6338 |
| Wheat | 3729 | 8915 |
| Crop Growth Stages | Image Acquisition Dates |
|---|---|
| leaf development | 13 June 2016 |
| stem elongation | 7 July 2016 |
| heading | 31 July 2016 |
| flowering | 24 August 2016 |
| Decomposition Method | Parameters |
|---|---|
| Cloude-Pottier (H/A/α) [39] | Entropy, Anisotropy, Alpha Angle |
| Freeman-Durden [40] | Surface, Double-Bounce and Volume Scattering |
| Van Zyl [38] | Surface, Double-Bounce and Volume Scattering |
| Yamaguchi [41] | Surface, Double-Bounce, Volume and Helix Scattering |
| Parameter | Value |
|---|---|
| Boosting type | GOSS |
| Maximum depth | 5 |
| Number of leaves | 100 |
| Learning rate | 0.1 |
| Objective | multiclass |
| Feature fraction | 1.0 |
| L1 regularization | 0 |
| L2 regularization | 0 |
| Min data in leaf | 20 |
| Number of boosting round (iterations) | 100 |
| Alfalfa | Maize | Potato | Sunflower | Wheat | |
|---|---|---|---|---|---|
| Backscatter Coefficient | |||||
| Alfalfa | 0.3 | 0.61 | 0.02 | 0.01 | 0.07 |
| Maize | 0.05 | 0.91 | 0.02 | 0.02 | 0.0 |
| Potato | 0.03 | 0.05 | 0.91 | 0.01 | 0.0 |
| Sunflower | 0.0 | 0.04 | 0.01 | 0.95 | 0.0 |
| Wheat | 0.01 | 0.0 | 0.0 | 0.0 | 0.99 |
| Cloude-Pottier Decomposition | |||||
| Alfalfa | 0.3 | 0.61 | 0.02 | 0.01 | 0.07 |
| Maize | 0.05 | 0.91 | 0.02 | 0.02 | 0.0 |
| Potato | 0.03 | 0.05 | 0.91 | 0.01 | 0.0 |
| Sunflower | 0.0 | 0.04 | 0.01 | 0.95 | 0.0 |
| Wheat | 0.01 | 0.0 | 0.0 | 0.0 | 0.99 |
| T Matrix | |||||
| Alfalfa | 0.3 | 0.61 | 0.02 | 0.01 | 0.07 |
| Maize | 0.05 | 0.91 | 0.02 | 0.02 | 0.0 |
| Potato | 0.03 | 0.05 | 0.91 | 0.01 | 0.0 |
| Sunflower | 0.0 | 0.04 | 0.01 | 0.95 | 0.0 |
| Wheat | 0.01 | 0.0 | 0.0 | 0.0 | 0.99 |
| C Matrix | |||||
| Alfalfa | 0.3 | 0.61 | 0.02 | 0.01 | 0.07 |
| Maize | 0.05 | 0.91 | 0.02 | 0.02 | 0.0 |
| Potato | 0.03 | 0.05 | 0.91 | 0.01 | 0.0 |
| Sunflower | 0.0 | 0.04 | 0.01 | 0.95 | 0.0 |
| Wheat | 0.01 | 0.0 | 0.0 | 0.0 | 0.99 |
| Freeman-Durden Decomposition | |||||
| Alfalfa | 0.3 | 0.61 | 0.02 | 0.01 | 0.07 |
| Maize | 0.05 | 0.91 | 0.02 | 0.02 | 0.0 |
| Potato | 0.03 | 0.05 | 0.91 | 0.01 | 0.0 |
| Sunflower | 0.0 | 0.04 | 0.01 | 0.95 | 0.0 |
| Wheat | 0.01 | 0.0 | 0.0 | 0.0 | 0.99 |
| Van Zyl Decomposition | |||||
| Alfalfa | 0.3 | 0.61 | 0.02 | 0.01 | 0.07 |
| Maize | 0.05 | 0.91 | 0.02 | 0.02 | 0.0 |
| Potato | 0.03 | 0.05 | 0.91 | 0.01 | 0.0 |
| Sunflower | 0.0 | 0.04 | 0.01 | 0.95 | 0.0 |
| Wheat | 0.01 | 0.0 | 0.0 | 0.0 | 0.99 |
| Yamaguchi Decomposition | |||||
| Alfalfa | 0.3 | 0.61 | 0.02 | 0.01 | 0.07 |
| Maize | 0.05 | 0.91 | 0.02 | 0.02 | 0.0 |
| Potato | 0.03 | 0.05 | 0.91 | 0.01 | 0.0 |
| Sunflower | 0.0 | 0.04 | 0.01 | 0.95 | 0.0 |
| Wheat | 0.01 | 0.0 | 0.0 | 0.0 | 0.99 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ustuner, M.; Balik Sanli, F. Polarimetric Target Decompositions and Light Gradient Boosting Machine for Crop Classification: A Comparative Evaluation. ISPRS Int. J. Geo-Inf. 2019, 8, 97. https://doi.org/10.3390/ijgi8020097
Ustuner M, Balik Sanli F. Polarimetric Target Decompositions and Light Gradient Boosting Machine for Crop Classification: A Comparative Evaluation. ISPRS International Journal of Geo-Information. 2019; 8(2):97. https://doi.org/10.3390/ijgi8020097
Chicago/Turabian StyleUstuner, Mustafa, and Fusun Balik Sanli. 2019. "Polarimetric Target Decompositions and Light Gradient Boosting Machine for Crop Classification: A Comparative Evaluation" ISPRS International Journal of Geo-Information 8, no. 2: 97. https://doi.org/10.3390/ijgi8020097
APA StyleUstuner, M., & Balik Sanli, F. (2019). Polarimetric Target Decompositions and Light Gradient Boosting Machine for Crop Classification: A Comparative Evaluation. ISPRS International Journal of Geo-Information, 8(2), 97. https://doi.org/10.3390/ijgi8020097