Dataset Reduction Techniques to Speed Up SVD Analyses on Big Geo-Datasets
Abstract
1. Introduction
2. Materials and Methods
2.1. Matrix Decomposition
2.2. Data Characteristics
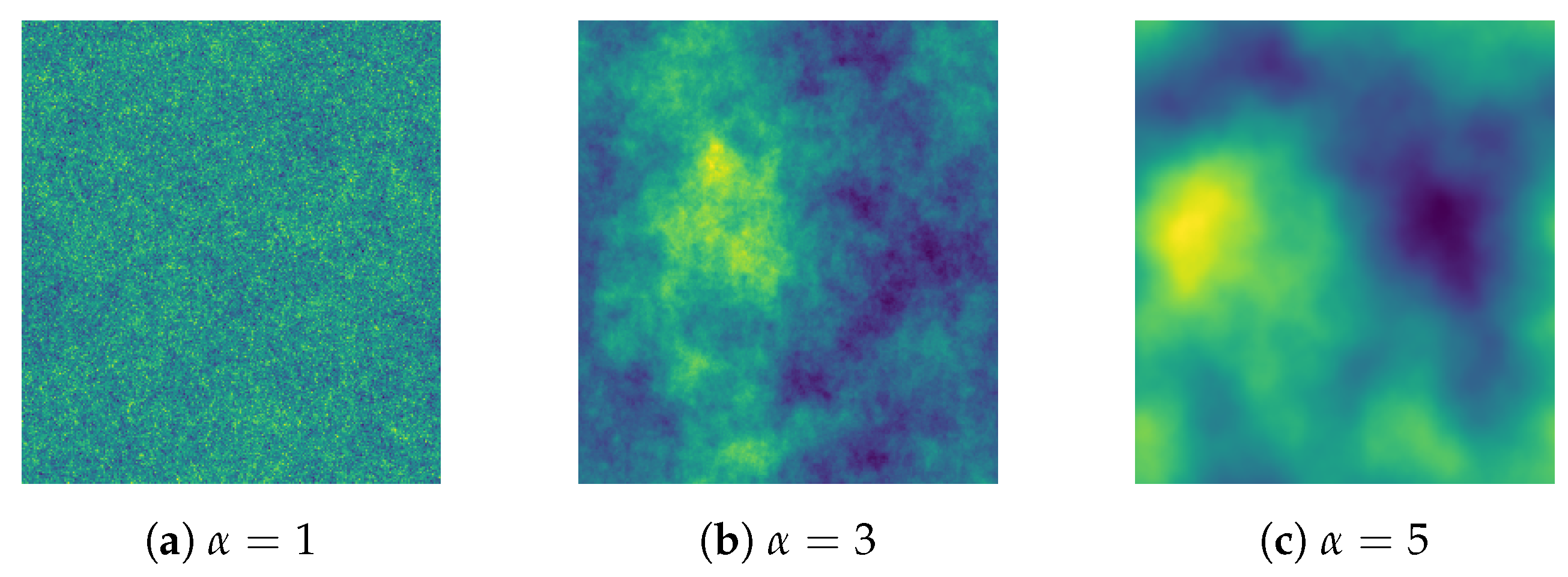
2.3. Simulated Spatio-Temporal Fields
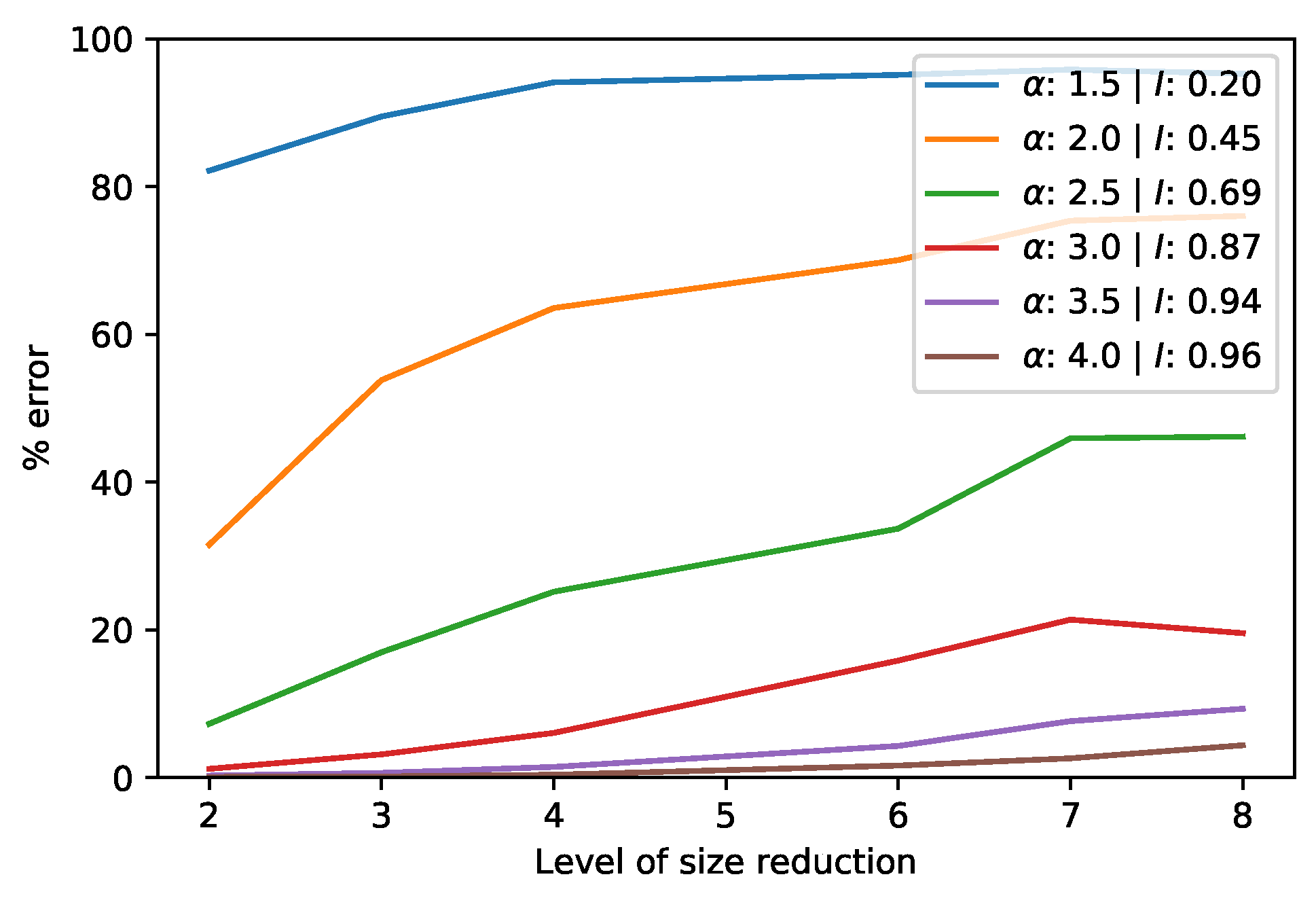
2.4. Measures of Autocorrelation
3. Results
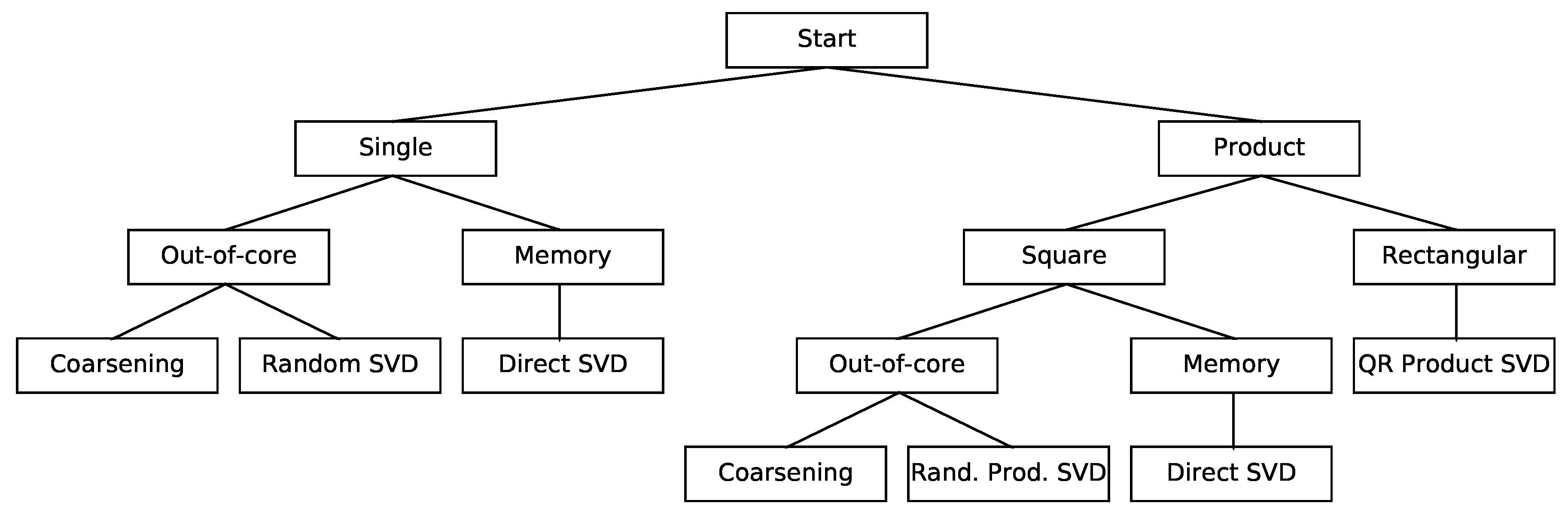
3.1. SVD of a Single Matrix
3.1.1. Approximate SVD of a Single Matrix via Coarsening
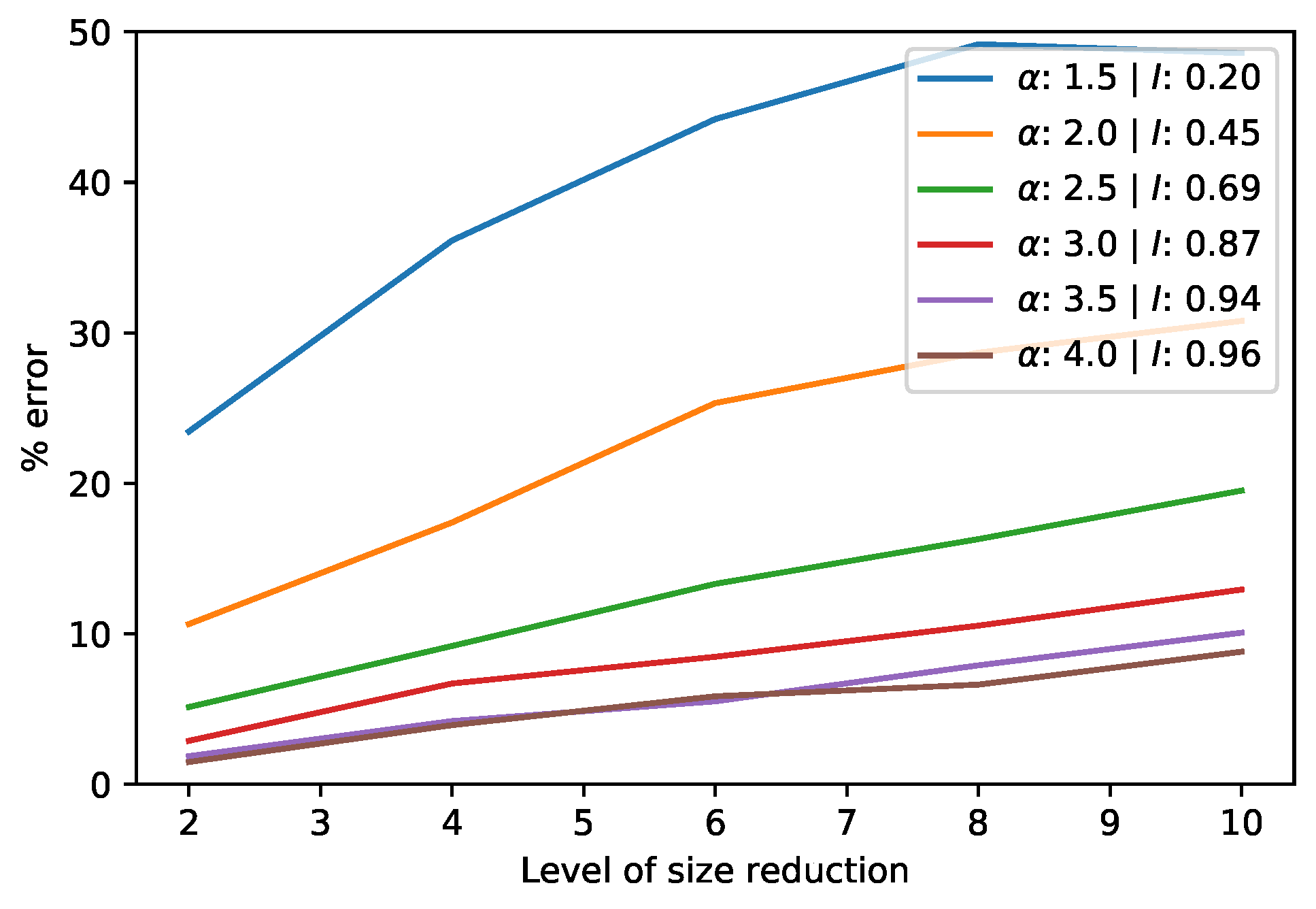
3.1.2. Approximate SVD of a Single Matrix via Dimensionality Reduction
3.1.3. Case Study of an SVD of a Single Matrix
3.2. Product SVD of Rectangular Matrices
3.2.1. Exact Product SVD of Rectangular Matrices via QR Decomposition
3.2.2. Case Study of a Product SVD of Rectangular Matrices
3.3. Product SVD of Square Matrices
3.3.1. Approximate Product SVD of Square Matrices via Coarsening
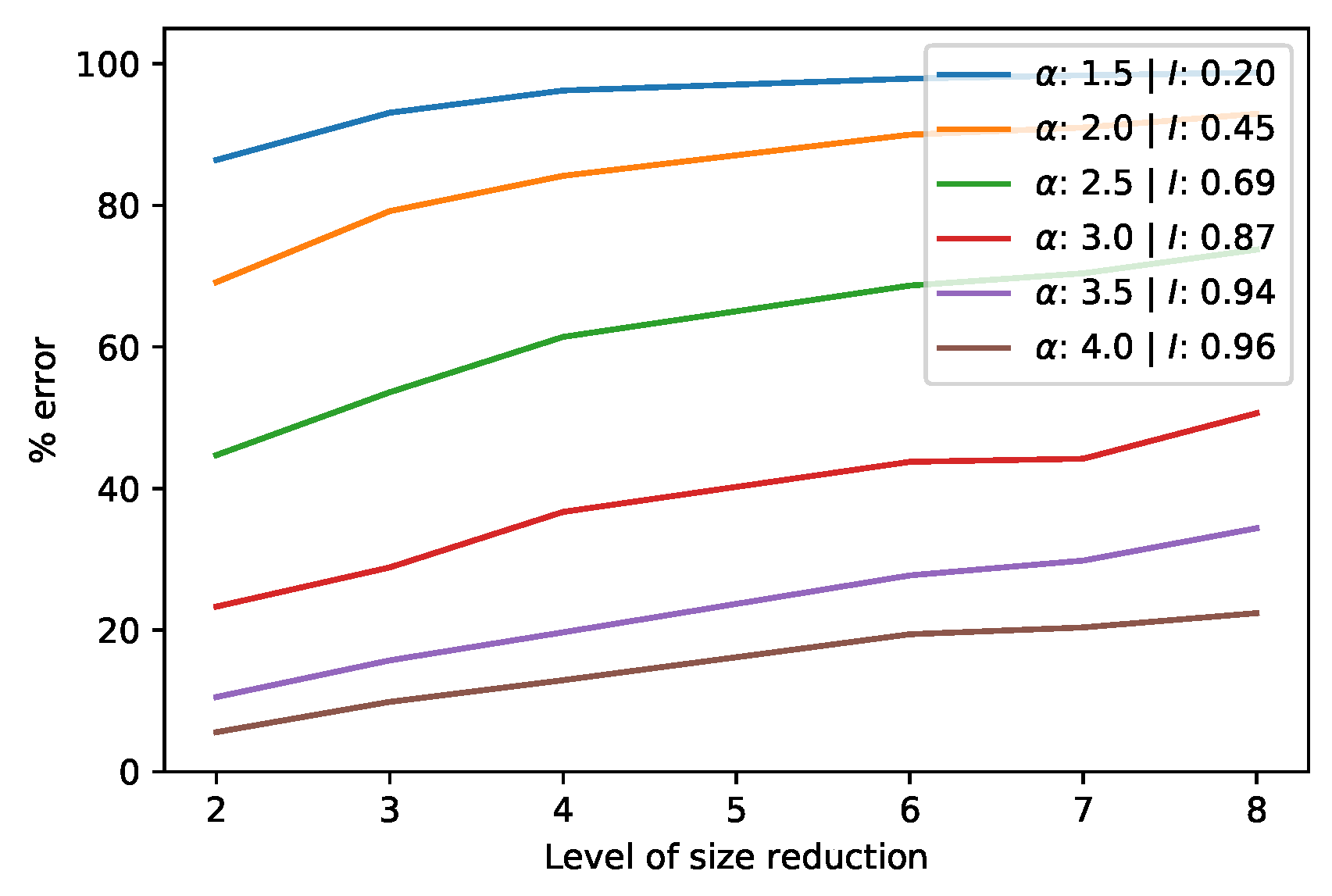
3.3.2. Approximate Product SVD of Square Matrices via Dimensionality Reduction
3.3.3. Case Study of a Product SVD of Square Matrices
4. Discussion
4.1. Further Work and Caveats
4.2. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SVD | Singular value decomposition |
| PLS | Partial least squares |
| MCA | Maximum covariance analysis |
| CCA | Canonical correlation analysis |
| MNF | Minimum noise fraction |
| PC | Principle component |
| EOF | Empirical orthogonal function |
| GRF | Gaussian random field |
| SI-x | Extended spring indices |
| ERA5 | European fifth generation reanalysis |
| JRA55 | Japanese 55-year reanalysis |
| HOSVD | Higher-order singular value decomposition |
References
- Golub, G.H.; Reinsch, C. Singular value decomposition and least squares solutions. Numer. Math. 1970, 14, 403–420. [Google Scholar] [CrossRef]
- Rajwade, A.; Rangarajan, A.; Banerjee, A. Image Denoising Using the Higher Order Singular Value Decomposition. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Khoshbin, F.; Bonakdari, H.; Ashraf Talesh, S.H.; Ebtehaj, I.; Zaji, A.H.; Azimi, H. Adaptive neuro-fuzzy inference system multi-objective optimization using the genetic algorithm/singular value decomposition method for modelling the discharge coefficient in rectangular sharp-crested side weirs. Eng. Optim. 2016, 48, 933–948. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Indahl, U.G.; Ødegård, J. Variable selection models for genomic selection using whole-genome sequence data and singular value decomposition. Genet. Sel. Evol. 2017, 49, 94. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Verdiguier, E.; Laparra, V.; Marí, J.M.; Chova, L.G.; Camps-Valls, G. Advanced Feature Extraction for Earth Observation Data Processing. In Comprehensive Remote Sensing, Volume 2: Data Processing and Analysis Methodology; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Izquierdo-Verdiguier, E.; Gómez-Chova, L.; Bruzzone, L.; Camps-Valls, G. Semisupervised kernel feature extraction for remote sensing image analysis. IEEE Trans. Geosci. Remote Sens. 2014, 52, 5567–5578. [Google Scholar] [CrossRef]
- Hansen, P.; Schjoerring, J. Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Munoz-Mari, J.; Gomez-Chova, L.; Amoros, J.; Izquierdo, E.; Camps-Valls, G. Multiset Kernel CCA for multitemporal image classification. In Proceedings of the MultiTemp 2013: 7th International Workshop on the Analysis of Multi-temporal Remote Sensing Images, Banff, AB, Canada, 25–27 June 2013; pp. 1–4. [Google Scholar]
- Nielsen, A.A. The regularized iteratively reweighted MAD method for change detection in multi-and hyperspectral data. IEEE Trans. Image Process. 2007, 16, 463–478. [Google Scholar] [CrossRef]
- Li, J.; Carlson, B.E.; Lacis, A.A. Application of spectral analysis techniques in the intercomparison of aerosol data. Part II: Using maximum covariance analysis to effectively compare spatiotemporal variability of satellite and AERONET measured aerosol optical depth. J. Geophys. Res. Atmos. 2014, 119, 153–166. [Google Scholar] [CrossRef]
- Li, J.; Carlson, B.E.; Lacis, A.A. Application of spectral analysis techniques to the intercomparison of aerosol data. Part IV: Synthesized analysis of multisensor satellite and ground-based AOD measurements using combined maximum covariance analysis. Atmos. Meas. Tech. 2014, 7, 2531–2549. [Google Scholar] [CrossRef]
- Eshel, G. Spatiotemporal Data Analysis; Princeton University Press: Princeton, NJ, USA, 2011. [Google Scholar]
- Von Storch, H.; Zwiers, F.W. Statistical Analysis in Climate Research; Cambridge University Press: Cambridge, UK, 1999. [Google Scholar]
- Björck, Å.; Golub, G.H. Numerical methods for computing angles between linear subspaces. Math. Comput. 1973, 27, 579–594. [Google Scholar] [CrossRef]
- Chan, T.F. An improved algorithm for computing the svd. ACM Trans. Math. Softw. 1982, 8, 72–83. [Google Scholar] [CrossRef]
- Bogaardt, L. Dataset Reduction Techniques to Speed Up Svd Analyses. 2018. Available online: https://github.com/phenology/ (accessed on 30 December 2018).
- Demirel, H.; Ozcinar, C.; Anbarjafari, G. Satellite Image Contrast Enhancement Using Discrete Wavelet Transform and Singular Value Decomposition. IEEE Geosci. Remote Sens. Lett. 2010, 7, 333–337. [Google Scholar] [CrossRef]
- Hannachi, A.; Jolliffe, I.T.; Stephenson, D.B. Empirical orthogonal functions and related techniques in atmospheric science: A review. Int. J. Climatol. 2007, 27, 1119–1152. [Google Scholar] [CrossRef]
- Martinsson, P.G. Randomized methods for matrix computations and analysis of high dimensional data. arXiv, 2016; arXiv:1607.01649. [Google Scholar]
- Eckart, C.; Young, G. The approximation of one matrix by another of lower rank. Psychometrika 1936, 1, 211–218. [Google Scholar] [CrossRef]
- Krige, D. A statistical approach to some basic mine valuation problems on the Witwatersrand. J. S. Afr. Inst. Min. Metall. 1951, 52, 119–139. [Google Scholar]
- Moran, P.A.P. Notes on continuous stochastic phenomena. Biometrika 1950, 37, 17–23. [Google Scholar] [CrossRef]
- Hubert, L.J.; Golledge, R.G.; Costanzo, C.M. Generalized procedures for evaluating spatial autocorrelation. Geogr. Anal. 1981, 13, 224–233. [Google Scholar] [CrossRef]
- Rey, S. PySAL. 2009–2013. Available online: http://pysal.readthedocs.io (accessed on 30 December 2018).
- Halko, N.; Martinsson, P.G.; Tropp, J.A. Finding structure with randomness: Probabilistic algorithms for constructing approximate matrix decompositions. SIAM Rev. 2011, 53, 217–288. [Google Scholar] [CrossRef]
- Li, H.; Kluger, Y.; Tygert, M. Randomized algorithms for distributed computation of principal component analysis and singular value decomposition. Adv. Comput. Math. 2018, 44, 1651. [Google Scholar] [CrossRef]
- Dee, D.P.; Uppala, S.M.; Simmons, A.J.; Berrisford, P.; Poli, P.; Kobayashi, S.; Andrae, U.; Balmaseda, M.A.; Balsamo, G.; Bauer, P.; et al. The ERA-Interim reanalysis: configuration and performance of the data assimilation system. Q. J. R. Meteorol. Soc. 2011, 137, 553–597. [Google Scholar] [CrossRef]
- Bretherton, C.S.; Smith, C.; Wallace, J.M. An intercomparison of methods for finding coupled patterns in climate data. J. Clim. 1992, 5, 541–560. [Google Scholar] [CrossRef]
- Tygert, M. Suggested during Personal Communication. 2017. Available online: http://tygert.com/ (accessed on 10 December 2018).
- Schwartz, M.D.; Ault, T.R.; Betancourt, J.L. Spring onset variations and trends in the continental united states: Past and regional assessment using temperature-based indices. Int. J. Climatol. 2013, 33, 2917–2922. [Google Scholar] [CrossRef]
- Izquierdo-Verdiguier, E.; Zurita-Milla, R.; Ault, T.R.; Schwartz, M.D. Using cloud computing to study trends and patterns in the extended spring indices. In Proceedings of the Third International Conference on Phenology, Kusadasi, Turkey, 5–8 October 2015; p. 51. [Google Scholar]
- Zurita-Milla, R.; Bogaardt, L.; Izquierdo-Verdiguier, E.; Gonçalves, R. Analyzing the cross-correlation between the extended spring indices and the AVHRR start of season phenometric. In Proceedings of the EGU General Assembly, Geophysical Research Abstracts, Vienna, Austria, 8–13 April 2018. [Google Scholar]
- Barnett, T.P.; Preisendorfer, R. Origins and levels of monthly and seasonal forecast skill for us surface air temperatures determined by canonical correlation analysis. Mon. Weather Rev. 1987, 115, 1825–1850. [Google Scholar] [CrossRef]
- Kobayashi, S.; Ota, Y.; Harada, Y.; Ebita, A.; Moriya, M.; Onoda, H.; Onogi, K.; Kamahori, H.; Kobayashi, C.; Endo, H.; et al. The JRA-55 reanalysis: general specifications and basic characteristics. J. Meteorol. Soc. Jpn. 2015, 93, 5–48. [Google Scholar] [CrossRef]
- Liu, Y.; Attema, J.; Moat, B.; Hazeleger, W. Synthesis and evaluation of historical meridional heat transport from midlatitudes towards the arctic. Clim. Dyn. 2018. submitted. [Google Scholar]
- Tucker, L.R. The extension of factor analysis to three-dimensional matrices. In Contributions to Mathematical Psychology; Gulliksen, H., Frederiksen, N., Eds.; Holt, Rinehart and Winston: Boston, MA, USA, 1964; pp. 110–127. [Google Scholar]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogaardt, L.; Goncalves, R.; Zurita-Milla, R.; Izquierdo-Verdiguier, E. Dataset Reduction Techniques to Speed Up SVD Analyses on Big Geo-Datasets. ISPRS Int. J. Geo-Inf. 2019, 8, 55. https://doi.org/10.3390/ijgi8020055
Bogaardt L, Goncalves R, Zurita-Milla R, Izquierdo-Verdiguier E. Dataset Reduction Techniques to Speed Up SVD Analyses on Big Geo-Datasets. ISPRS International Journal of Geo-Information. 2019; 8(2):55. https://doi.org/10.3390/ijgi8020055
Chicago/Turabian StyleBogaardt, Laurens, Romulo Goncalves, Raul Zurita-Milla, and Emma Izquierdo-Verdiguier. 2019. "Dataset Reduction Techniques to Speed Up SVD Analyses on Big Geo-Datasets" ISPRS International Journal of Geo-Information 8, no. 2: 55. https://doi.org/10.3390/ijgi8020055
APA StyleBogaardt, L., Goncalves, R., Zurita-Milla, R., & Izquierdo-Verdiguier, E. (2019). Dataset Reduction Techniques to Speed Up SVD Analyses on Big Geo-Datasets. ISPRS International Journal of Geo-Information, 8(2), 55. https://doi.org/10.3390/ijgi8020055




