Forest above Ground Biomass Inversion by Fusing GLAS with Optical Remote Sensing Data
Abstract
:1. Introduction
2. Study Area and Experimental Data
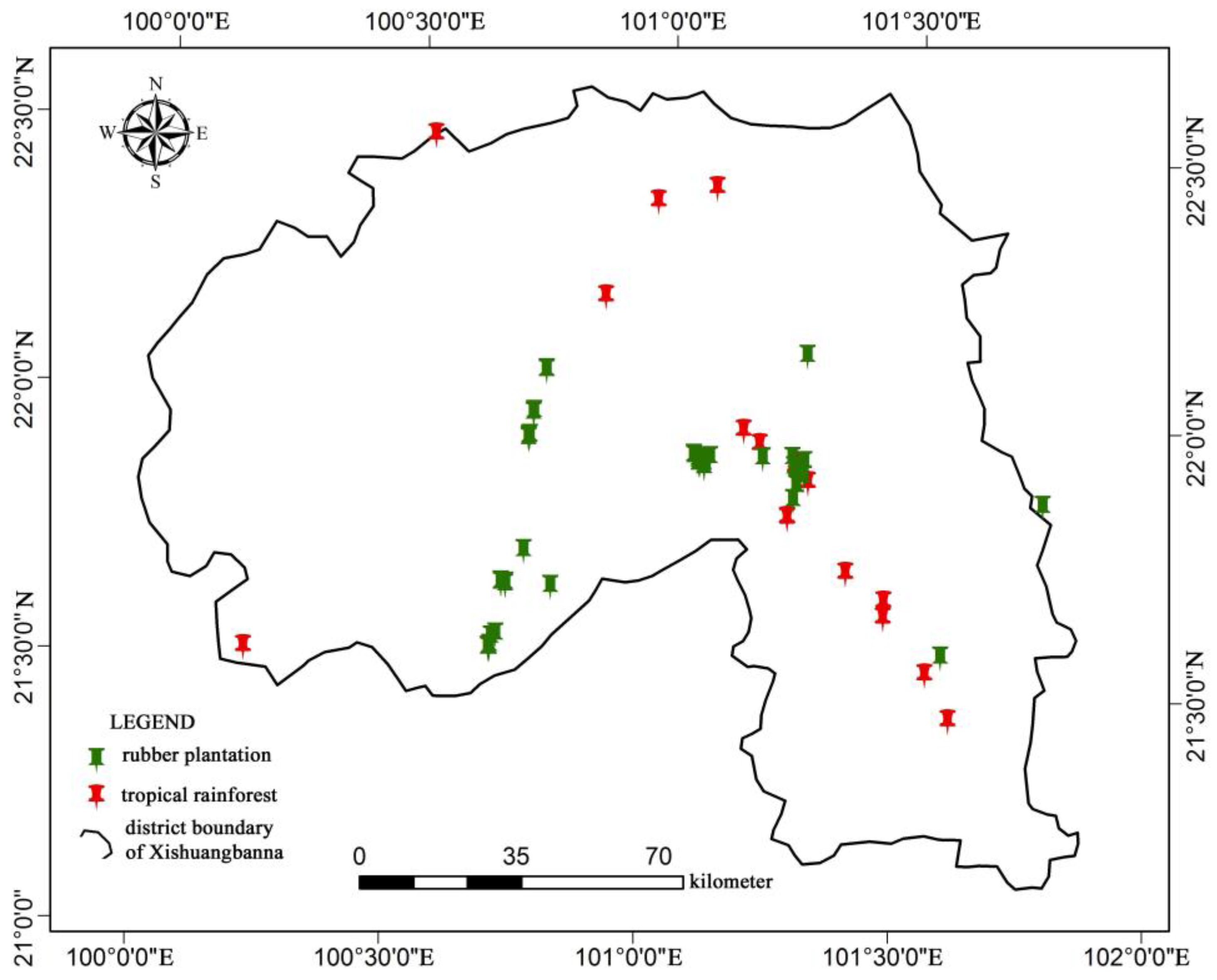
2.1. Study Area
2.2. GLAS Data
2.3. MODIS BRDF Data
2.4. Landsat TM Data
2.5. ASTER GDEM Data
2.6. Ground Measured Data
3. Methods
3.1. GLAS Data Processing
3.1.1. Pre-Processing
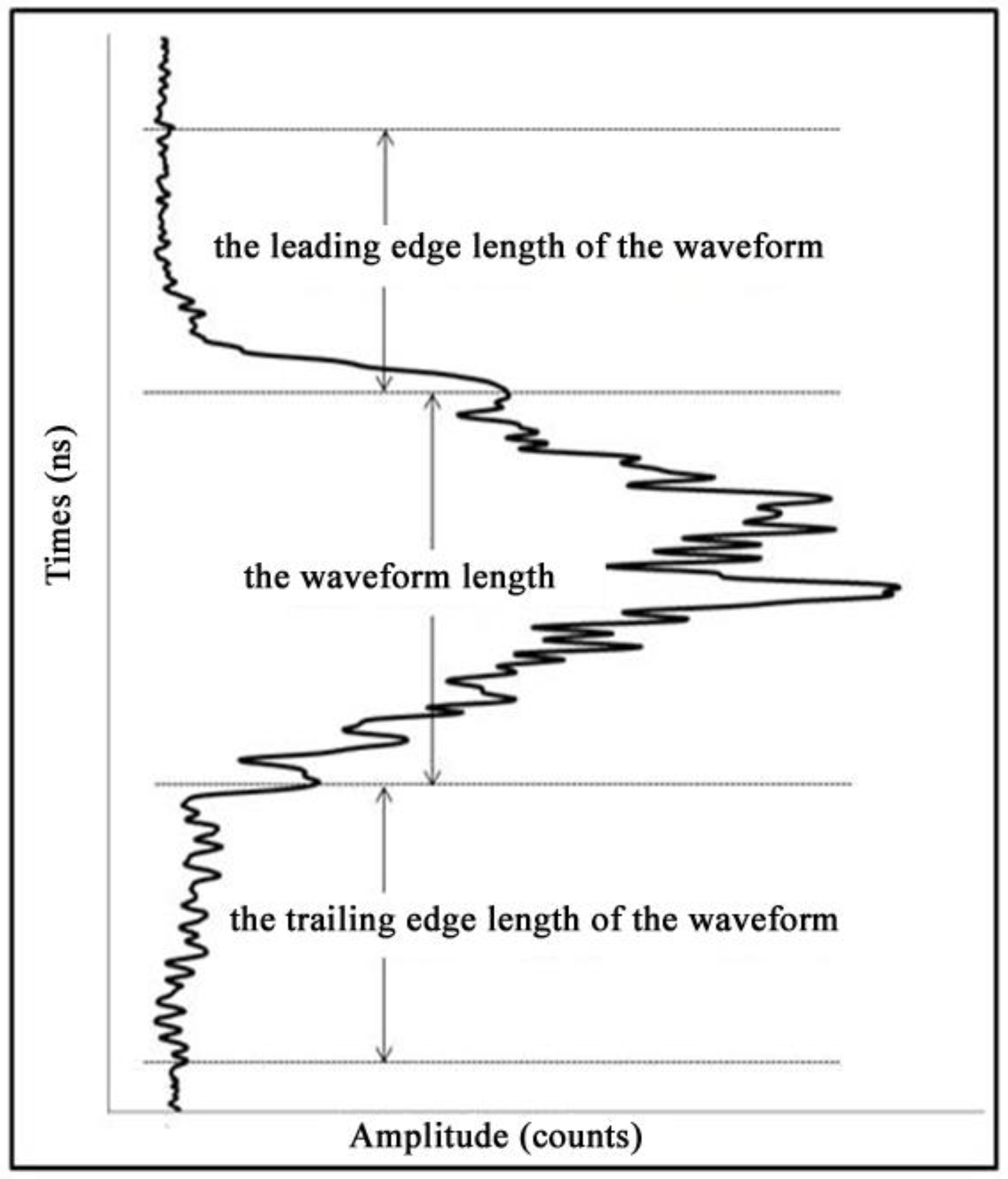
3.1.2. The GLAS Waveform Characteristics Extraction
3.1.3. Terrain Correction
3.2. MODIS Data Pre-Processing
3.3. Fusion GLAS with MODIS to Estimate Tree Height
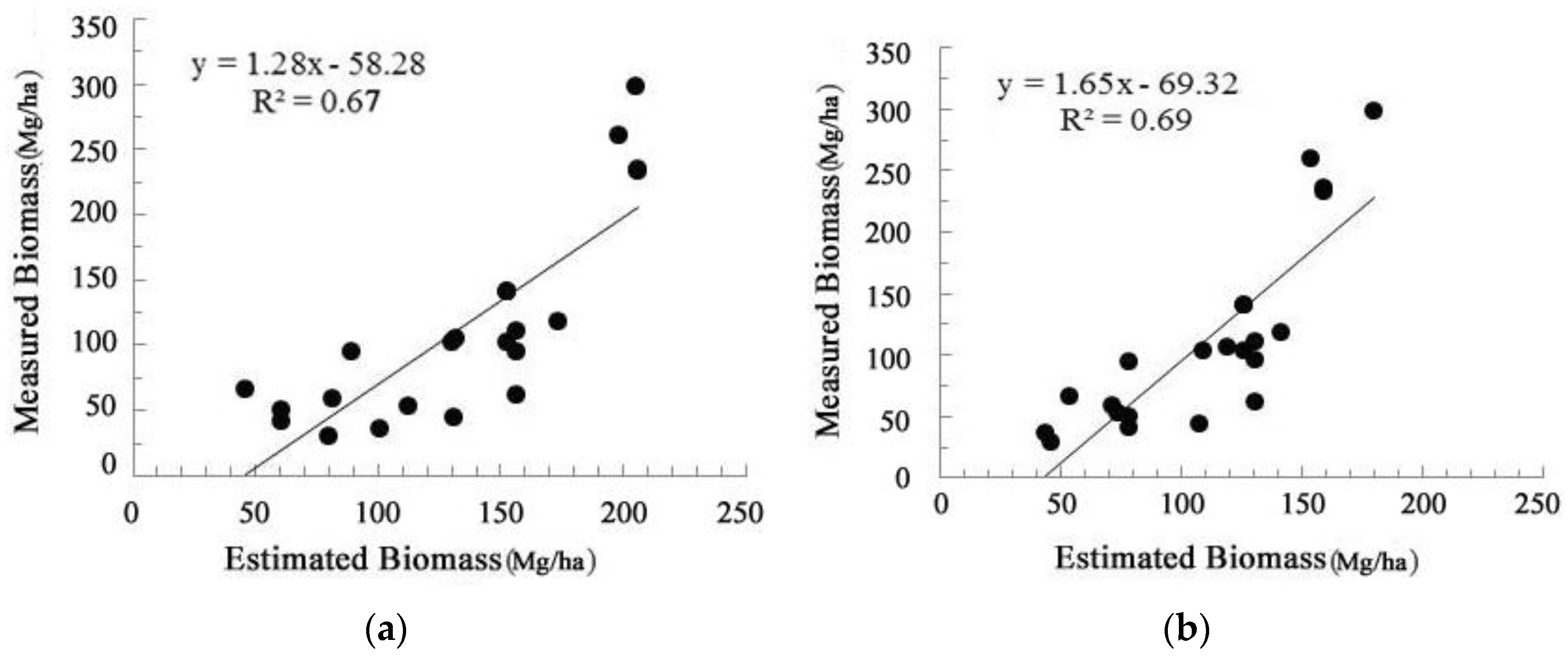
3.4. Biomass Modeling
4. Result Analysis
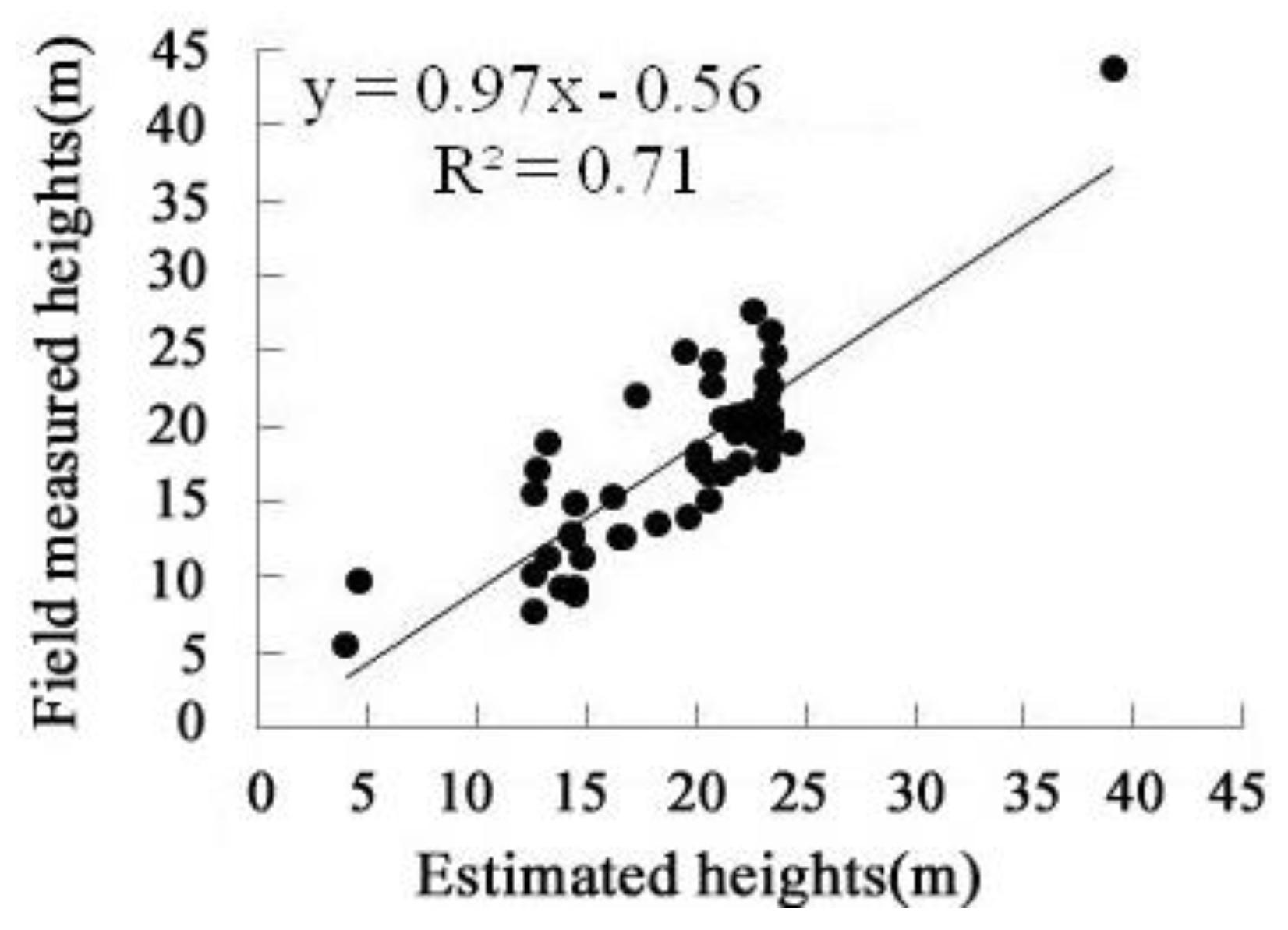
4.1. Continuous Vegetation Heights
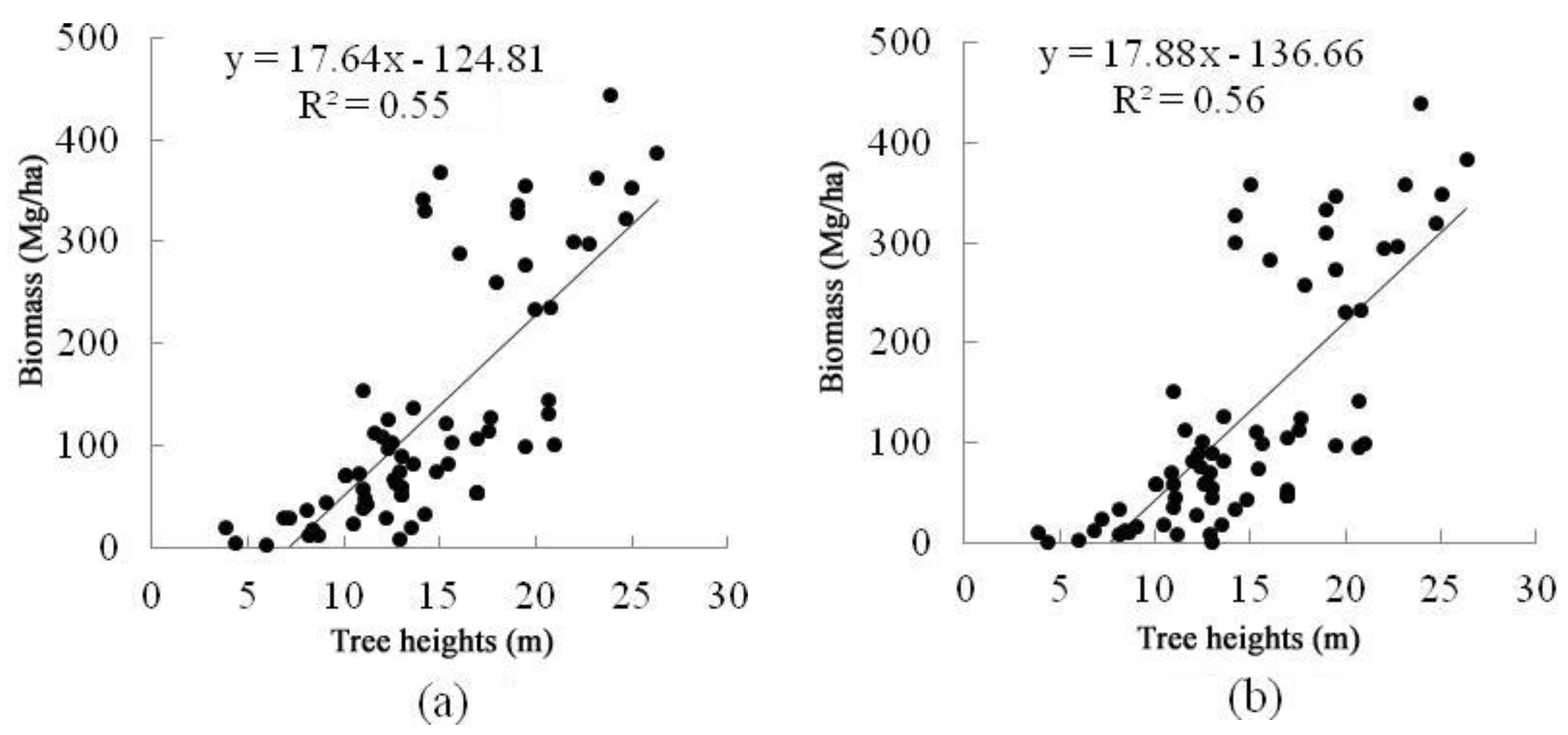
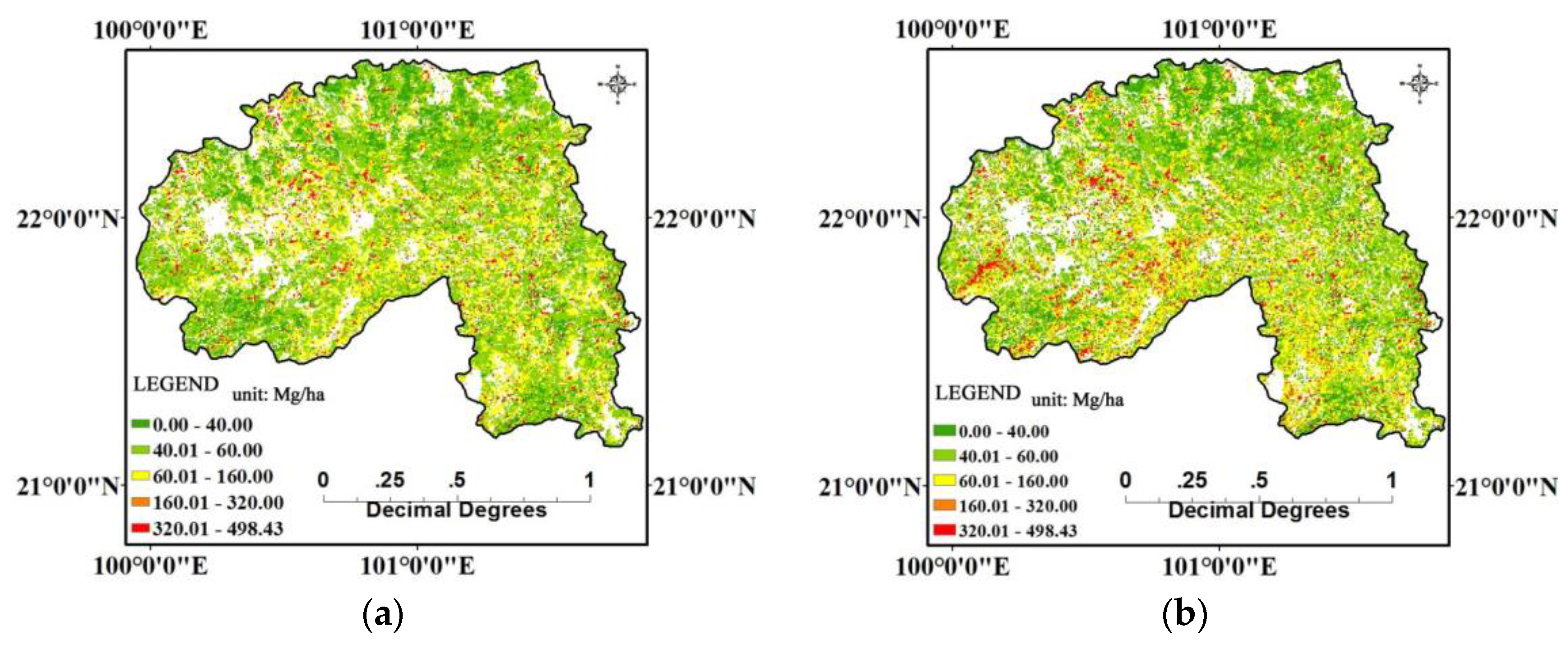
4.2. Forest AGB Estimation Results
5. Discussion and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yang, H.X.; Wu, B.; Zhang, J.T.; Lin, D.R.; Chang, S.L. Progress of research into carbon fixation and storage of forest ecosystems. J. Beijing Norm. Univ. (Nat. Sci.) 2005, 41, 172–177. (In Chinese) [Google Scholar]
- Jingyun, F.; Guohua, L.; Songling, X. Biomass and net production of forest vegetation in China. Acta Ecol. Sin. 1996, 16, 497–508. [Google Scholar]
- Nascimento, H.E.; Laurance, W.F. Total aboveground biomass in central Amazonian rainforests: A landscape-scale study. For. Ecol. Manag. 2002, 168, 311–321. [Google Scholar] [CrossRef]
- Pan, Y.; Birdsey, R.A.; Fang, J.; Houghton, R.; Kauppi, P.E.; Kurz, W.A.; Hayes, D. A large and persistent carbon sink in the world’s forests. Science 2001, 333, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.W.; Wang, X.K.; Wu, G. Biomass and Production of Forest Ecosystems in China; Science Press: Beijing, China, 1999. (In Chinese) [Google Scholar]
- Wang, L.H.; Xing, Y.Q. Remote sensing estimation of natural forest biomass based on an artificial neural network. Ying Yong Sheng Tai Xue Bao 2008, 19, 261–266. (In Chinese) [Google Scholar] [PubMed]
- Wagner, W.; Luckman, A.; Vietmeier, J.; Tansey, K.; Balzter, H.; Schmullius, C.; Yu, J.J. Large-scale mapping of boreal forest in SIBERIA using ERS tandem coherence and JERS backscatter data. Remote Sens. Environ. 2003, 85, 125–144. [Google Scholar] [CrossRef]
- Cloude, S.R.; Papathanassiou, K.P. Three-stage inversion process for polarimetric SAR interferometry. IEEE Proc. Radar Sonar Navig. 2003, 150, 125–134. [Google Scholar] [CrossRef]
- Wang, C.; Glenn, N.F. Integrating LiDAR intensity and elevation data for terrain characterization in a forested area. Geosci. Remote Sens. Lett. IEEE 2009, 6, 463–466. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Harding, D.J.; Keller, M.; Cohen, W.B.; Carabajal, C.C.; Del Bom Espirito Santo, F.; de Oliveira, R. Estimates of forest canopy height and aboveground biomass using ICESat. Geophys. Res. Lett. 2005, 32. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Hudak, A.T.; Cohen, W.B.; Acker, S.A. Geographic variability in Lidar predictions of forest stand structure in the Pacific Northwest. Remote Sens. Environ. 2005, 95, 532–548. [Google Scholar] [CrossRef]
- Sun, G.; Ranson, K.J. Modeling lidar returns from forest canopies. IEEE Trans. Geosci. Remote Sens. 2000, 38, 2617–2626. [Google Scholar]
- Sun, G.; Ranson, K.J.; Kimes, D.S.; Blair, J.B.; Kovacs, K. Forest vertical structure from GLAS: An evaluation using LVIS and SRTM data. Remote Sens. Environ. 2008, 112, 107–117. [Google Scholar] [CrossRef]
- Pang, Y.; Zhao, F.; Li, Z.Y.; Zhou, S.F.; Deng, G.; Liu, Q.W.; Chen, E.X. Forest height inversion using airborne Lidar technology. J. Remote Sens. 2008, 12, 152–158. (In Chinese) [Google Scholar]
- Means, J.E.; Acker, S.A.; Harding, D.J.; Blair, J.B.; Lefsky, M.A.; Cohen, W.B.; McKee, W.A. Use of large-footprint scanning airborne Lidar to estimate forest stand characteristics in the Western Cascades of Oregon. Remote Sens. Environ. 1999, 67, 298–308. [Google Scholar] [CrossRef]
- Ranson, K.J.; Sun, G.; Kovacs, K.; Kharuk, V.I. Landcover attributes from ICESat GLAS data in central Siberia. In Proceedings of the Geoscience and Remote Sensing Symposium, Anchorage, AK, USA, 20–24 September 2004; pp. 753–756.
- Nelson, R.; Ranson, K.J.; Sun, G.; Kimes, D.S.; Kharuk, V.; Montesano, P. Estimating Siberian timber volume using MODIS and ICESat/GLAS. Remote Sens. Environ. 2009, 113, 691–701. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Turner, D.P.; Guzy, M.; Cohen, W.B. Combining lidar estimates of aboveground biomass and Landsat estimates of stand age for spatially extensive validation of modeled forest productivity. Remote Sens. Environ. 2005, 95, 549–558. [Google Scholar] [CrossRef]
- Huang, K.; Pang, Y.; Shu, Q.; Fu, T. Aboveground forest biomass estimation using ICESat GLAS in Yunnan, China. J. Remote Sens. 2013, 17, 165–179. (In Chinese) [Google Scholar]
- Lv, X.; Tang, J.; He, Y.; Duan, W.; Song, J.; Xu, H.; Zhu, S. Biomass and its allocation in tropical seasonal rain forest in Xishuangbanna, Southwest China. J. Plant Ecol. 2007, 31, 11–22. (In Chinese) [Google Scholar]
- Tang, J.W.; Pang, J.P.; Chen, M.Y.; Guo, X.M.; Zeng, R. Biomass and its estimation model of rubber plantations in Xishuangbanna, Southwest China. Chin. J. Ecol. 2009, 28, 1942–1948. (In Chinese) [Google Scholar]
- Yang, C.; Liu, J.; Luo, J. Correlation analysis of Landsat TM data and its derived data, meteorological data and topographic data with the biomass of different aged tropical forests. Acta Phytoecol. Sin. 2003, 28, 862–867. (In Chinese) [Google Scholar]
- Cheng, F. Forest Biomass Estimation by Using Multi-source Remote Sensing Data in Yunnan. Master’s Thesis, Yunnan Normal University, Kunming, China, 2011. [Google Scholar]
- Li, Z.J.; Ma, Y.X.; Li, H.M.; Peng, M.C.; Liu, W.J. Relation of land use and cover change to topography in Xishuangbanna, Southwest China. J. Plant Ecol. 2008, 32, 1091–1103. (In Chinese) [Google Scholar]
- Wang, C.; Tang, F.; Li, L. Wavelet analysis for waveform decomposition of ICESat/GLAS data and its application in tree height estimation. IEEE Geosci. Remote Sens. Lett. 2013, 10, 115–119. [Google Scholar] [CrossRef]
- Deng, F.; Chen, M.; Plummer, S.; Chen, M.; Pisek, J. Algorithm for global leaf area index retrieval using satellite imagery. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2219–2229. [Google Scholar] [CrossRef]
- Han, T.T.; Xi, X.H.; Wang, C. Forest leaf area index inversion based on TM data in Xishuangbanna Area, China. Remote Sens. Inf. 2014, 29, 29–32. (In Chinese) [Google Scholar]
- Yang, T.; Wang, C.; Li, G.C.; Luo, S.Z.; Xi, X.H.; Gao, S.; Zeng, H.C. Forest height mapping of China using GLAS and MODIS data. China Sci. D Ser. 2014, 57, 1–11. [Google Scholar] [CrossRef]
- Jiang, X.G.; Wang, C.; Wang, C. Optimum band selection of hyperspectral remote sensing data. Arid Land Geogr. 2000, 23, 214–220. (In Chinese) [Google Scholar]
- Zhu, H. Reviews on forest classification of Xishuangbanna in south of Yunnan. Acta Bot. Yunnanica 2007, 29, 377–387. (In Chinese) [Google Scholar]
- Li, G.F.; Ren, H. Biomass and net primary productivity of the forests in diffferent climatic zones of China. Trop. Geogr. 2004, 24, 306–310. (In Chinese) [Google Scholar] [CrossRef]
- Zhang, X.Y.; Xu, Z.L.; Wang, J.N. Study on forest carbon storage trends and sink enhancement potential in Xishuangbanna. Ecol. Environ. Sci. 2011, 20, 397–402. (In Chinese) [Google Scholar]
- Luo, S.Z.; Wang, C.; Li, G.C.; Xi, X.H. Retrieving leaf area index using ICESat/GLAS full-waveform data. Remote Sens. Lett. 2013, 4, 745–753. [Google Scholar] [CrossRef]

| No. | Parameters | Model | |
|---|---|---|---|
| No. 1 | 0.725 | ||
| No. 2 | 0.726 | ||
| No. 3 | 0.728 | ||
| No. 4 | 0.729 | ||
| No. 5 | 0.750 | ||
| No. 6 | 0.729 | ||
| No. 7 | 0.730 | ||
| No. 8 | 0.756 | ||
| No. 9 | 0.757 |
| Parameters | Function | No. | Models | R2 | F Test |
|---|---|---|---|---|---|
| Height vs. Biomass | Polynomial Power function | No. 1 | 0.60 | 56.73 | |
| No. 2 | 0.61 | 59.23 | |||
| No. 3 | 0.61 | 59.25 | |||
| Exponential function | No. 4 | 0.60 | 59.03 | ||
| No. 5 | 0.57 | 51.40 | |||
| No. 6 | 0.61 | 58.84 | |||
| Heights & LAI vs. Biomass | Polynomial | No. 7 | 0.68 | 79.20 | |
| No. 8 | 0.70 | 87.29 | |||
| Power function | No. 9 | 0.72 | 99.63 | ||
| No. 10 | 0.73 | 100.62 | |||
| No. 11 | 0.70 | 87.60 | |||
| Logarithmic function | No. 12 | 0.67 | 78.43 | ||
| No. 13 | 0.71 | 94.50 | |||
| Exponential function | No. 14 | 0.70 | 88.12 | ||
| No. 15 | 0.71 | 95.07 |
| Model | ME | RMSE |
|---|---|---|
| Canopy Heights vs. Biomass (Mg/ha) | 45.60 | 52.79 |
| Canopy Heights & LAI vs. Biomass (Mg/ha) | 32.45 | 38.20 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, X.; Han, T.; Wang, C.; Luo, S.; Xia, S.; Pan, F. Forest above Ground Biomass Inversion by Fusing GLAS with Optical Remote Sensing Data. ISPRS Int. J. Geo-Inf. 2016, 5, 45. https://doi.org/10.3390/ijgi5040045
Xi X, Han T, Wang C, Luo S, Xia S, Pan F. Forest above Ground Biomass Inversion by Fusing GLAS with Optical Remote Sensing Data. ISPRS International Journal of Geo-Information. 2016; 5(4):45. https://doi.org/10.3390/ijgi5040045
Chicago/Turabian StyleXi, Xiaohuan, Tingting Han, Cheng Wang, Shezhou Luo, Shaobo Xia, and Feifei Pan. 2016. "Forest above Ground Biomass Inversion by Fusing GLAS with Optical Remote Sensing Data" ISPRS International Journal of Geo-Information 5, no. 4: 45. https://doi.org/10.3390/ijgi5040045
APA StyleXi, X., Han, T., Wang, C., Luo, S., Xia, S., & Pan, F. (2016). Forest above Ground Biomass Inversion by Fusing GLAS with Optical Remote Sensing Data. ISPRS International Journal of Geo-Information, 5(4), 45. https://doi.org/10.3390/ijgi5040045







