Bile Acid Analogs with Anti-Germination Activities for Prophylaxis of Clostridioides difficile Infection Alter Bile Acid Homeostasis in the Enterohepatic Cycle
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of CamSA and CA-Quin
2.3. Animals
2.4. Anti-Germinant Treatment and Organ Harvest
2.5. RNA Extraction
2.6. Sequencing
2.7. RT-PCR and qPCR
2.8. LC-MS of Bile Species
2.9. Differential Gene Expression Analysis
2.10. Enrichment Analysis
2.11. Statistical Analysis
3. Results
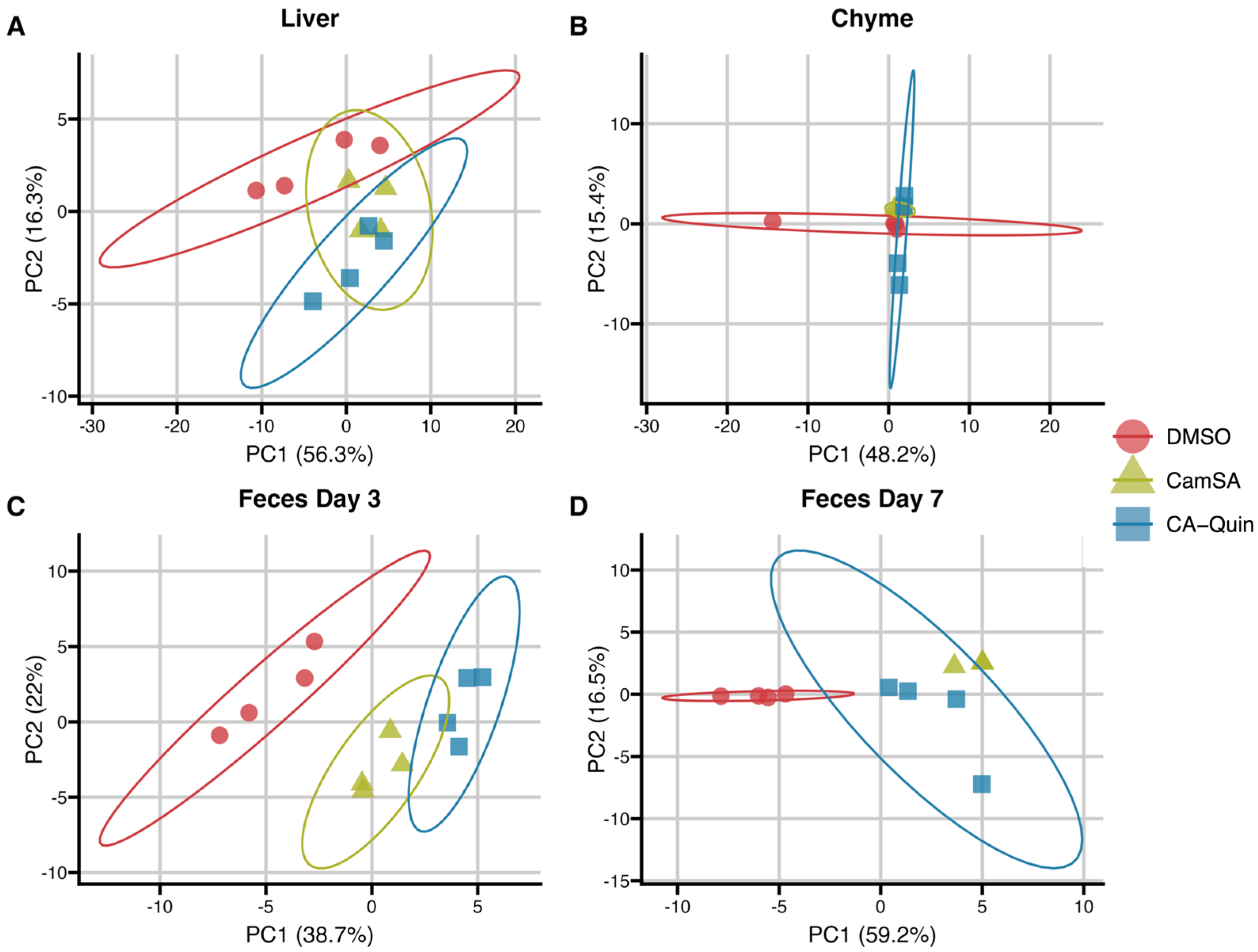
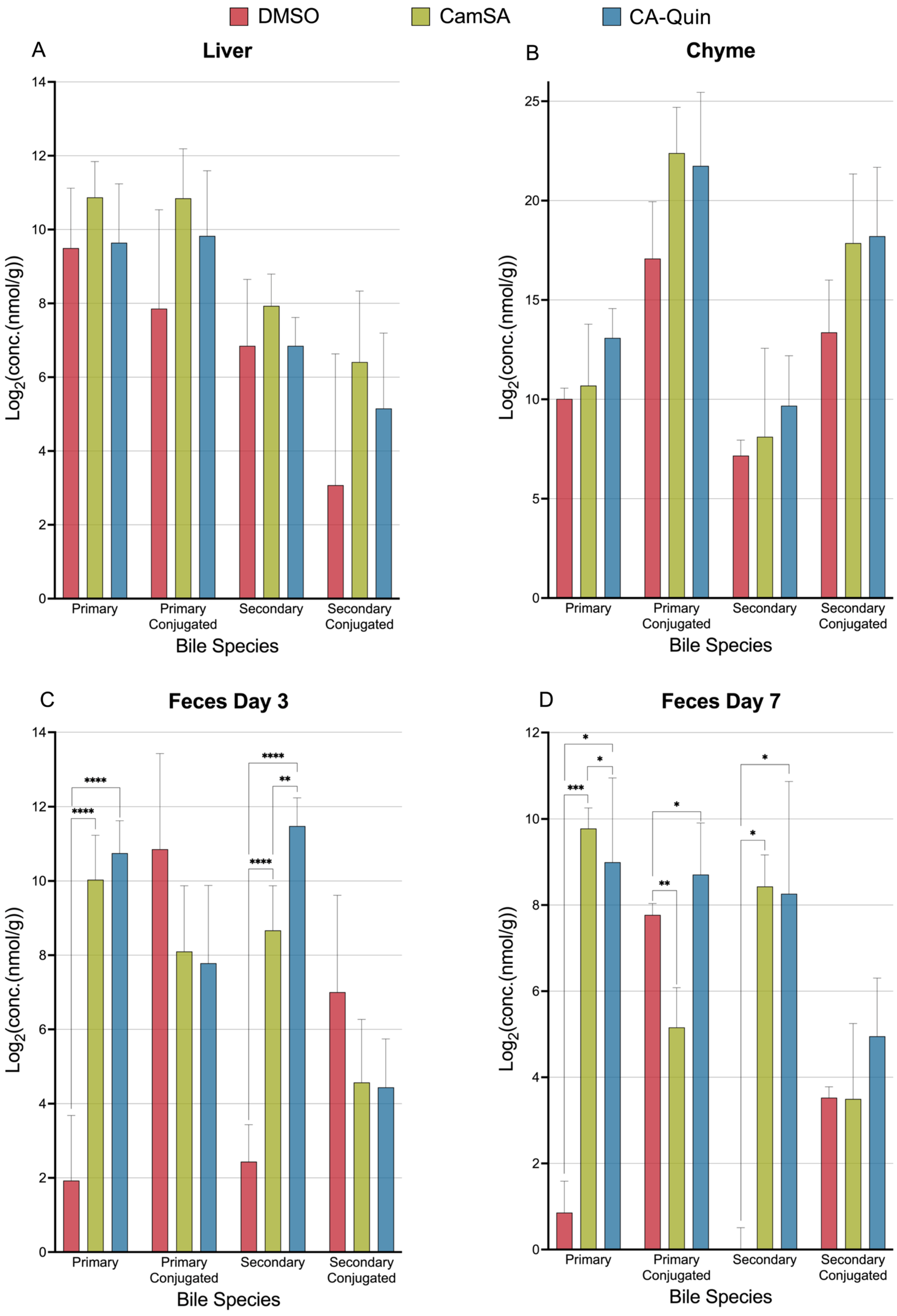
3.1. Overall Alterations of Bile Acids by CamSA and CA-Quin in the Liver, Chyme, and Fecal Samples
Alternation of Individual BA Levels y CamSA and CA-Quin Treatment
3.2. Liver Transcriptomics
Genes and Transcriptome Patterns Associated with Drug Treatments
3.3. Impact of Drug Treatment on Key Pathways in the Liver
3.4. qPCR Quantification of BA Metabolism and Transporter Genes in the Liver and Ileum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 12-keto-LCA | 2-keto-lithocholic acid |
| 3-keto,7α,12α(OH)2 | 3-keto-7α,12α-dihydroxy-5β-cholan-24-oic acid |
| 3-keto-LCA | 3-keto-lithocholic acid |
| 7-keto-DCA | 7-keto-deoxycholic acid |
| 7-keto-LCA | 7-keto-lithocholic acid |
| C4 | 7α-hydroxy-4-cholesten-3-one |
| allo-isoLCA | allo-isolithocholic acid |
| α-MCA | alpha-muricholic acid |
| β-MCA | beta-muricholic acid |
| BA | bile acid |
| CDCA | chenodeoxycholic acid |
| CDCA-3-S | chenodeoxycholic acid-3-sulfate |
| CA | cholic acid |
| CA-3-S | cholic acid-3-sulfate |
| CA-7-S | cholic acid-7-sulfate |
| CDI | Clostridioides difficile infection |
| DCA | deoxycholic acid |
| DCA-3-S | deoxycholic acid-3-sulfate |
| DMSO | dimethyl sulfoxide |
| EHC | enterohepatic circulation |
| Gβ-MCA | glyco-beta-muricholic acid |
| GCDCA | glycochenodeoxycholic acid |
| GCA | glycocholic acid |
| GDCA | glycodeoxycholic acid |
| GHCA | glycohyocholic acid |
| GHDCA | glycohyodeoxycholic acid |
| GLCA | glycolithocholic acid |
| GUDCA | glycoursodeoxycholic acid |
| HCA | hyocholic acid |
| HDCA | hyodeoxycholic acid |
| isoDCA | isodeoxycholic acid |
| isoLCA | isolithocholic acid |
| LC-MS | liquid chromatography- mass spectrometry |
| LCA | lithocholic acid |
| LCA-3-S | lithocholic acid-3-sulfate |
| mSA | metanilic acid |
| MDCA | murideoxycholic acid |
| ω-MCA | omega-muricholic acid |
| PCA | principal component analysis |
| Tα-MCA | tauro-alpha-muricholic acid |
| Tβ-MCA | tauro-beta-muricholic acid |
| Tω-MCA | tauro-omega-muricholic acid |
| TCDCA | taurochenodeoxycholic acid |
| TCA | taurocholic acid |
| TDCA | taurodeoxycholic acid |
| THDCA | taurohyodeoxycholic acid |
| TLCA | taurolithocholic acid |
| TUDCA | tauroursodeoxycholic acid |
| UDCA | ursodeoxycholic acid |
| UDCA-3-S | ursodeoxycholic acid-3-sulfate |
References
- Mada, P.K.; Alam, M.U. Clostridioides Difficile Infection; StatPearls: Treasure Island, FL, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK431054/ (accessed on 4 October 2024).
- Research, M.F. Difficile Infection. Available online: https://www.mayoclinic.org/diseases-conditions/c-difficile/symptoms-causes/syc-20351691 (accessed on 4 October 2024).
- Prevention, U.S. Diff (Clostridioides Difficile). Available online: https://www.cdc.gov/c-diff/about/index.html (accessed on 4 October 2024).
- Dicks, L.M.T.; Mikkelsen, L.S.; Brandsborg, E.; Marcotte, H. Clostridium difficile, the Difficult “Kloster” Fuelled by Antibiotics. Curr. Microbiol. 2019, 76, 774–782. [Google Scholar] [CrossRef]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, e1–e48. [Google Scholar] [CrossRef]
- Sorg, J.A.; Sonenshein, A.L. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 2009, 191, 1115–1117. [Google Scholar] [CrossRef] [PubMed]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef]
- Poland, J.C.; Flynn, C.R. Bile Acids, Their Receptors, and the Gut Microbiota. Physiology 2021, 36, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S.; Klaassen, C.D. Molecular Regulation of Bile Acid Homeostasis. Drug Metab. Dispos. 2022, 50, 425–455. [Google Scholar] [CrossRef]
- Alvarez, Z.; Abel-Santos, E. Potential Use of Inhibitors of Bacteria Spore Germination in the Prophylactic Treatment of anthrax and Clostridium difficile-Associated Disease. Expert Rev. Anti-Infect. Ther. 2007, 5, 783–792. [Google Scholar] [CrossRef]
- Sharma, S.; Phan, J.; Abel-Santos, E.; Firestine, S. 5,6-Fused Heterocycle Cholate Derivatives as Spore Germination Inhibitors of Clostridioides difficile. ChemRxiv 2023. [Google Scholar] [CrossRef]
- Sharma, S.K.; Schilke, A.R.; Phan, J.R.; Yip, C.; Sharma, P.V.; Abel-Santos, E.; Firestine, S.M. The design, synthesis, and inhibition of Clostridioides difficile spore germination by acyclic and bicyclic tertiary amide analogs of cholate. Eur. J. Med. Chem. 2023, 261, 115788. [Google Scholar] [CrossRef]
- Sharma, S.K.; Yip, C.; Simon, M.P.; Phan, J.; Abel-Santos, E.; Firestine, S.M. Studies on the Importance of the 7α-, and 12α- hydroxyl groups of N-Aryl-3α,7α,12α-trihydroxy-5β-cholan-24-amides on their Antigermination Activity Against a Hypervirulent Strain of Clostridioides (Clostridium) difficile. Bioorg. Med. Chem. 2021, 52, 116503. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Yip, C.; Esposito, E.X.; Sharma, P.V.; Simon, M.P.; Abel-Santos, E.; Firestine, S.M. The Design, Synthesis, and Characterizations of Spore Germination Inhibitors Effective against an Epidemic Strain of Clostridium difficile. J. Med. Chem. 2018, 61, 6759–6778. [Google Scholar] [CrossRef]
- Phan, J.R.; Do, D.M.; Truong, M.C.; Ngo, C.; Phan, J.H.; Sharma, S.K.; Schilke, A.; Mefferd, C.C.; Villarama, J.V.; Lai, D.; et al. An Aniline-Substituted Bile Salt Analog Protects both Mice and Hamsters from Multiple Clostridioides difficile Strains. Antimicrob. Agents Chemother. 2022, 66, e01435-21. [Google Scholar] [CrossRef]
- Howerton, A.; Seymour, C.O.; Murugapiran, S.K.; Liao, Z.; Phan, J.R.; Estrada, A.; Wagner, A.J.; Mefferd, C.C.; Hedlund, B.P.; Abel-Santos, E. Effect of the Synthetic Bile Salt Analog CamSA on the Hamster Model of Clostridium difficile Infection. Antimicrob. Agents Chemother. 2018, 62, 10–1128. [Google Scholar] [CrossRef]
- Howerton, A.; Patra, M.; Abel-Santos, E. Fate of Ingested Clostridium difficile Spores in Mice. PLoS ONE 2013, 8, e72620. [Google Scholar] [CrossRef]
- Howerton, A.; Patra, M.; Abel-Santos, E. A New Strategy for the Prevention of Clostridium difficile Infection. J. Infect. Dis. 2013, 207, 1498–1504. [Google Scholar] [CrossRef] [PubMed]
- Howerton, A.; Ramirez, N.; Abel-Santos, E. Mapping Interactions between Germinants and Clostridium difficile Spores. J. Bacteriol. 2011, 193, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.; Phan, J.R.; Abel-Santos, E. Mechanism of germination inhibition of Clostridioides difficile spores by an aniline substituted cholate derivative (CaPA). bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.; Okada, N.C.; Howerton, A.; Amei, A.; Abel-Santos, E. Pharmacokinetics of Camsa, a Potential Prophylactic Compound Against Clostridioides difficile Infections. Biochem. Pharmacol. 2021, 183, 114314. [Google Scholar] [CrossRef]
- Sattar, A.; Thommes, P.; Payne, L.; Warn, P.; Vickers, R.J. SMT19969 for Clostridium difficile infection (CDI): In Vivo Efficacy Compared with Fidaxomicin and Vancomycin in the Hamster Model of CDI. J. Antimicrob. Chemother. 2015, 70, 1757–1762. [Google Scholar] [CrossRef]
- Kakiyama, G.; Marques, D.; Martin, R.; Takei, H.; Rodriguez-Agudo, D.; LaSalle, S.A.; Hashiguchi, T.; Liu, X.; Green, R.; Erickson, S.; et al. Insulin resistance dysregulates CYP7B1 leading to oxysterol accumulation: A pathway for NAFL to NASH transition. J. Lipid Res. 2020, 61, 1629–1644. [Google Scholar] [CrossRef]
- Wang, Y.; Tai, Y.L.; Zhao, D.; Zhang, Y.; Yan, J.; Kakiyama, G.; Wang, X.; Gurley, E.C.; Liu, J.; Liu, J.; et al. Berberine Prevents Disease Progression of Nonalcoholic Steatohepatitis through Modulating Multiple Pathways. Cells 2021, 10, 210. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 7 September 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Putri, G.H.; Anders, S.; Pyl, P.T.; Pimanda, J.E.; Zanini, F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 2022, 38, 2943–2945. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing, R Version 4.4.2 (31 October 2024); R Foundation for Statistical Computing: Vienna, Austria, 2024.
- Team, P. RStudio: Integrated Development Environment for R, 2024.9.1.394; Posit Software, PBC: Boston, MA, USA, 2024.
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Yan, L. ggvenn: Draw Venn Diagram by ‘ggplot2’, R Package Version 0.1.19; The R Foundation for Statistical Computing: Vienna, Austria, 2023. Available online: https://cran.r-project.org/web/packages/ggvenn/ggvenn.pdf (accessed on 7 September 2025).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Gu, Z. Complex heatmap visualization. Imeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Maintainer, D.T. KEGGREST: Client-Side REST Access to the Kyoto Encyclopedia of Genes and Genomes (KEGG), R Package Version 1.46.0; The R Foundation for Statistical Computing: Vienna, Austria, 2024.
- Pagès, H.; Carlson, M.; Falcon, S.; Li, N. AnnotationDbi: Manipulation of SQLite-Based Annotations in Bioconductor, R Package Version 1.68.0; The R Foundation for Statistical Computing: Vienna, Austria, 2024.
- Carlson, M. Org.Mm.eg.db: Genome Wide Annotation for Mouse, R Package Version 3.20.0; The R Foundation for Statistical Computing: Vienna, Austria, 2024.
- Slowikowski, K. Ggrepel: Automatically Position Non-Overlapping Text Labels with ‘ggplot2’, R Package Version 0.9.6; The R Foundation for Statistical Computing: Vienna, Austria, 2024.
- Dawson, C. Ggprism: A ‘ggplot2’ Extension Inspired by ‘GraphPad Prism’, R Package Version 1.0.7; The R Foundation for Statistical Computing: Vienna, Austria, 2025.
- Schauberger, P.; Walker, A. openxlsx: Read, Write and Edit xlsx Files, R Package Version 4.2.8.1; The R Foundation for Statistical Computing: Vienna, Austria, 2025.
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. vegan: Community Ecology Package; R Package Version 2.7-2; The R Foundation for Statistical Computing: Vienna, Austria, 2025. [Google Scholar]
- Davison, A.H.D. Bootstrap Methods and Their Applications; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar]
- Canty, A.R.B. Boot: Bootstrap Function; R Package Version 1.3-32; The R Foundation for Statistical Computing: Vienna, Austria, 2025. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 2025, 53, D672–D677. [Google Scholar] [CrossRef]
- Weizmann Institute of Science. SLC51B. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SLC51B (accessed on 4 October 2024).
- Ramirez, N.; Liggins, M.; Abel-Santos, E. Kinetic evidence for the presence of putative germination receptors in Clostridium difficile spores. J. Bacteriol. 2010, 192, 4215–4222. [Google Scholar] [CrossRef]
- McMillan, A.S.; Theriot, C.M. Bile acids impact the microbiota, host, and C. difficile dynamics providing insight into mechanisms of efficacy of FMTs and microbiota-focused therapeutics. Gut Microbes 2024, 16, 2393766. [Google Scholar] [CrossRef]
- Li, W.; Chen, H.; Tang, J. Interplay between Bile Acids and Intestinal Microbiota: Regulatory Mechanisms and Therapeutic Potential for Infections. Pathogens 2024, 13, 702. [Google Scholar] [CrossRef]
- Fuchs, C.D.; Simbrunner, B.; Baumgartner, M.; Campbell, C.; Reiberger, T.; Trauner, M. Bile acid metabolism and signalling in liver disease. J. Hepatol. 2025, 82, 134–153. [Google Scholar] [CrossRef]
- Gou, H.; Zeng, R.; Lau, H.C.H.; Yu, J. Gut microbial metabolites: Shaping future diagnosis and treatment against gastrointestinal cancer. Pharmacol. Res. 2024, 208, 107373. [Google Scholar] [CrossRef] [PubMed]
- Mullish, B.H.; Allegretti, J.R. The contribution of bile acid metabolism to the pathogenesis of Clostridioides difficile infection. Ther. Adv. Gastroenterol. 2021, 14, 17562848211017725. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vakeesan, N.; Heredia, E.; Hassan, C.; Jiang, Y.; Sharma, S.; Su, L.; Zhou, H.; Firestine, S.; Abel-Santos, E.; Liu, W. Bile Acid Analogs with Anti-Germination Activities for Prophylaxis of Clostridioides difficile Infection Alter Bile Acid Homeostasis in the Enterohepatic Cycle. Biomolecules 2025, 15, 1672. https://doi.org/10.3390/biom15121672
Vakeesan N, Heredia E, Hassan C, Jiang Y, Sharma S, Su L, Zhou H, Firestine S, Abel-Santos E, Liu W. Bile Acid Analogs with Anti-Germination Activities for Prophylaxis of Clostridioides difficile Infection Alter Bile Acid Homeostasis in the Enterohepatic Cycle. Biomolecules. 2025; 15(12):1672. https://doi.org/10.3390/biom15121672
Chicago/Turabian StyleVakeesan, Nivisa, Efren Heredia, Chandler Hassan, Yang Jiang, Shiv Sharma, Lianyong Su, Huiping Zhou, Steven Firestine, Ernesto Abel-Santos, and Wanqing Liu. 2025. "Bile Acid Analogs with Anti-Germination Activities for Prophylaxis of Clostridioides difficile Infection Alter Bile Acid Homeostasis in the Enterohepatic Cycle" Biomolecules 15, no. 12: 1672. https://doi.org/10.3390/biom15121672
APA StyleVakeesan, N., Heredia, E., Hassan, C., Jiang, Y., Sharma, S., Su, L., Zhou, H., Firestine, S., Abel-Santos, E., & Liu, W. (2025). Bile Acid Analogs with Anti-Germination Activities for Prophylaxis of Clostridioides difficile Infection Alter Bile Acid Homeostasis in the Enterohepatic Cycle. Biomolecules, 15(12), 1672. https://doi.org/10.3390/biom15121672






