Mutagenesis of DsbAss is Crucial for the Signal Recognition Particle Mechanism in Escherichia coli: Insights from Molecular Dynamics Simulations
Abstract
:1. Introduction
2. Materials and Methods
2.1. E. Coli Strain, Plasmid, Enzymes, and Kits
2.2. Isolation of Total RNA and Construction of Recombinant pTzOGH-1 Plasmid
2.3. Construction of Recombinant pOGH-1 to -8 Expression Plasmids
2.4. Expression and Subcellular Fractionation of pOGH-1 to -8 Constructs
2.5. Protein and Western Blot Analysis
2.6. Molecular Modeling Studies
2.7. Molecular Dynamics Simulation Protocol
2.8. MM-GBSA Binding Free Energy Calculations
3. Results
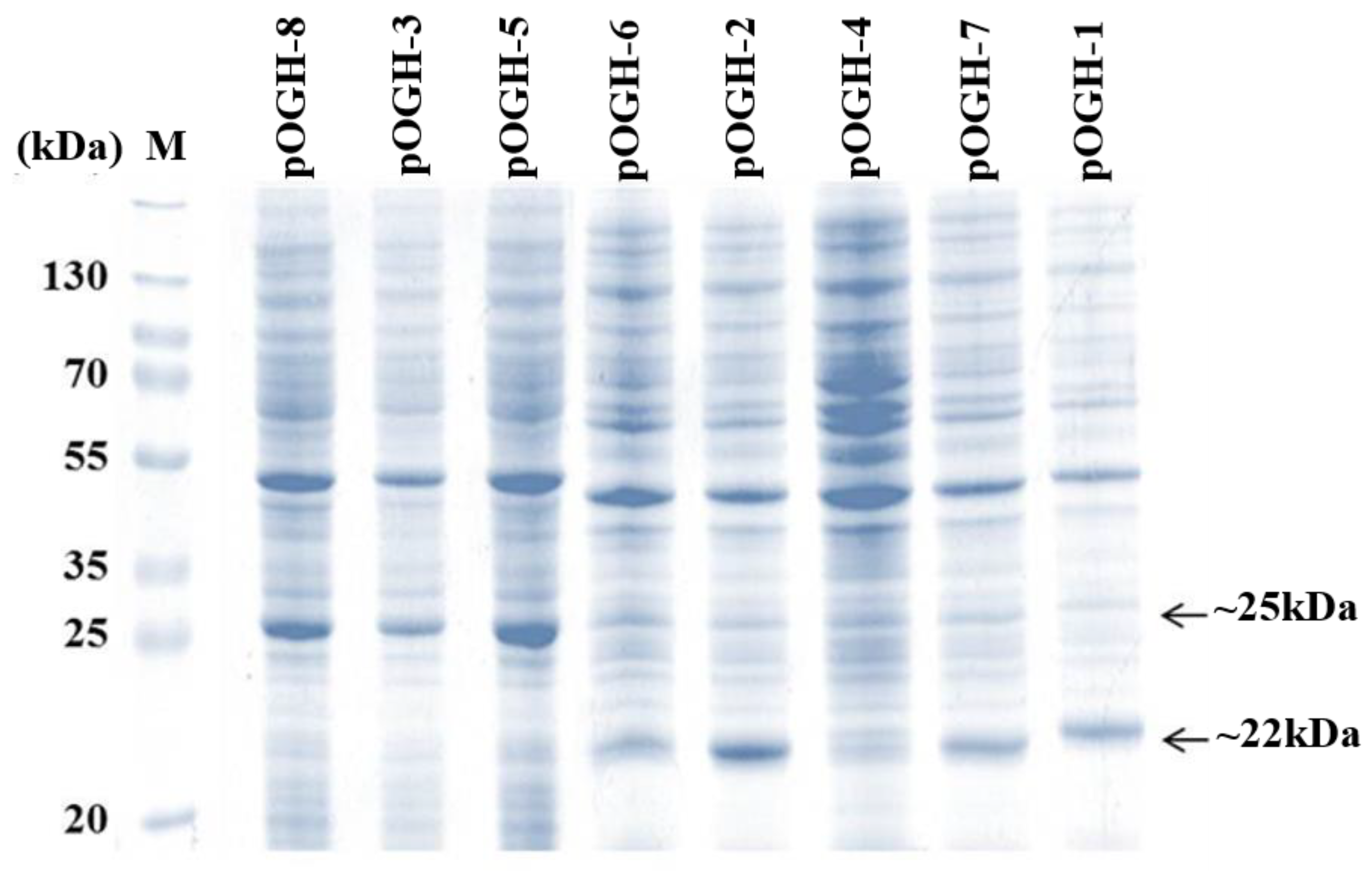
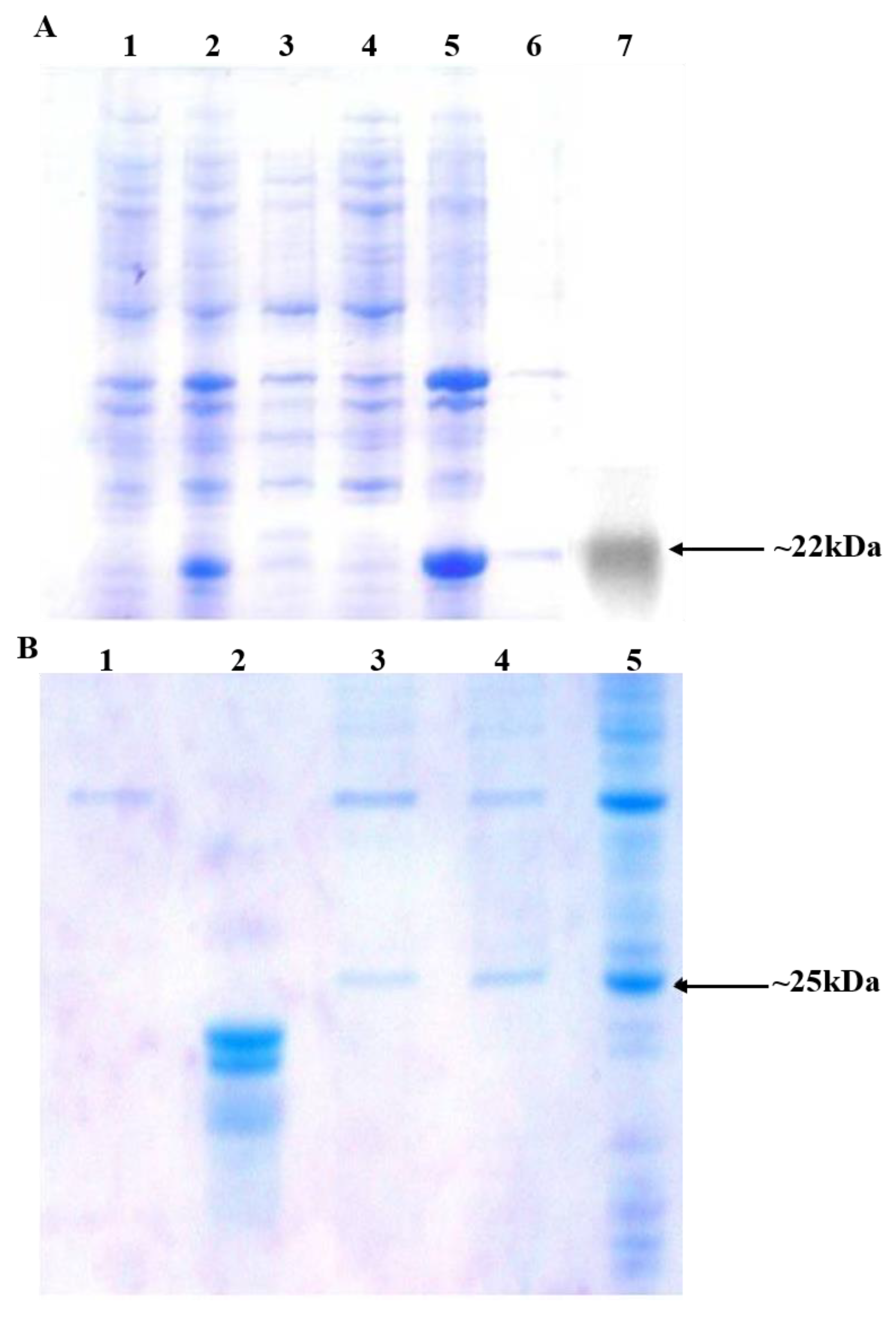
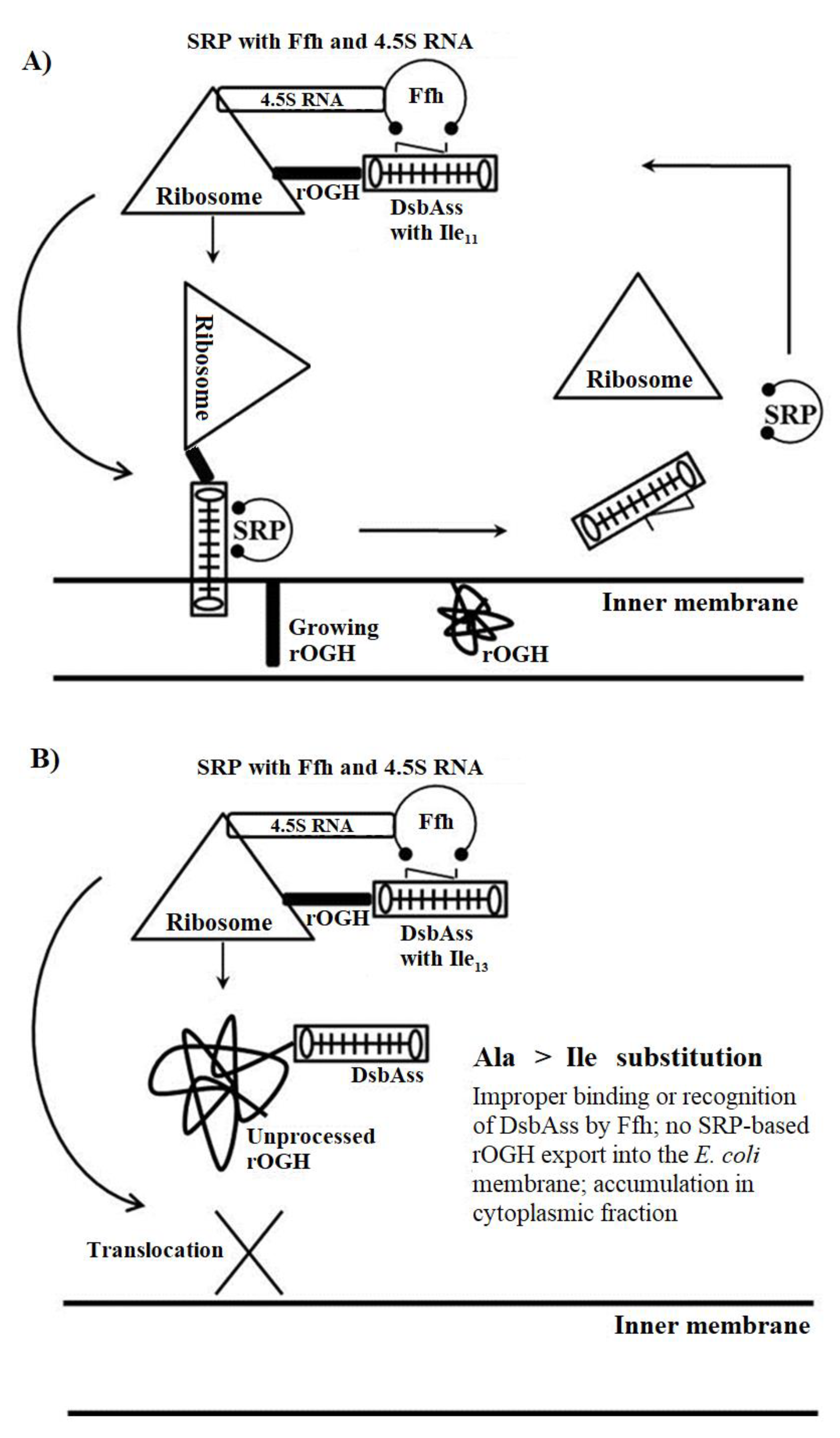
3.1. rOGH Expression Analysis by Mutant Signal Sequences
3.2. Impact of Ala Substitutions in DsbAss H-Domain
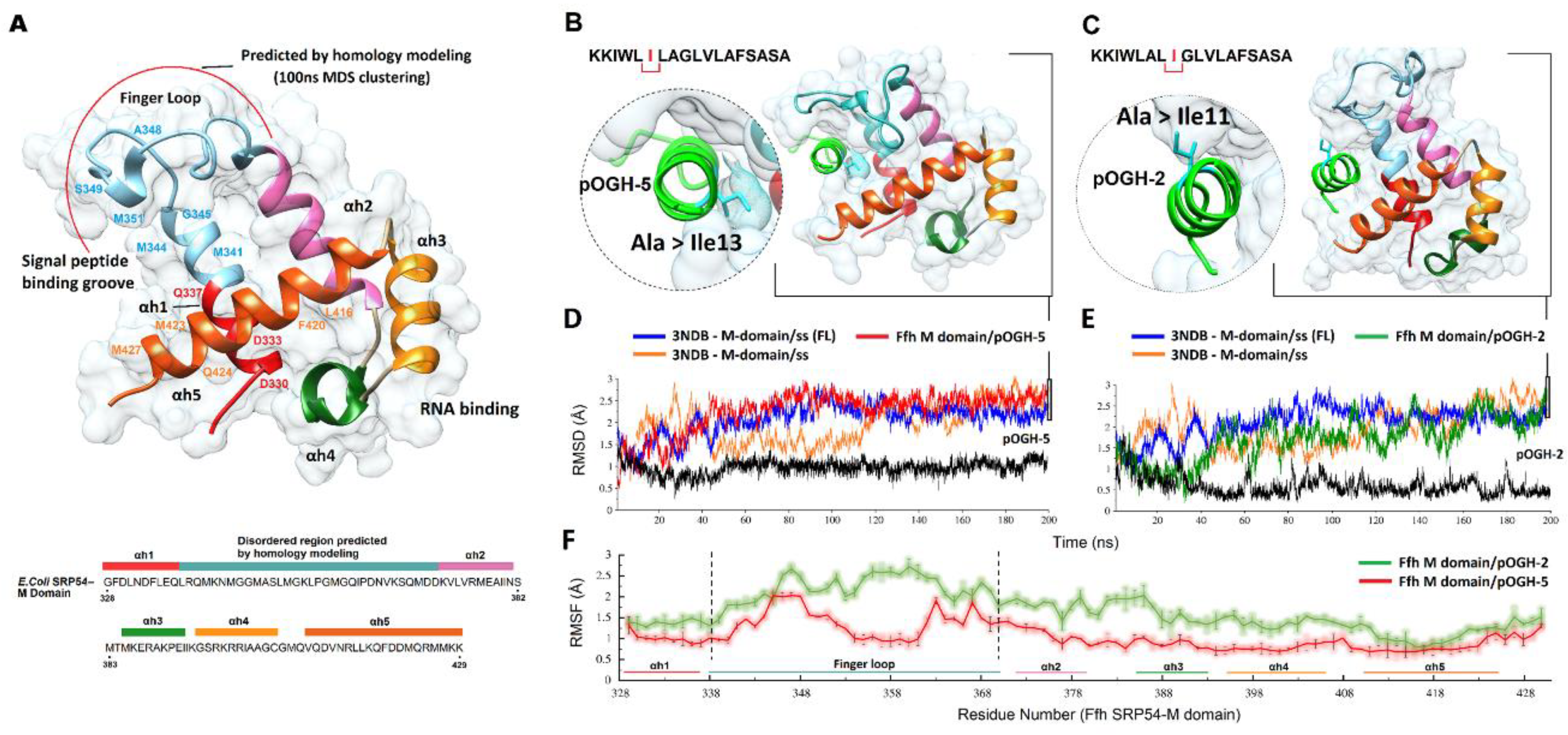
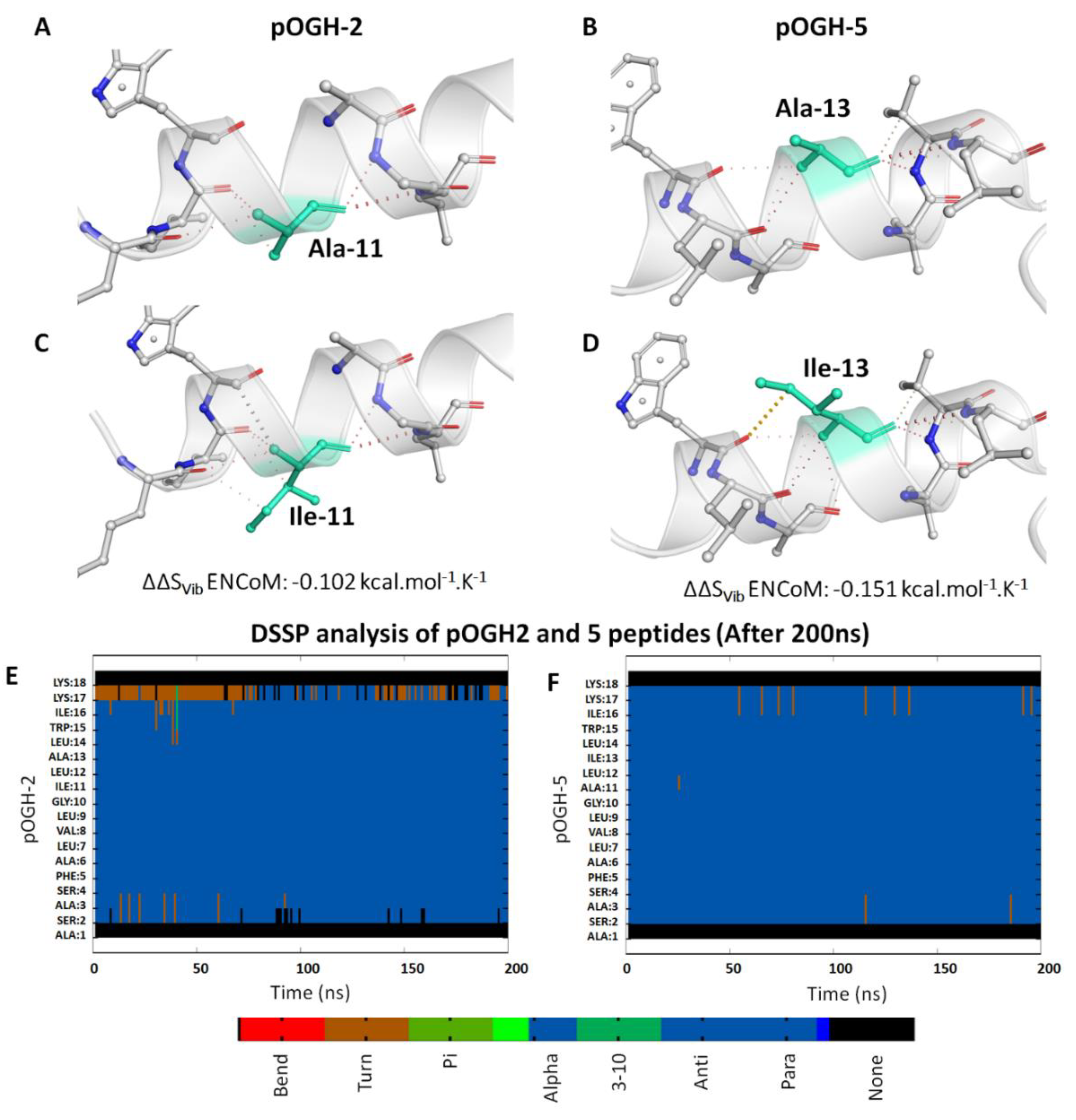
3.3. Molecular Modeling Analysis
3.4. Binding Free Energy Calculations Using the MM-GBSA Method
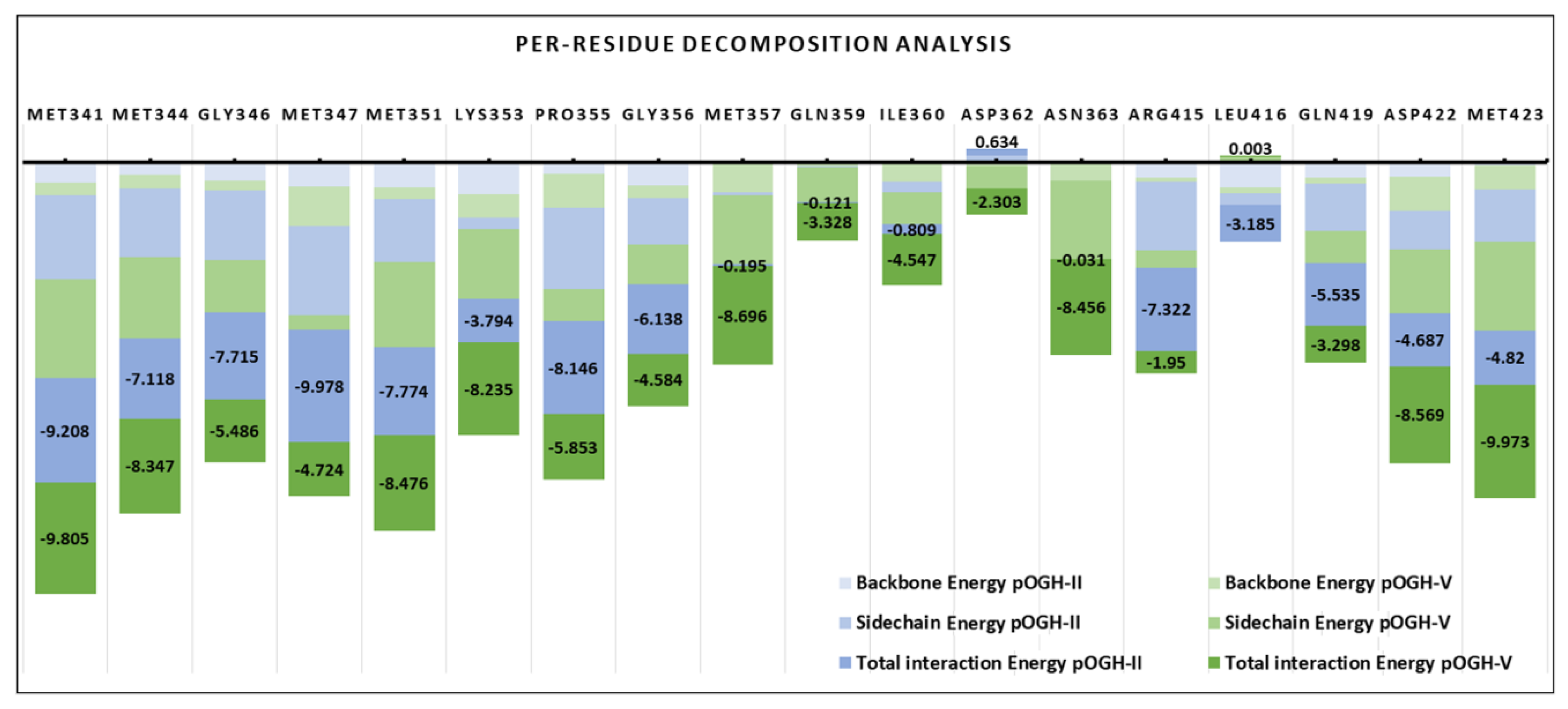
3.5. Per-Residue Decomposition Analysis Using MM-GBSA Method
3.6. pOGH Construct Substitution with Lys → Arg at N Terminal of DsbAss
3.7. Influence of Substitution with Ser → Cys at C Terminal of DsbAss on rOGH Translocation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef]
- Neu, H.C. The crisis in antibiotic resistance. Science 1992, 257, 1064–1073. [Google Scholar] [CrossRef]
- The Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 31 December 2014).
- Cegelski, L.; Marshall, G.R.; Eldridge, G.R.; Hultgren, S.J. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008, 6, 17–27. [Google Scholar] [CrossRef]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heras, B.; Shouldice, S.R.; Totsika, M.; Scanlon, M.J.; Schembri, M.A.; Martin, J.L. DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 2009, 7, 215–225. [Google Scholar] [CrossRef]
- Shouldice, S.R.; Heras, B.; Walden, P.M.; Totsika, M.; Schembri, M.A.; Martin, J.L. Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid. Redox Signal. 2011, 14, 1729–1760. [Google Scholar] [CrossRef]
- Smith, R.; Paxman, J.; Scanlon, M.; Heras, B. Targeting bacterial Dsb proteins for the development of anti-virulence agents. Molecules 2016, 21, 811. [Google Scholar] [CrossRef]
- Grauschopf, U.; Winther, J.R.; Korber, P.; Zander, T.; Dallinger, P.; Bardwell, J.C. Why is DsbA such an oxidizing disulfide catalyst? Cell 1995, 83, 947–955. [Google Scholar] [CrossRef] [Green Version]
- Heras, B.; Scanlon, M.J.; Martin, J.L. Targeting virulence not viability in the search for future antibacterials. Brit. J. Clin. Pharmacol. 2015, 79, 208–215. [Google Scholar] [CrossRef] [Green Version]
- Kamitani, S.; Akiyama, Y.; Ito, K. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 1992, 11, 57. [Google Scholar] [CrossRef]
- Totsika, M.; Heras, B.; Wurpel, D.J.; Schembri, M.A. Characterization of two homologous disulfide bond systems involved in virulence factor biogenesis in uropathogenic Escherichia coli CFT073. J. Bacteriol. 2009, 191, 3901–3908. [Google Scholar] [CrossRef]
- Yu, J.; Edwards-Jones, B.; Neyrolles, O.; Kroll, J.S. Key Role for DsbA in Cell-to-Cell Spread ofShigella flexneri, Permitting Secretion of Ipa Proteins into Interepithelial Protrusions. Infect. Immun. 2000, 68, 6449–6456. [Google Scholar] [CrossRef] [PubMed]
- Peek, J.A.; Taylor, R.K. Characterization of a periplasmic thiol: Disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 1992, 89, 6210–6214. [Google Scholar] [CrossRef]
- Stenson, T.H.; Weiss, A.A. DsbA and DsbC are required for secretion of pertussis toxin by Bordetella pertussis. Infect. Immun. 2002, 70, 2297–2303. [Google Scholar] [CrossRef] [PubMed]
- Luirink, J.; Dobberstein, B. Mammalian and Escherichia coli signal recognition particles. Mol. Microbiol. 1994, 11, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Phillips, G.J.; Silhavy, T.J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 1992, 359, 744–746. [Google Scholar] [CrossRef]
- Soares, C.R.; Gomide, F.I.; Ueda, E.K.; Bartolini, P. Periplasmic expression of human growth hormone via plasmid vectors containing the λPL promoter: Use of HPLC for product quantification. Protein Eng. 2003, 16, 1131–1138. [Google Scholar] [CrossRef]
- Akopian, D.; Shen, K.; Zhang, X.; Shan, S.-O. Signal recognition particle: An essential protein-targeting machine. Annu. Rev. Biochem. 2013, 82, 693–721. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial secretion systems—An overview. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Nilsson, I.; Lara, P.; Hessa, T.; Johnson, A.E.; von Heijne, G.; Karamyshev, A.L. The code for directing proteins for translocation across ER membrane: SRP cotranslationally recognizes specific features of a signal sequence. J. Mol. Biol. 2015, 427, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- Schierle, C.F.; Berkmen, M.; Huber, D.; Kumamoto, C.; Boyd, D.; Beckwith, J. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 2003, 185, 5706–5713. [Google Scholar] [CrossRef]
- Lee, H.C.; Bernstein, H.D. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA 2001, 98, 3471–3476. [Google Scholar] [CrossRef] [PubMed]
- Mergulhao, F.; Summers, D.K.; Monteiro, G.A. Recombinant protein secretion in Escherichia coli. Biotechnol. Adv. 2005, 23, 177–202. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, P. Expressing genes in different Escherichia coli compartments. Curr. Opin. Biotechnol. 2000, 11, 450–454. [Google Scholar] [CrossRef]
- Durrani, F.G.; Gul, R.; Sadaf, S.; Akhtar, M.W. Expression and rapid purification of recombinant biologically active ovine growth hormone with DsbA targeting to Escherichia coli inner membrane. Appl. Microbiol. Biotechnol. 2015, 99, 6791–6801. [Google Scholar] [CrossRef]
- Hainzl, T.; Huang, S.; Sauer-Eriksson, A.E. Interaction of signal-recognition particle 54 GTPase domain and signal-recognition particle RNA in the free signal-recognition particle. Proc. Natl. Acad. Sci. USA 2007, 104, 14911–14916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doudna, J.A.; Batey, R.T. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 2004, 73, 539–557. [Google Scholar] [CrossRef]
- Walter, P.; Johnson, A.E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994, 10, 87–119. [Google Scholar] [CrossRef]
- Chu, F.; Shan, S.-o.; Moustakas, D.T.; Alber, F.; Egea, P.F.; Stroud, R.M.; Walter, P.; Burlingame, A.L. Unraveling the interface of signal recognition particle and its receptor by using chemical cross-linking and tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 16454–16459. [Google Scholar] [CrossRef] [Green Version]
- Römisch, K.; Webb, J.; Lingelbach, K.; Gausepohl, H.; Dobberstein, B. The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J. Cell Biol. 1990, 111, 1793–1802. [Google Scholar] [CrossRef]
- Lütcke, H.; High, S.; Römisch, K.; Ashford, A.J.; Dobberstein, B. The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J. 1992, 11, 1543. [Google Scholar] [CrossRef] [PubMed]
- High, S.; Dobberstein, B. The signal sequence interacts with the methionine-rich domain of the 54-kD protein of signal recognition particle. J. Cell Biol. 1991, 113, 229–233. [Google Scholar] [CrossRef]
- Janda, C.Y.; Li, J.; Oubridge, C.; Hernández, H.; Robinson, C.V.; Nagai, K. Recognition of a signal peptide by the signal recognition particle. Nature 2010, 465, 507–510. [Google Scholar] [CrossRef]
- Keenan, R.J.; Freymann, D.M.; Walter, P.; Stroud, R.M. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 1998, 94, 181–191. [Google Scholar] [CrossRef]
- Schaffitzel, C.; Oswald, M.; Berger, I.; Ishikawa, T.; Abrahams, J.P.; Koerten, H.K.; Koning, R.I.; Ban, N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature 2006, 444, 503. [Google Scholar] [CrossRef]
- Focia, P.J.; Shepotinovskaya, I.V.; Seidler, J.A.; Freymann, D.M. Heterodimeric GTPase core of the SRP targeting complex. Science 2004, 303, 373–377. [Google Scholar] [CrossRef]
- Egea, P.F.; Stroud, R.M.; Walter, P. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005, 15, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Wild, K.; Bange, G.; Motiejunas, D.; Kribelbauer, J.; Hendricks, A.; Segnitz, B.; Wade, R.C.; Sinning, I. Structural basis for conserved regulation and adaptation of the signal recognition particle targeting complex. J. Mol. Biol. 2016, 428, 2880–2897. [Google Scholar] [CrossRef]
- Cleverley, R.M.; Gierasch, L.M. Mapping the signal sequence-binding site on SRP reveals a significant role for the NG domain. J. Biol. Chem. 2002, 277, 46763–46768. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; Kuriyan, J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. USA 2005, 102, 6679–6685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klepeis, J.L.; Lindorff-Larsen, K.; Dror, R.O.; Shaw, D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 2009, 19, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Dodson, G.G.; Lane, D.P.; Verma, C.S. Molecular simulations of protein dynamics: new windows on mechanisms in biology. EMBO Rep. 2008, 9, 144–150. [Google Scholar] [CrossRef] [Green Version]
- Dagliyan, O.; Proctor, E.A.; D’Auria, K.M.; Ding, F.; Dokholyan, N.V. Structural and dynamic determinants of protein-peptide recognition. Structure 2011, 19, 1837–1845. [Google Scholar] [CrossRef]
- Ulmschneider, J.P.; Ulmschneider, M.B. Molecular Dynamics Simulations Are Redefining Our View of Peptides Interacting with Biological Membranes. Acc. Chem. Res. 2018, 51, 1106–1116. [Google Scholar] [CrossRef]
- Kaderbhai, N.N.; Harding, V.; Kaderbhai, M.A. Signal peptidase I-mediated processing of an engineered mammalian cytochrome b5 precursor is an exocytoplasmic post-translocational event in Escherichia coli. Mol. Membr. Biol. 2008, 25, 388–399. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Batey, R.T.; Sagar, M.B.; Doudna, J.A. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 2001, 307, 229–246. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. 2014, 1137, 1–15. [Google Scholar]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast interaction refinement in molecular docking. Prot. Struct. Funct. Bioinf. 2007, 69, 139–159. [Google Scholar] [CrossRef]
- Comeau, S.R.; Gatchell, D.W.; Vajda, S.; Camacho, C.J. ClusPro: An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 2004, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Lensink, M.F.; Wodak, S.J. Docking and scoring protein interactions: CAPRI 2009. Prot. Struct. Funct. Bioinf. 2010, 78, 3073–3084. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Hainzl, T.; Huang, S.; Meriläinen, G.; Brännström, K.; Sauer-Eriksson, A.E. Structural basis of signal-sequence recognition by the signal recognition particle. Nat. Struct. Mol. Biol. 2011, 18, 389. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Jabbar, B.; Rafique, S.; Salo-Ahen, O.M.; Ali, A.; Munir, M.; Idrees, M.; Mirza, M.U.; Vanmeert, M.; Shah, S.Z.; Jabbar, I. Antigenic Peptide Prediction From E6 and E7 Oncoproteins of HPV Types 16 and 18 for Therapeutic Vaccine Design Using Immunoinformatics and MD Simulation Analysis. Front. Immun. 2018, 9. [Google Scholar] [CrossRef]
- Mirza, M.U.; Rafique, S.; Ali, A.; Munir, M.; Ikram, N.; Manan, A.; Salo-Ahen, O.M.; Idrees, M. Towards peptide vaccines against Zika virus: Immunoinformatics combined with molecular dynamics simulations to predict antigenic epitopes of Zika viral proteins. Sci. Rep. 2016, 6, 37313. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Roe, D.R.; Cheatham III, T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Gohlke, H.; Kiel, C.; Case, D.A. Insights into protein–protein binding by binding free energy calculation and free energy decomposition for the Ras–Raf and Ras–RalGDS complexes. J. Mol. Biol. 2003, 330, 891–913. [Google Scholar] [CrossRef]
- Hayes, J.M.; Archontis, G. MM-GB (PB) SA calculations of protein-ligand binding free energies. In Molecular Dynamics-Studies of Synthetic and Biological Macromolecules; InTech: The Philippines, 2012. [Google Scholar]
- Nagai, K.; Oubridge, C.; Kuglstatter, A.; Menichelli, E.; Isel, C.; Jovine, L. Structure, function and evolution of the signal recognition particle. EMBO J. 2003, 22, 3479–3485. [Google Scholar] [CrossRef]
- Frappier, V.; Chartier, M.; Najmanovich, R.J. ENCoM server: Exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Res. 2015, 43, W395–W400. [Google Scholar] [CrossRef]
- Massova, I.; Kollman, P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000, 18, 113–135. [Google Scholar] [CrossRef]
- Lee, P.A.; Tullman-Ercek, D.; Georgiou, G. The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 2006, 60, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Kadokura, H.; Beckwith, J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell 2009, 138, 1164–1173. [Google Scholar] [CrossRef]
- Coulthurst, S.J.; Lilley, K.S.; Hedley, P.E.; Liu, H.; Toth, I.K.; Salmond, G.P. DsbA plays a critical and multifaceted role in the production of secreted virulence factors by the phytopathogen Erwinia carotovora subsp. atroseptica. J. Biol. Chem. 2008, 283, 23739–23753. [Google Scholar] [CrossRef] [PubMed]
- Kadokura, H.; Tian, H.; Zander, T.; Bardwell, J.C.; Beckwith, J. Snapshots of DsbA in action: Detection of proteins in the process of oxidative folding. Science 2004, 303, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K.; Mukhopadhyay, R.; Mukhija, R.; Krishnan, A.; Garg, L.; Panda, A.K. Optimization of inclusion body solubilization and renaturation of recombinant human growth hormone from Escherichia coli. Prot. Exp. Purif. 2000, 18, 182–192. [Google Scholar] [CrossRef]
- Sadaf, S.; Khan, M.; Wilson, D.; Akhtar, M. Molecular cloning, characterization, and expression studies of water buffalo (Bubalus bubalis) somatotropin. Biochemistry 2007, 72, 162–169. [Google Scholar] [CrossRef]
- Huang, J.-F.; Xu, Y.-M.; Hao, D.-M.; Huang, Y.-B.; Liu, Y.; Chen, Y. Structure-guided de novo design of α-helical antimicrobial peptide with enhanced specificity. Pure Appl. Chem. 2010, 82, 243–257. [Google Scholar] [CrossRef]
- Bensing, B.A.; Siboo, I.R.; Sullam, P.M. Glycine residues in the hydrophobic core of the GspB signal sequence route export toward the accessory Sec pathway. J. Bacteriol. 2007, 189, 3846–3854. [Google Scholar] [CrossRef] [PubMed]
- Jomaa, A.; Boehringer, D.; Leibundgut, M.; Ban, N. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 2016, 7, 10471. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, P.; Draycheva, A.; Vogt, A.; Petriman, N.-A.; Sturm, L.; Drepper, F.; Warscheid, B.; Wintermeyer, W.; Koch, H.-G. Ribosome binding induces repositioning of the signal recognition particle receptor on the translocon. J. Cell Biol. 2015, 211, 91–104. [Google Scholar] [CrossRef] [Green Version]
| Primer Name | Nucleotide Sequence |
|---|---|
| FP-1 | 5’-CAT ATG AAA AAG ATT TGG CTG GCG CTG GCT GGT TTA GTT TTA GCG TTT AGC GCA TCG GCG GCC TTC CCA GCC ATG TCC-3’ |
| FP-2 | 5’-CAT ATG AAA AAG ATT TGG CTG ATT CTG GCT GGT TTA GTT TTA GCG TTT AGC GCA TCG GCG GCC TTC CCA GCC ATG TCC-3’ |
| FP-3 | 5’-CAT ATG AAA AAG ATT TGG CTG ATT CTG ATT GGT TTA GTT TTA ATT TTT AGC ATT TCG GCG GCC TTC CCA GCC ATG TCC-3’ |
| FP-4 | 5’-CAT ATG AAA AAG ATT TGG CTG ATT CTG ATT GGT TTA GTT TTA GCG TTT AGC GCA TCG GCG GCC TTC CCA GCC ATG TCC-3’ |
| FP-5 | 5’-CAT ATG AAA AAG ATT TGG CTG GCG CTG GCT GGT TTA GTT TTA ATT TTT AGC GCA TCG GCG GCC TTC CCA GCC ATG TCC-3’ |
| FP-6 | 5’-CAT ATG AAA AAG ATT TGG CTG GCG CTG GCT GGT TTA GTT TTA ATT TTT AGC ATT TCG GCG GCC TTC CCA GCC ATG TCC-3’ |
| FP-7 | 5’-CAT ATG AGAAGG ATT TGG CTG GCG CTG GCT GGT TTA GTT TTA GCG TTT AGC GCA TCG GCG GCC TTC CCA GCC ATG TCC-3’ |
| FP-8 | 5’-CAT ATG AAA AAG ATT TGG CTG GCG CTG GCT GGT TTA GTT TTA GCG TTT TGT GCA TGT GCG GCC TTC CCA GCC ATG TCC-3’ |
| pOGH Constructs | DsbA Signal Sequence | Description | Hydropathy Index | Approximate Size of Expressed rOGH (kDa) |
|---|---|---|---|---|
| −1 |  | Native DsbAss | 1.389 | 22 |
| −2 | 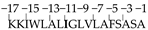 | −11 Ala → Ile | 1.539 | 22 |
| −3 | KKIWLILIGLVLIFSISA | −3, −6, −11 and −13 Ala → Ile | 1.989 | 25 |
| −4 | KKIWLILIGLVLAFSASA | −11 and −13 Ala → Ile | 1.689 | UD |
| −5 | KKIWLILAGLVLAFSASA | −13 Ala → Ile | 1.539 | 25 |
| −6 | KKIWLALAGLVLIFSISA | −3 and −6 Ala → Ile | 1.689 | 22 |
| −7 | RRIWLALAGLVLAFSASA | −17 and −18 Lys → Arg | 1.321 | 22 |
| −8 | KKIWLALAGLVLAFCACA | −2 and −4 Ser → Cys | 1.531 | 25 |
| Contributions (Total Binding Free Energy) | pOGH-2 | pOGH-5 | 3NDB (without F-loop) | 3NDB (with F-loop) |
|---|---|---|---|---|
| ΔEele | −117.62 | −135.43 | −89.36 | −118.44 |
| ΔEvdw | −87.36 | −99.48 | −79.45 | −95.01 |
| ΔEMM | −204.98 | −234.91 | −168.81 | −213.45 |
| ΔGp | 95.59 | 109.7 | 89.4 | 104.8 |
| ΔGnp | −14.85 | −15.41 | −14.47 | −18.76 |
| ΔGsol | 80.74 | 94.29 | 74.93 | 86.04 |
| ΔGtol | −124.24 | −140.62 | −93.88 | −127.41 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durrani, F.G.; Gul, R.; Mirza, M.U.; Kaderbhai, N.N.; Froeyen, M.; Saleem, M. Mutagenesis of DsbAss is Crucial for the Signal Recognition Particle Mechanism in Escherichia coli: Insights from Molecular Dynamics Simulations. Biomolecules 2019, 9, 133. https://doi.org/10.3390/biom9040133
Durrani FG, Gul R, Mirza MU, Kaderbhai NN, Froeyen M, Saleem M. Mutagenesis of DsbAss is Crucial for the Signal Recognition Particle Mechanism in Escherichia coli: Insights from Molecular Dynamics Simulations. Biomolecules. 2019; 9(4):133. https://doi.org/10.3390/biom9040133
Chicago/Turabian StyleDurrani, Faiza Gul, Roquyya Gul, Muhammad Usman Mirza, Naheed Nazly Kaderbhai, Matheus Froeyen, and Mahjabeen Saleem. 2019. "Mutagenesis of DsbAss is Crucial for the Signal Recognition Particle Mechanism in Escherichia coli: Insights from Molecular Dynamics Simulations" Biomolecules 9, no. 4: 133. https://doi.org/10.3390/biom9040133
APA StyleDurrani, F. G., Gul, R., Mirza, M. U., Kaderbhai, N. N., Froeyen, M., & Saleem, M. (2019). Mutagenesis of DsbAss is Crucial for the Signal Recognition Particle Mechanism in Escherichia coli: Insights from Molecular Dynamics Simulations. Biomolecules, 9(4), 133. https://doi.org/10.3390/biom9040133






