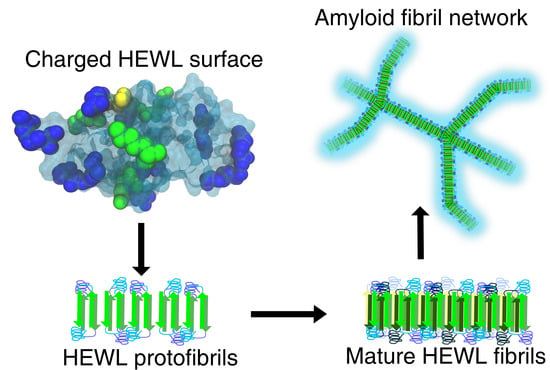
The Role of Buffers in Wild-Type HEWL Amyloid Fibril Formation Mechanism
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Buffers
2.3. Fibril Preparation
2.4. UV Measurements
2.5. Fluorescence Quenching
2.6. Circular Dichroism (CD)
2.7. Differential Scanning Calorimetry (DSC)
3. Results
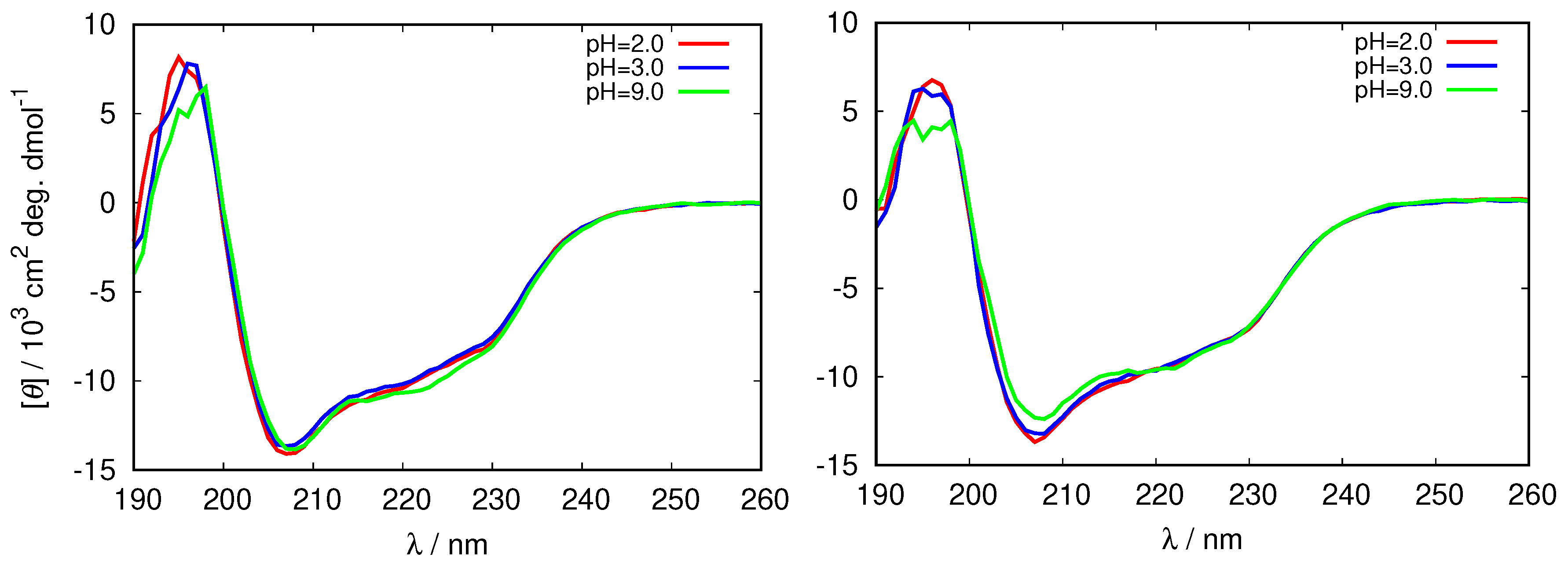
3.1. HEWL Molecular Structure and Stability Depend on the Buffer Identity
3.2. The Amyloid Fibril Formation
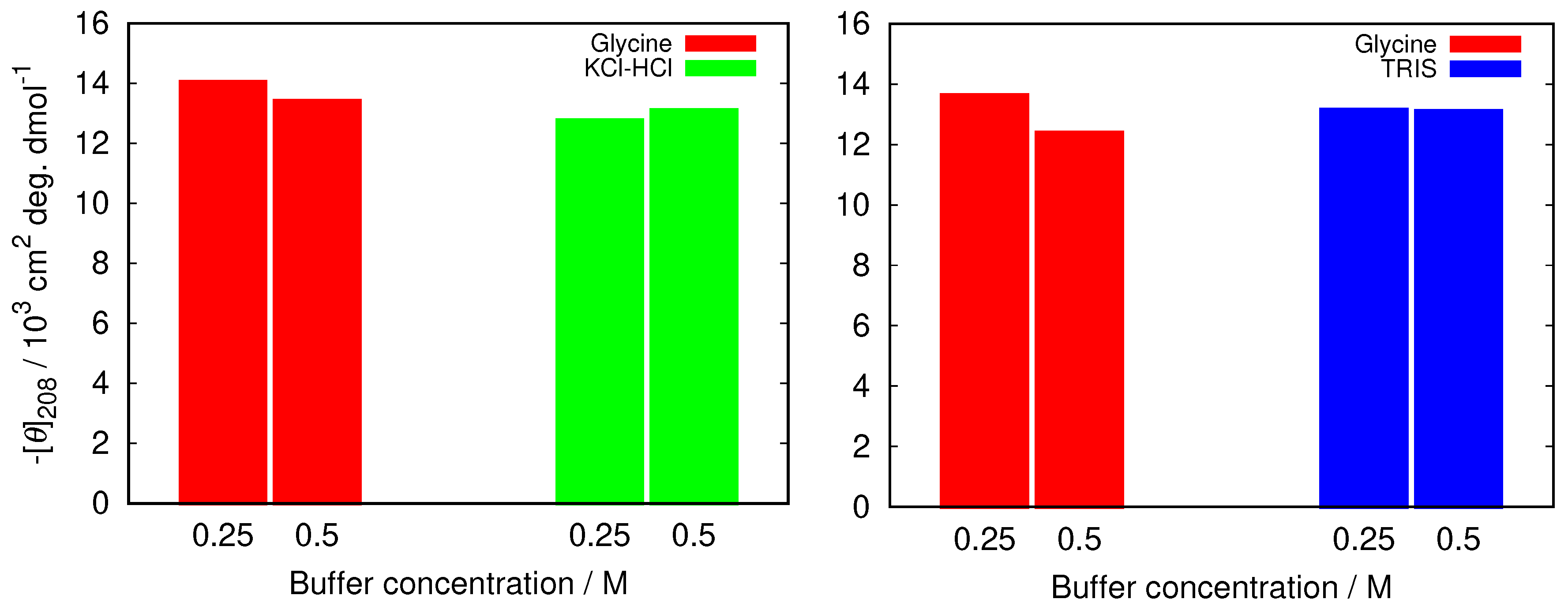
3.3. Electrostatic Screening Facilitates Fibrillization
3.4. Binding Inert Molecules Inhibits HEWL Fibrillization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HEWL | Hen egg-white lysozyme |
| CD | Circular dichroism |
| DSC | Differential scanning calorimetry |
| ThT | Thioflavin T |
| TRIS | tris(hydroxymethyl)aminomethane |
| HEPES | 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid |
| ANS | 8-(Phenylamino)-1-naphthalenesulfonic acid |
| PEG | Polyethylene glycol |
References
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddel, N.; Ramponi, G.; Dobson, C.; Stefani, M. Inherent Toxicity of Aggregates Implies a Common Mechanism for Protein Misfolding Diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, R.; Ravi, V.K.; Kumar, S.; Kumar, M.; Chandra, N. Lysozyme: A model protein for amyloid research. Adv. Protein Chem. Struct. Biol. 2011, 84, 63–111. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Poirier, M.A. Protein Aggregation and Neurodegenerative Disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef] [PubMed]
- Colvin, M.T.; Silvers, R.; Ni, Q.Z.; Can, T.V.; Sergeyev, I.; Rosay, M.; Donovan, K.J.; Michael, B.; Wall, J.; Linse, S.; et al. Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J. Am. Chem. Soc. 2016, 138, 9663–9674. [Google Scholar] [CrossRef] [PubMed]
- Trexler, A.J.; Nilsson, M.R. The Formation of Amyloid Fibrils from Proteins in the Lysozyme Family. Curr. Protein Pept. Sci. 2007, 8, 537–557. [Google Scholar] [CrossRef] [PubMed]
- Sivalingam, V.; Prasanna, N.L.; Sharma, N.; Prasad, A.; Patel, B.K. Wild-type hen egg white lysozyme aggregation in vitro can form self-seeding amyloid conformational variants. Biophys. Chem. 2016, 219, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Bayro, M.J.; Debelouchina, G.T.; Eddy, M.T.; Birkett, N.R.; MacPhee, C.E.; Rosay, M.; Maas, W.E.; Dobson, C.M.; Griffin, R.G. Intermolecular Structure Determination of Amyloid Fibrils with Magic-Angle Spinning and Dynamic Nuclear Polarization NMR. J. Am. Chem. Soc. 2011, 133, 13967–13974. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Do, T.D.; Hayden, E.Y.; Teplow, D.B.; Bowers, M.T.; Shea, J.E. Aggregation of Chameleon Peptides: Implications of α-Helicity in Fibril Formation. J. Phys. Chem. B 2016, 120, 5874–5883. [Google Scholar] [CrossRef]
- Do, T.D.; Chamas, A.; Zheng, X.; Barnes, A.; Chang, D.; Veldstra, T.; Takhar, H.; Dressler, N.; Trapp, B.; Miller, K.; et al. Elucidation of the Aggregation Pathways of Helix-Turn-Helix Peptides: Stabilization at the Turn Region Is Critical for Fibril Formation. Biochemistry 2015, 54, 4050–4062. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, P.; Boserman, P.; van der Vegt, N.F.A.; Shea, J.E. Trimethylamine N-oxide Counteracts Urea Denaturation by Inhibiting Protein-Urea Preferential Interaction. J. Am. Chem. Soc. 2018, 140, 483–492. [Google Scholar] [CrossRef]
- Schwierz, N.; Horinek, D.; Sivan, U.; Netz, R.R. Reversed Hofmeister series—The rule rather than the exception. Curr. Opin. Colloid Interface Sci. 2016, 23, 10–18. [Google Scholar] [CrossRef]
- Bastos-González, D.; Pérez-Fuentes, L.; Drummondb, C.; Faraudo, J. Ions at interfaces: The central role of hydration and hydrophobicity. Curr. Opin. Colloid Interface Sci. 2016, 23, 19–28. [Google Scholar] [CrossRef]
- Kuehner, D.E.; Engmann, J.; Fergg, F.; Wernick, M.; Blanch, H.W.; Prausnitz, J.M. Lysozyme Net Charge and Ion Binding in Concentrated Aqueous Electrolyte Solutions. J. Phys. Chem. B 1999, 103, 1368–1374. [Google Scholar] [CrossRef]
- Camilloni, C.; Rocco, A.G.; Eberini, I.; Gianazza, E.; Broglia, R.; Tiana, G. Urea and Guanidinium Chloride Denature Protein L in Different Ways in Molecular Dynamics Simulations. Biophys. J. 2008, 94, 4654–4661. [Google Scholar] [CrossRef] [PubMed]
- Brandts, J.F.; Hunt, L. Thermodynamics of protein denaturation. III. Denaturation of ribonuclease in water and in aqueous urea and aqueous ethanol mixtures. J. Am. Chem. Soc. 1967, 89, 4826–4838. [Google Scholar] [CrossRef] [PubMed]
- Satish, L.; Millan, S.; Das, S.; Jena, S.; Sahoo, H. Thermal Aggregation of Bovine Serum Albumin in Conventional Buffers: An Insight into Molecular Level Interactions. J. Solut. Chem. 2017, 46, 831–848. [Google Scholar] [CrossRef]
- Zbacnik, T.; Holcomb, R.; Katayama, D.; Murphy, B.; Payne, R.; Coccaro, R.; Evans, G.; Matsuura, J.; Henry, C.; Manning, M. Role of Buffers in Protein Formulations. J. Pharm. Sci. 2017, 106, 713–733. [Google Scholar] [CrossRef]
- Salis, A.; Monduzzi, M. Not only pH. Specific buffer effects in biological systems. Curr. Opin. Colloid Interface Sci. 2016, 23, 1–9. [Google Scholar] [CrossRef]
- Katayama, D.; Nayar, R.; Chou, D.; Valente, J.; Cooper, J.; Henry, C.; Velde, D.V.; Villarete, L.; Liu, C.; Manning, M. Effect of buffer species on the thermally induced aggregation of interferon-tau. J. Pharm. Sci. 2006, 95, 1212–1226. [Google Scholar] [CrossRef]
- Metrick, M.; Temple, J.; MacDonald, G. The effects of buffers and pH on the thermal stability, unfolding and substrate binding of RecA. Biophys. Chem. 2013, 184, 29–36. [Google Scholar] [CrossRef]
- Roberts, D.; Keeling, R.; Tracka, M.; van der Walle, C.; Uddin, S.; Warwicker, J.; Curtis, R. Specific Ion and Buffer Effects on Protein-Protein Interactions of a Monoclonal Antibody. Mol. Pharm. 2015, 12, 179–193. [Google Scholar] [CrossRef]
- Cugia, F.; Monduzzi, M.; Ninham, B.W.; Salis, A. Interplay of ion specificity, pH and buffers: Insights from electrophoretic mobility and pH measurements of lysozyme solutions. RSC Adv. 2013, 3, 5882–5888. [Google Scholar] [CrossRef]
- Baldwin, R.L. How Hofmeister ion interactions affect protein stability. Biophys. J. 1996, 71, 2056–2063. [Google Scholar] [CrossRef]
- Vrbka, L.; Jungwirth, P.; Bauduin, P.; Touraud, D.; Kunz, W. Specific Ion Effects at Protein Surfaces: A Molecular Dynamics Study of Bovine Pancreatic Trypsin Inhibitor and Horseradish Peroxidase in Selected Salt Solutions. J. Phys. Chem. B 2006, 110, 7036–7043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cremer, P.S. The inverse and direct Hofmeister series for lysozyme. Proc. Natl. Acad. Sci. USA 2009, 106, 15249–15253. [Google Scholar] [CrossRef] [PubMed]
- Metrick, M.A.; MacDonald, G. Hofmeister ion effects on the solvation and thermal stability of model proteins lysozyme and myoglobin. Colloids Surf. A Physicochem. Eng. Asp. 2015, 469, 242–251. [Google Scholar] [CrossRef]
- Garajova, K.; Balogova, A.; Dusekovs, E.; Sedlakova, D.; Sedlak, E.; Varhac, R. Correlation of lysozyme activity and stability in the presence of Hofmeister series anions. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Poniková, S.; Antošová, A.; Demjén, E.; Sedláková, D.; Marek, J.; Varhač, R.; Gažová, Z.; Sedlák, E. Lysozyme stability and amyloid fibrillization dependence on Hofmeister anions in acidic pH. J. Biol. Inorg. Chem. 2015, 20, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.R.H.; Wilkins, D.K.; Chung, E.W.; Pitkeathly, M.C.; Chamberlain, A.K.; Zurdo, J.; Robinson, C.V.; Dobson, C.M. Formation and Seeding of Amyloid Fibrils form Wild-type Hen Lysozyme and a Peptide Fragment from the β-Domain. J. Mol. Biol. 2000, 300, 541–549. [Google Scholar] [CrossRef]
- Wong, A.G.; Wu, C.; Hannaberry, E.; Watson, M.D.; Shea, J.E.; Raleigh, D.P. Analysis of the Amyloidogenic Potential of Pufferfish (Takifugu rubripes) Islet Amyloid Polypeptide Highlights the Limitations of Thioflavin-T Assays and the Difficulties in Defining Amyloidogenicity. Biochemistry 2016, 55, 510–518. [Google Scholar] [CrossRef]
- Groenning, M.; Norrman, M.; Flink, J.M.; van de Weert, M.; Bukrinsky, J.T.; Schluckebier, G.; Frokjaer, S. Binding mode of Thioflavin T in insulin amyloid fibrils. J. Struct. Biol. 2007, 159, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Hawe, A.; Sutter, M.; Jiskoot, W. Extrinsic Fluorescent Dyes as Tools for Protein Characterization. Pharm. Res. 2008, 25, 1487–1499. [Google Scholar] [CrossRef]
- Kelly, S.M.; Price, N.C. The Use of Circular Dichroism in the Investigation of Protein Structure and Function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.; Goto, Y.; Refregiers, M.; Kardos, J. Accuarate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.H.; Prenner, E.J. Differential scanning calorimetry: An invaluable tool for a detailed thermodynamic characterization of macromolecules and their interactions. J. Pharm. Bioall. Sci. 2011, 3, 39–59. [Google Scholar] [CrossRef]
- Phillips, D.C. The Hen Egg-White Lysozyme Molecule. Proc. Natl. Acad. Sci. USA 1967, 57, 483–495. [Google Scholar] [CrossRef]
- Wetter, L.R.; Deutsch, H.F. Immunological Studies on Egg White Proteins: IV. Immunochemical and Physical Studies of Lysoyme. J. Biol. Chem. 1951, 192, 237–242. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Artymiuk, P.; Blake, C.C.F.; Rice, D.W.; Wilson, K.S. The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Å Resolution. Acta Cryst. 1982, B38, 778–783. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New Ways to Boost Molecular Dynamics Simulation. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Case, D.; Babin, V.; Berryman, J.; Betz, R.; Cai, Q.; Cerutti, D.; Cheatham, T.E., III; Darden, T.; Duke, R.; Gohlke, H.; et al. Amber 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Pannuru, P.; Rani, A.; Venkatesu, P.; Lee, M.J. The effects of biological buffers TRIS, TAPS, TES on the stability of lysozyme. Int. J. Biol. Macromol. 2018, 112, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Klotz, I.M.; Hunston, D.L. Properties of graphical representations of multiple classes of binding sites. Biochemistry 1971, 10, 3065–3069. [Google Scholar] [CrossRef] [PubMed]
- Danilenko, A.; Dianova, V.; Braudo, E.; Henning, T.; Mothes, R.; Schwenke, K. A novel approach to the evaluation of hydrophobicity of food proteins. Nahrung 1998, 42, 179–182. [Google Scholar] [CrossRef]
- Frid, P.; Anisimov, S.V.; Popovic, N. Congo red and protein aggregation in neurodegenerative diseases. Brain Res. Rev. 2006, 53, 135–160. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhao, C.; Lin, W.; Hu, R.; Wang, Q.; Chen, H.; Li, L.; Chen, S.; Zheng, J. Binding characteristics between polyethylene glycol (PEG) and proteins in aqueous solution. J. Mater. Chem. B 2014, 2, 2983–2992. [Google Scholar] [CrossRef]
- Kirincic, S.; Klofutar, C. Viscosity of aqueous solutions of poly(ethylene glycol)s at 298.15 K. Fluid Phase Equilib. 1999, 155, 311–325. [Google Scholar] [CrossRef]
- Felice, F.G.D.; Vieira, M.N.N.; Nazareth, M.; Meirelles, L.; Morozova-Roche, L.A.; Dobson, C.M.; Ferreira, S.T. Formation of amyloid aggregates from human lysozyme and its disease-associated variants using hydrostatic pressure. FASEB J. 2004, 18, 1099–1101. [Google Scholar] [CrossRef]
- Aso, Y.; Shiraki, K.; Takagi, M. Systematic Analysis of Aggregates from 38 Kinds of Non Disease-Related Proteins: Identifying the Intrinsic Propensity of Polypeptides to Form Amyloid Fibrils. Biosci. Biotechnol. Biochem. 2007, 71, 1313–1321. [Google Scholar] [CrossRef]
- Mishra, R.; Soergjerd, K.; Nystroem, S.; Nordigården, A.; Yu, Y.C.; Hammarstroem, P. Lysozyme Amyloidogenesis Is Accelerated by Specific Nicking and Fragmentation but Decelerated by Intact Protein Binding and Conversion. J. Mol. Biol. 2007, 366, 1029–1044. [Google Scholar] [CrossRef]
- Petkova, A.; Leapman, R.; Guo, Z.; Yau, W.M.; Mattson, M.; Tycko, R. Self-Propagating, Molecular-Level Polymorphism in Alzheimer’s β-Amyloid Fibrils. Science 2005, 307, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.K.; Liebman, S.W. “Prion-proof” for [PIN+]: Infection with In Vitro-made Amyloid Aggregates of Rnq1p-(132-405) Induces [PIN+]. J. Mol. Biol. 2007, 365, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, J.K.; Bhat, R. A mechanistic analysis of the increase in the thermal stability of proteins in aqueous carboxylic acid salt solutions. Protein Sci. 1999, 8, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Garidel, P.; Blume, A.; Wagner, M. Prediction of colloidal stability of high concentration protein formulations. Pharm. Dev. Technol. 2015, 20, 367–374. [Google Scholar] [CrossRef]
- Gupta, B.S.; Taha, M.; Lee, M.J. Buffers more than buffering agent: introducing a new class of stabilizers for the protein BSA. Phys. Chem. Chem. Phys. 2015, 17, 1114–1133. [Google Scholar] [CrossRef]
- Hamada, H.; Arakawa, T.; Shiraki, K. Effect of Additives on Protein Aggregation. Curr. Pharm. Biotechnol. 2009, 10, 400–407. [Google Scholar] [CrossRef]
- Hill, S.E.; Robinson, J.; Matthews, G.; Muschol, M. Amyloid Protofibrils of Lysozyme Nucleate and Grow Via Oligomer Fusion. Biophys. J. 2009, 96, 3781–3790. [Google Scholar] [CrossRef]
- Vieira, M.N.N.; Figueroa-Villar, J.D.; Meirelles, M.N.L.; Ferreira, S.T.; Felice, F.G.D. Small molecule inhibitors of lysozyme amyloid aggregation. Cell Biochem. Biophys. 2006, 44, 549–553. [Google Scholar] [CrossRef]
- Yamin, G.; Munishkina, L.A.; Karymov, M.A.; Lyubchenko, Y.L.; Uversky, V.N.; Fink, A.L. Forcing Nonamyloidogenic α-Synuclein To Fibrillate. Biochemistry 2005, 44, 9096–9107. [Google Scholar] [CrossRef]
- Hatters, D.M.; Minton, A.P.; Howlett, G.J. Macromolecular Crowding Accelerates Amyloid Formation by Human Apolipoprotein C-II*. J. Biol. Chem. 2002, 277, 7824–7830. [Google Scholar] [CrossRef]
- Zhou, B.R.; Zhou, Z.; Hu, Q.L.; Chen, J.; Liang, Y. Mixed macromolecular crowding inhibits amyloid formation of hen egg white lysozyme. Biochim. Biophys. Acta 2008, 1784, 472–480. [Google Scholar] [CrossRef] [PubMed]
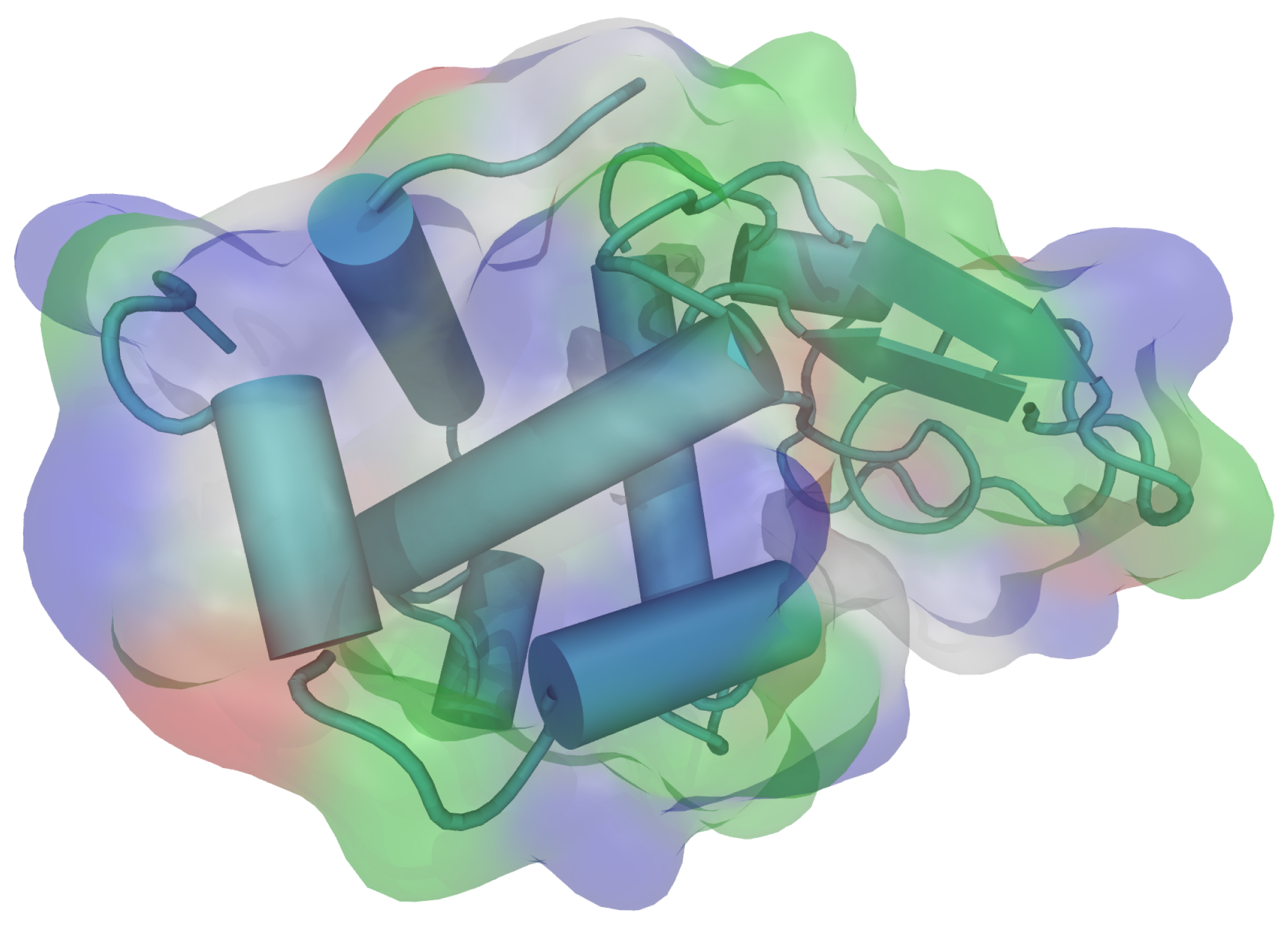
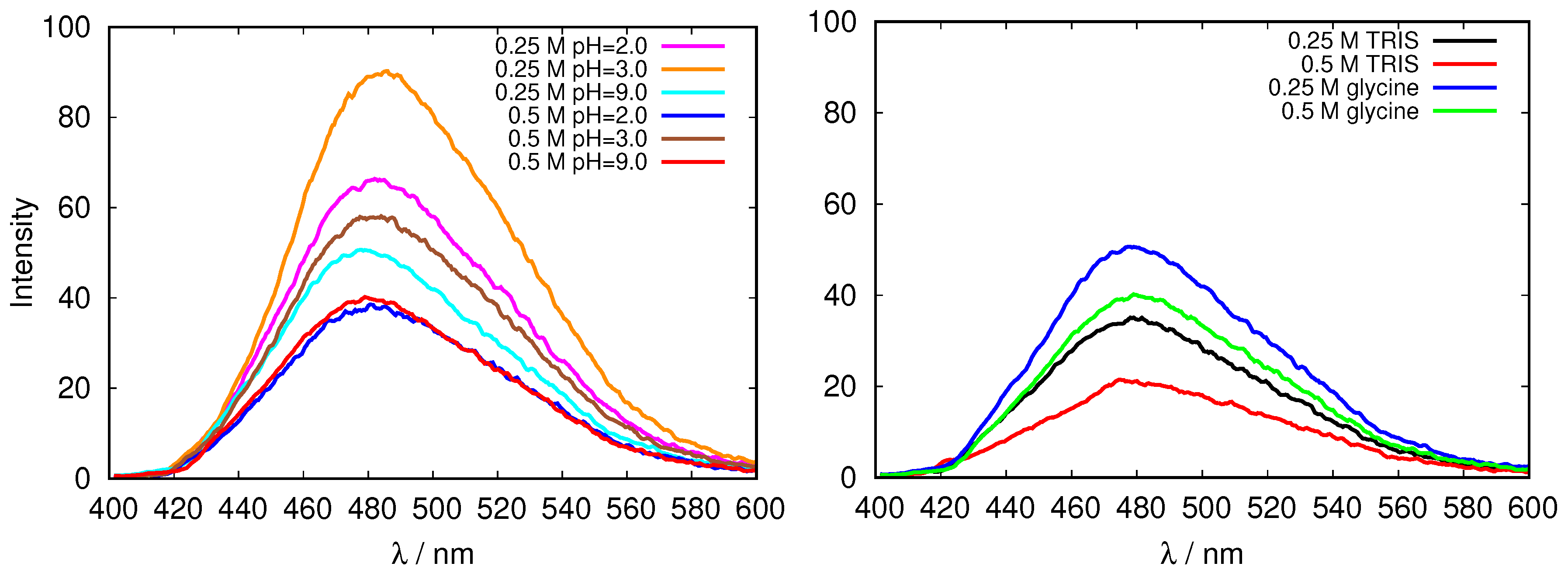
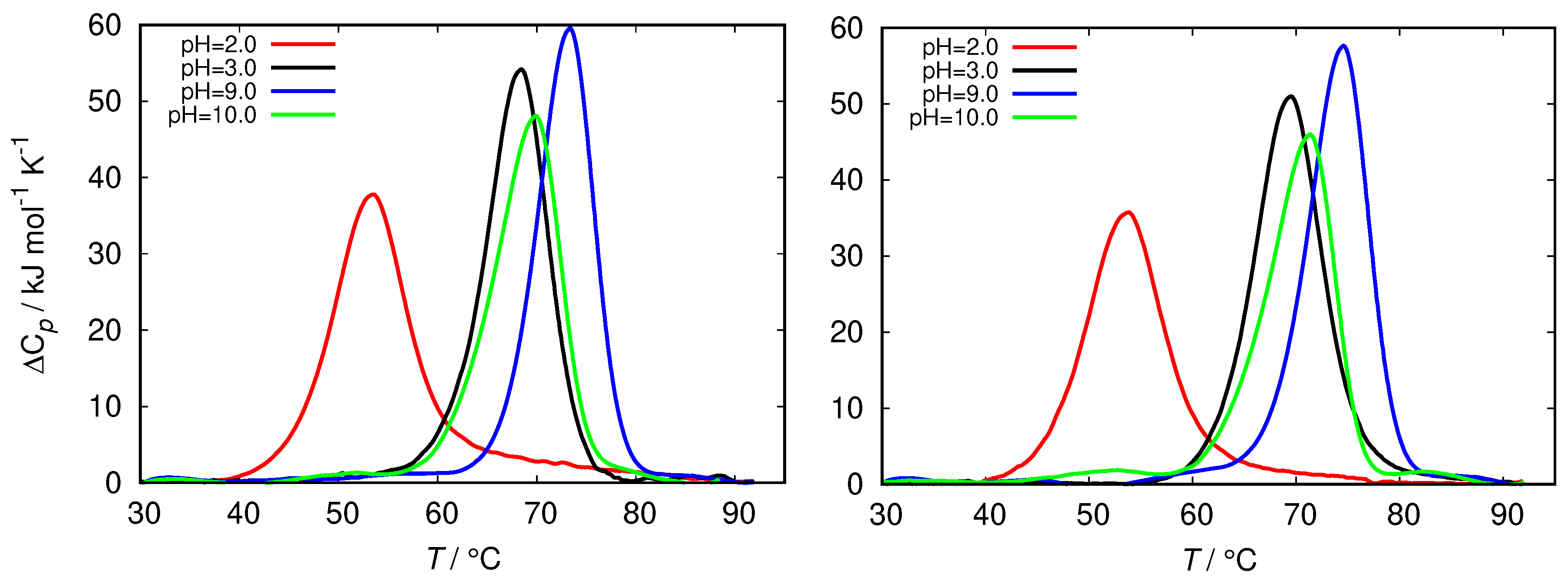
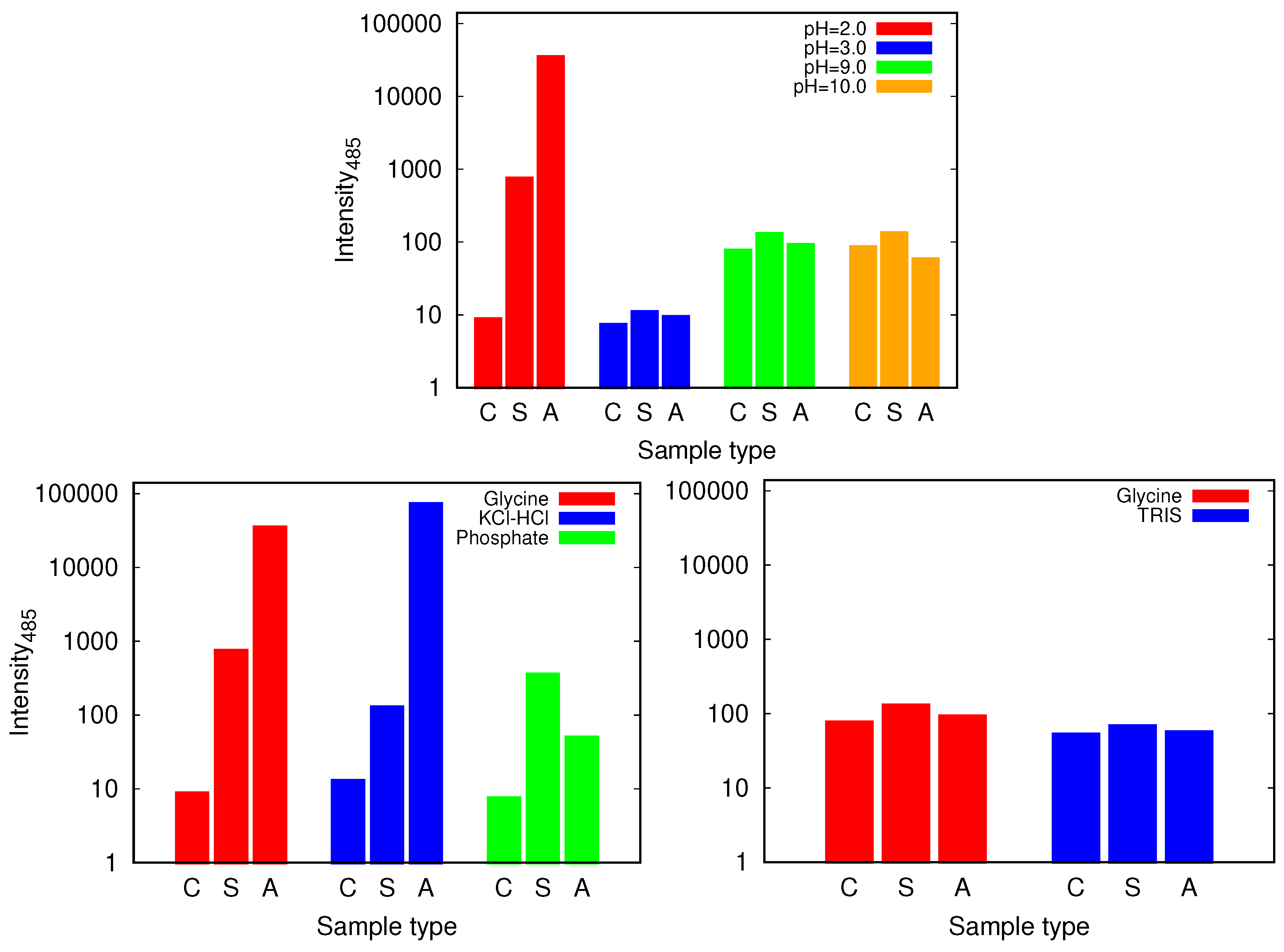
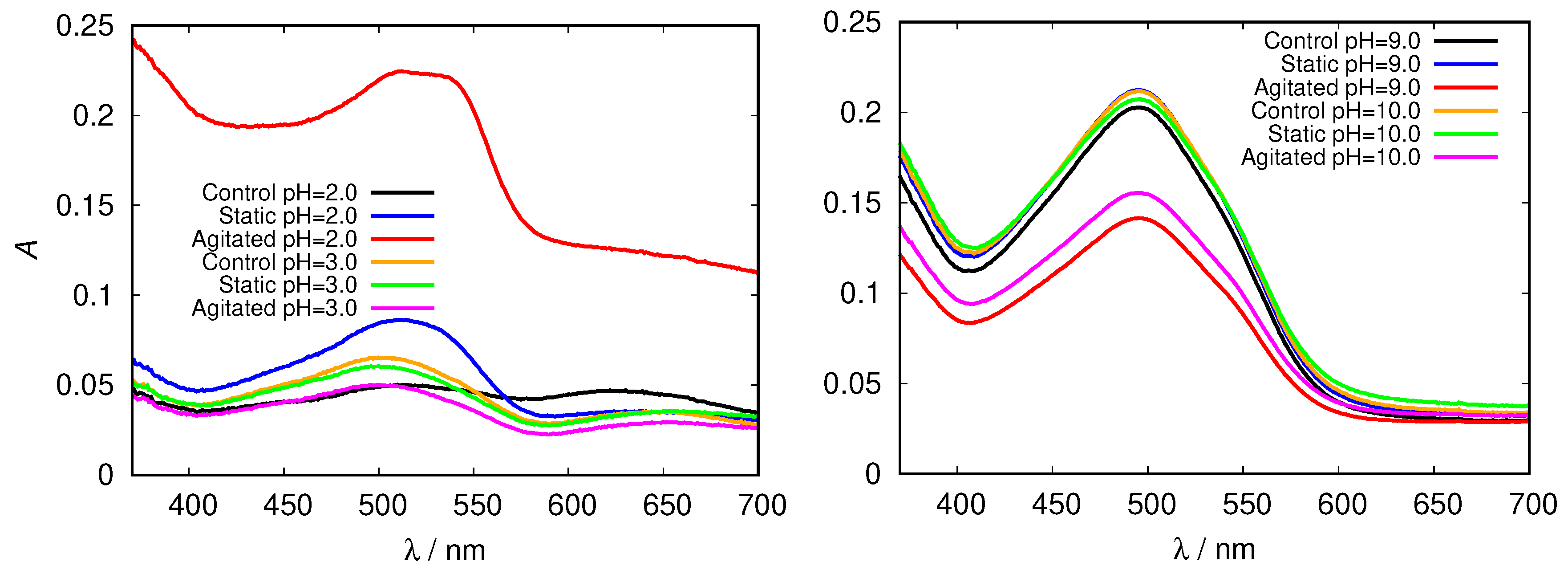
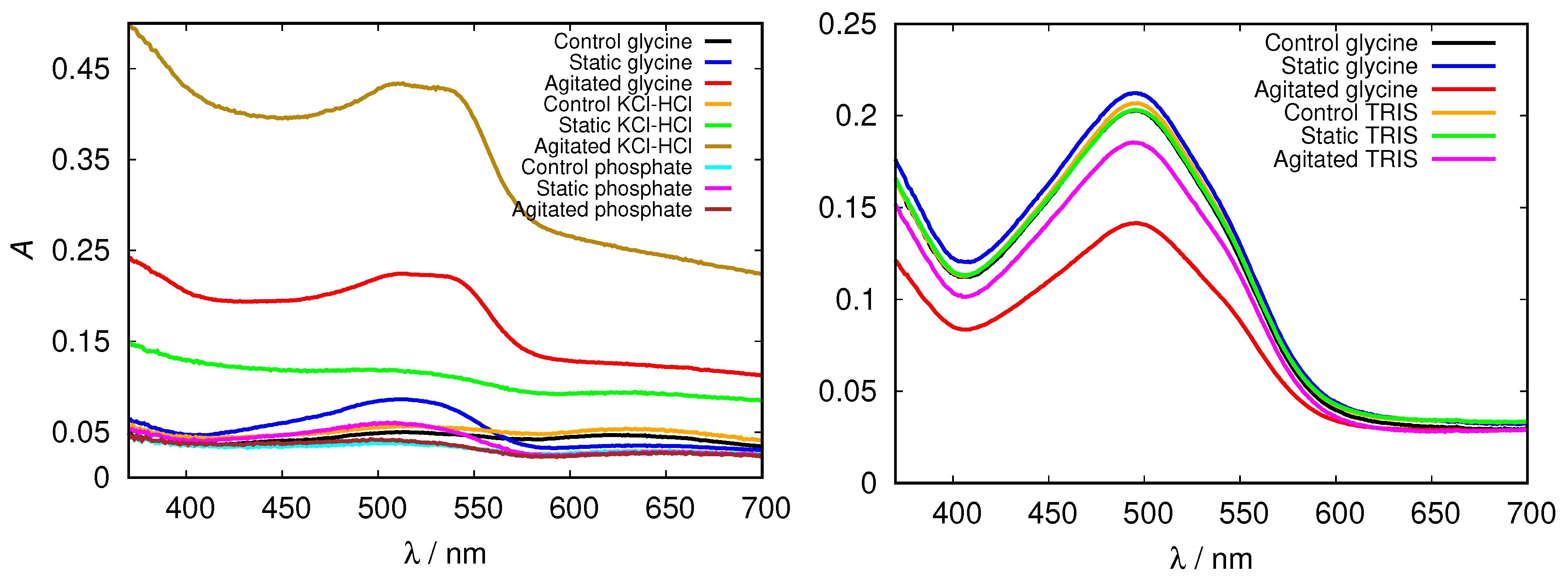
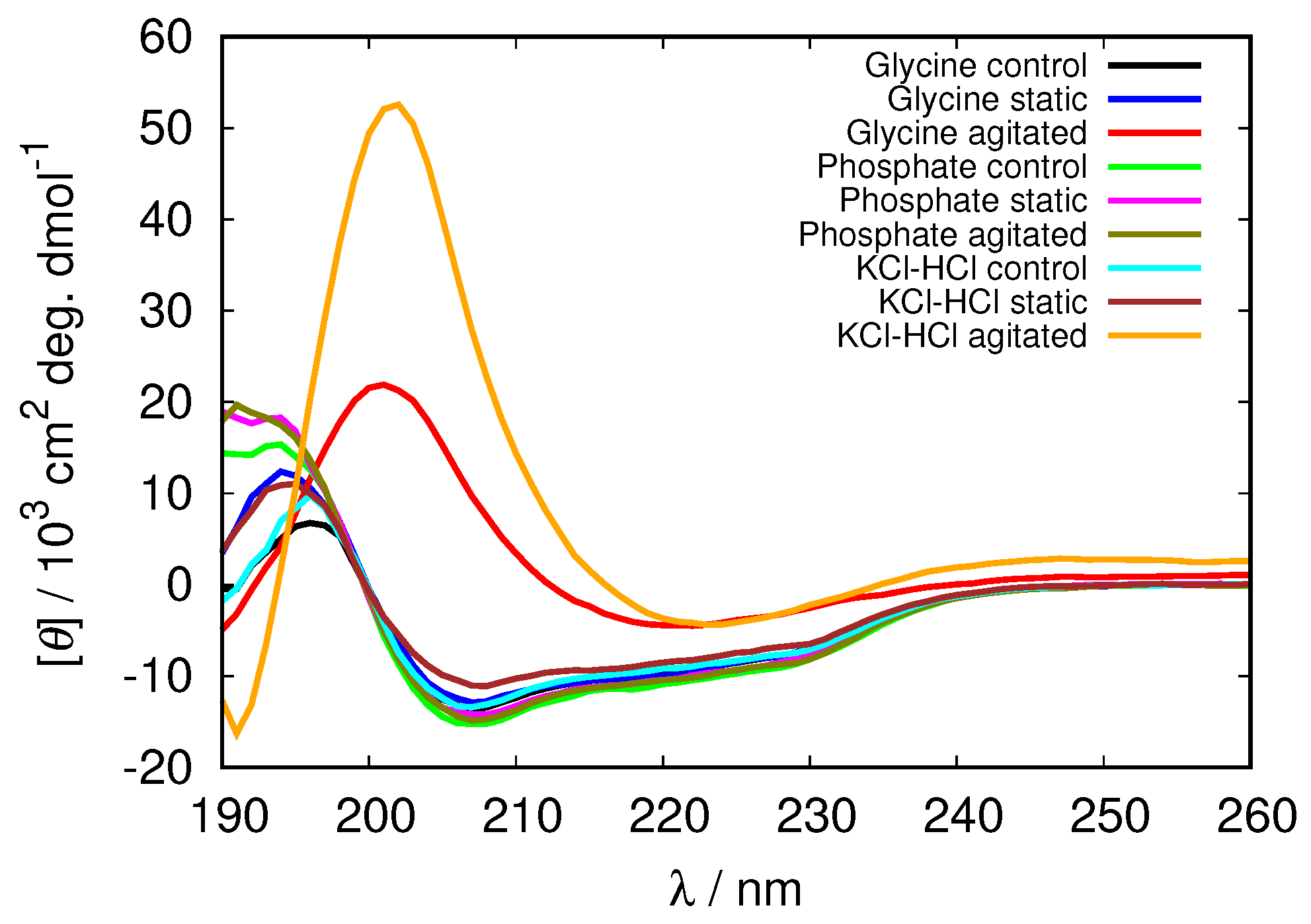
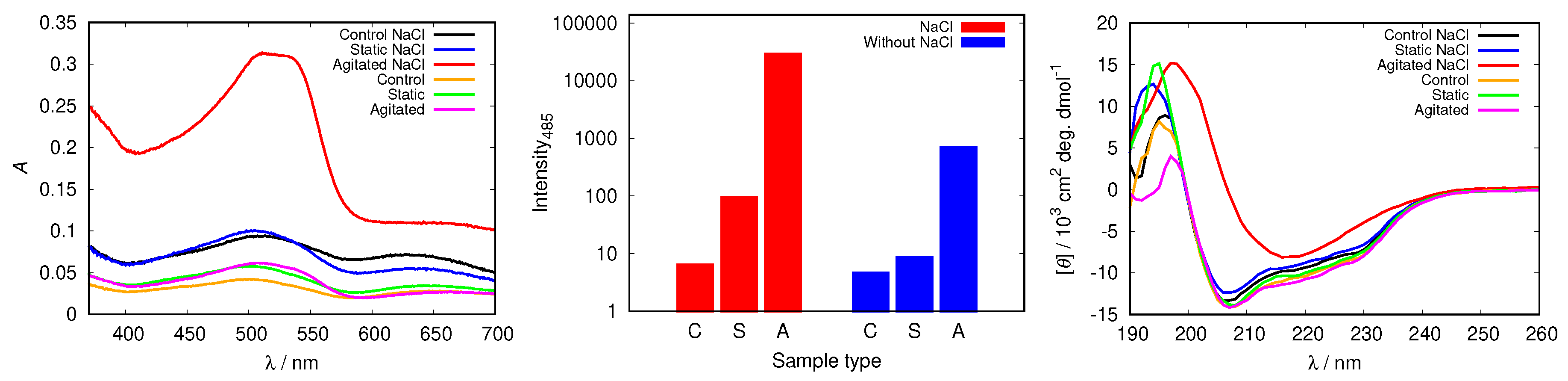

| pH Value | Net Charge | Positive Amino Acids | Negative Amino Acids |
|---|---|---|---|
| 2.0 | +18.0 | 11 Arg, 6 Lys, 1 His | / |
| 3.0 | +18.0 | 11 Arg, 6 Lys, 1 His | / |
| 4.5 | +8.0 | 11 Arg, 6 Lys, 1 His | 7 Asp, 2 Glu, 1 Leu |
| 7.0 | +7.0 | 11 Arg, 6 Lys | 7 Asp, 2 Glu, 1 Leu |
| 7.5 | +7.0 | 11 Arg, 6 Lys | 7 Asp, 2 Glu, 1 Leu |
| 8.0 | +7.0 | 11 Arg, 6 Lys | 7 Asp, 2 Glu, 1 Leu |
| 9.0 | +7.0 | 11 Arg, 6 Lys | 7 Asp, 2 Glu, 1 Leu |
| 10.0 | +7.0 | 11 Arg, 6 Lys | 7 Asp, 2 Glu, 1 Leu |
| Solution | -Helix | -Antiparallel Sheet | -Parallel Sheet | Turn |
|---|---|---|---|---|
| 0.5 M glycine, pH = 2.0 | 24 | 12 | 3 | 16 |
| 0.25 M glycine, pH = 2.0 | 29 | 8 | 3 | 11 |
| 0.5 M glycine, pH = 3.0 | 23 | 15 | 4 | 13 |
| 0.25 M glycine, pH = 3.0 | 28 | 9 | 2 | 10 |
| 0.5 M glycine, pH = 9.0 | 28 | 11 | 4 | 15 |
| 0.25 M glycine, pH = 9.0 | 30 | 10 | 1 | 11 |
| 0.5 M glycine, pH = 10.0 | 28 | 14 | 2 | 18 |
| 0.25 M glycine, pH = 10.0 | 31 | 12 | 1 | 13 |
| 0.5 M TRIS, pH = 7.0 | 24 | 12 | 5 | 14 |
| 0.25 M TRIS, pH = 7.0 | 28 | 10 | 1 | 11 |
| 0.5 M TRIS, pH = 7.5 | 25 | 9 | 4 | 16 |
| 0.25 M TRIS, pH = 7.5 | 29 | 10 | 1 | 13 |
| 0.5 M TRIS, pH = 8.0 | 25 | 10 | 5 | 15 |
| 0.25 M TRIS, pH = 8.0 | 29 | 14 | 3 | 14 |
| 0.5 M TRIS, pH = 9.0 | 31 | 13 | 2 | 17 |
| 0.25 M TRIS, pH = 9.0 | 30 | 14 | 0 | 14 |
| 0.25 M cacodylate, pH = 7.0 | 24 | 13 | 5 | 16 |
| 0.5 M cacodylate, pH = 7.0 | 27 | 15 | 2 | 16 |
| 0.1 M KCl-HCl, pH = 2.0 | 24 | 11 | 3 | 16 |
| 0.5 M KCl-HCl, pH = 2.0 | 23 | 12 | 2 | 15 |
| 0.25 M KCl-HCl, pH = 2.0 | 24 | 15 | 3 | 15 |
| 0.5 M Phosphate, pH = 2.0 | 30 | 7 | 2 | 15 |
| 0.5 M Phosphate, pH = 7.0 | 25 | 11 | 4 | 16 |
| Solution | -Helix | -Antiparallel Sheet | -Parallel Sheet | Turn |
|---|---|---|---|---|
| 0.5 M glycine (C) | 24 | 12 | 3 | 16 |
| 0.5 M glycine (S) | 23 | 10 | 5 | 15 |
| 0.5 M glycine (A) | 2 | 87 | 0 | 11 |
| 0.25 M glycine (C) | 29 | 8 | 3 | 11 |
| 0.25 M glycine (S) | 28 | 4 | 6 | 12 |
| 0.25 M glycine (A) | 33 | 15 | 0 | 12 |
| 0.1 M KCl-HCl (C) | 24 | 11 | 3 | 16 |
| 0.1 M KCl-HCl (S) | 22 | 13 | 3 | 15 |
| 0.1 M KCl-HCl (A) | 28 | 18 | 10 | 11 |
| 0.25 M KCl-HCl (C) | 24 | 15 | 3 | 15 |
| 0.25 M KCl-HCl (S) | 21 | 12 | 5 | 16 |
| 0.25 M KCl-HCl (A) | 0 | 99 | 0 | 2 |
| 0.5 M KCl-HCl (C) | 23 | 12 | 2 | 15 |
| 0.5 M KCl-HCl (S) | 19 | 18 | 5 | 14 |
| 0.5 M KCl-HCl (A) | 0 | 93 | 0 | 7 |
| 0.5 M phosphate (C) | 30 | 7 | 2 | 15 |
| 0.5 M phosphate (S) | 28 | 6 | 3 | 16 |
| 0.5 M phosphate (A) | 30 | 6 | 2 | 15 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brudar, S.; Hribar-Lee, B. The Role of Buffers in Wild-Type HEWL Amyloid Fibril Formation Mechanism. Biomolecules 2019, 9, 65. https://doi.org/10.3390/biom9020065
Brudar S, Hribar-Lee B. The Role of Buffers in Wild-Type HEWL Amyloid Fibril Formation Mechanism. Biomolecules. 2019; 9(2):65. https://doi.org/10.3390/biom9020065
Chicago/Turabian StyleBrudar, Sandi, and Barbara Hribar-Lee. 2019. "The Role of Buffers in Wild-Type HEWL Amyloid Fibril Formation Mechanism" Biomolecules 9, no. 2: 65. https://doi.org/10.3390/biom9020065
APA StyleBrudar, S., & Hribar-Lee, B. (2019). The Role of Buffers in Wild-Type HEWL Amyloid Fibril Formation Mechanism. Biomolecules, 9(2), 65. https://doi.org/10.3390/biom9020065