Paraoxonase 3: Structure and Its Role in Pathophysiology of Coronary Artery Disease
Abstract
:1. Introduction
2. Paraoxonases (PONs)
3. Paraoxonase Gene Cluster
4. Evolution of PON Genes
5. Paraoxonase 3 (PON3) 3.1.1.2
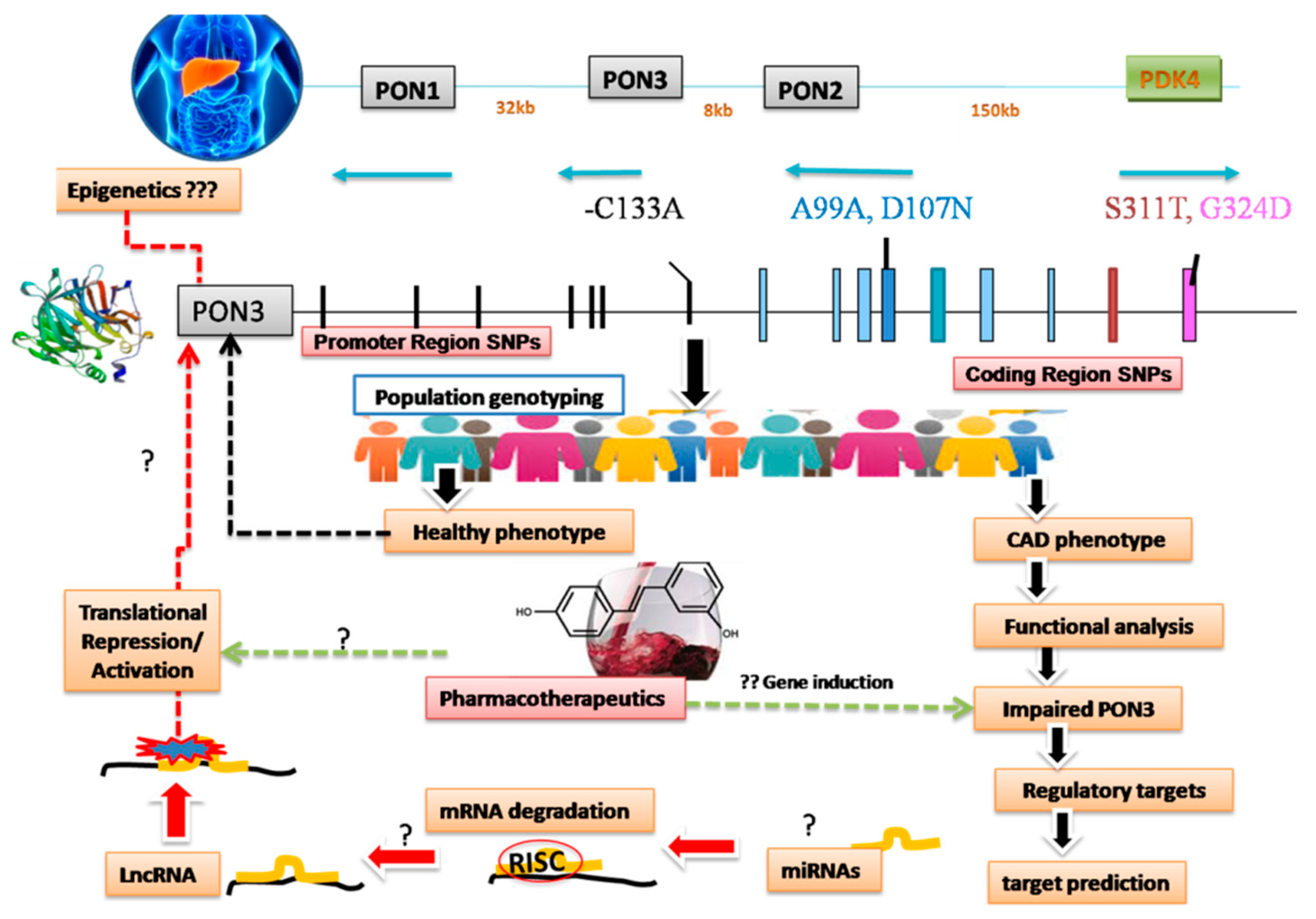
6. PON3: Single Nucleotides Variants (SNPs) and Haplotypes
7. Substrate Specificities and PON3 Status
8. Amino Acid Sequence Similarity
9. Paraoxonase3 Structure: (PDB 1v04)
10. PON3 Concentration
11. Histological Distribution
12. Antioxidant Potential
13. PON3 Regulation in Various Diseases
14. PON3 Gene Expression
15. Role of Environmental Factors
16. Summary and Conclusions
17. Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Lopez, A.D. Alternative projections of mortality and disability by cause 1990–2020: Global Burden of Disease Study. Lancet 1997, 349, 1498–1504. [Google Scholar] [CrossRef]
- Kullo, I.J.; Ding, K. Mechanisms of Disease: The genetic basis of coronary heart disease. Nat. Clin. Pract. Neurol. 2007, 4, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Enas, E.; Garg, A.; Davidson, M.A.; Nair, V.M.; Huet, B.A.; Yusuf, S. Coronary heart disease and its risk factors in first-generation immigrant Asian Indians to the United States of America. Indian Hear. J. 1996, 48, 3433–3453. [Google Scholar]
- Enas, E.A.; Yusuf, S.; Sharma, S. Coronary artery disease in South Asians. Second meeting of the International Working Group, Anaheim, California. Indian Heart J. 1998, 50, 105–113. [Google Scholar]
- McKeigue, P.M.; Ferrie, J.E.; Pierpoint, T.; Marmot, M.G.; Marmot, M. Association of early-onset coronary heart disease in South Asian men with glucose intolerance and hyperinsulinemia. Circulation 1993, 87, 152–161. [Google Scholar] [CrossRef] [Green Version]
- Janus, E.D.; Postiglione, A.; Singh, R.B.; Lewis, B. The modernization of Asia. Implications for coronary heart disease. Council on Arteriosclerosis of the International Society and Federation of Cardiology. Circulation 1996, 94, 2671–2673. [Google Scholar] [CrossRef]
- Ross, R. The Pathogenesis of Atherosclerosis—An Update. N. Engl. J. Med. 1986, 314, 488–500. [Google Scholar] [CrossRef]
- Oliver, M. Pioneer research in Britain into atherosclerosis and coronary heart disease—An historical review. Atherosclerosis 2000, 150, 1–12. [Google Scholar] [CrossRef]
- Steinberg, D.; Parthasarathy, S.; Carew, T.E.; Khoo, J.C.; Witztum, J.L. Beyond cholesterol. Modifications of low-density lipoprotein that increase its atherogenicity. N. Engl. J. Med. 1989, 320, 915–924. [Google Scholar]
- Mahdi, G.; Srikanth, K.; Dmitry, L. Role of Oxidized Lipids in Atherosclerosis, Oxidative Stress and Diseases. In: Dr. Volodymyr Lushchak (Ed.). InTech, 2012. Available online: http://www.intechopen.com/books/oxidative-stress-and-diseases/role-of-oxidized-lipids-in-atherosclerosis (accessed on 3 October 2019).
- Wang, Q. Advances in the genetic basis of coronary artery disease. Curr. Atheroscler. Rep. 2005, 7, 235–241. [Google Scholar] [CrossRef] [Green Version]
- Tanne, D.; Yaari, S.; Goldbourt, U. High-density lipoprotein cholesterol and risk of ischemic stroke mortality. A 21-year follow-up of 8586 men from the Israeli Ischemic Heart Disease Study. Stroke 1997, 28, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Assmann, G.; Schulte, H.; Von Eckardstein, A.; Huang, Y. High-density lipoprotein cholesterol as a predictor of coronary heart disease risk. The PROCAM experience and pathophysiological implications for reverse cholesterol transport. Atheroscler 1996, 124, 124. [Google Scholar] [CrossRef]
- Draganov, D.I.; Stetson, P.L.; Watson, C.E.; Billecke, S.S.; La Du, B.N. Rabbit Serum Paraoxonase 3 (PON3) Is a High-Density Lipoprotein-associated Lactonase and Protects Low Density Lipoprotein against Oxidation. J. Boil. Chem. 2000, 275, 33435–33442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, A.D.; Berliner, J.A.; Hama, S.Y.; La Du, B.N.; Faull, K.F.; Fogelman, A.M.; Navab, M. Protective effect of high-density lipoprotein associated paraoxonase inhibition of the biological activity of minimally oxidized low density lipoprotein. J. Clin. Investig. 1995, 95, 774–782. [Google Scholar] [CrossRef]
- Navab, M.; Van Lenten, B.J.; Reddy, S.T.; Fogelman, A.M. High-density lipoprotein and the dynamics of atherosclerotic lesions. Circulation 2001, 104, 2386–2387. [Google Scholar] [CrossRef] [Green Version]
- Mackness, B.; Davies, G.K.; Turkie, W.; Lee, E.; Roberts, D.H.; Hill, E.; Roberts, C.; Durrington, P.N.; Mackness, M.I. Paraoxonase status in coronary heart disease: Are activity and concentration more important than genotype? Arterioscler. Thromb. Vasc. Boil. 2001, 21, 1451–1457. [Google Scholar] [CrossRef] [Green Version]
- Pandey, U.; Kumari, R.; Nath, B.; Ganesh, S.; Banerjee, I.; Hasan, O.M.; Midha, T.; Pandey, S. Association of angiotensin-converting enzyme, methylene tetrahydrofolate reductase and paraoxonase gene polymorphism and coronary artery disease in an Indian population. Cardiol. J. 2011, 18, 385–394. [Google Scholar]
- Davies, H.G.; Richter, R.J.; Keifer, M.; Broomfield, C.A.; Sowalla, J.; Furlong, C.E. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat. Genet. 1996, 14, 334–336. [Google Scholar] [CrossRef]
- Durrington, P.N.; Mackness, B.; Mackness, M.I. Paraoxonase and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 473–480. [Google Scholar] [CrossRef] [Green Version]
- Rowles, J.; Scherer, S.W.; Xi, T.; Majer, M.; Nickle, D.C.; Rommens, J.M.; Popov, K.M.; Harris, R.A.; Riebow, N.L.; Xia, J.; et al. Cloning and Characterization of PDK4 on 7q21.3 Encoding a Fourth Pyruvate Dehydrogenase Kinase Isoenzyme in Human. J. Boil. Chem. 1996, 271, 22376–22382. [Google Scholar] [CrossRef] [Green Version]
- Primo-Parmo, S.L.; Sorenson, R.C.; Teiber, J.; Du, B.N. The Human Serum Paraoxonase/Arylesterase Gene (PON1) Is One Member of a Multigene Family. Genomics 1996, 33, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Clendenning, J.B.; Humbert, R.; Green, E.D.; Wood, C.; Traver, D.; Furlong, C.E. Structural organisation of human PON1 gene. Genomics 1996, 35, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Mackness, M.I.; Durrington, P.N.; Mackness, B. How high-density lipoprotein protects against the effects of lipid peroxidation. Curr. Opin. Lipidol. 2000, 11, 383–388. [Google Scholar] [CrossRef]
- Lu, H.; Zhu, J.; Zang, Y.; Ze, Y.; Qin, J. Cloning, purification, and refolding of human paraoxonase-3 expressed in Escherichia coli and its characterization. Protein Expr. Purif. 2006, 46, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.C.; Suzuki, S.M.; Cole, T.B.; Park, S.S.; Richter, R.J.; Furlong, C.E. Engineered recombinant human paraoxonase (19rHuPON1) purified from Escherichia. coli protects against organophosphate poisoning. Proc. Natl. Acad. Sci. USA 2008, 105, 12780–12784. [Google Scholar] [CrossRef] [Green Version]
- Otto, T.C.; Harsch, C.K.; Yeung, D.T.; Magliery, T.J.; Cerasoli, D.M.; Lenz, D.E. Dramatic differences in organophosphorus hydrolase activity between human and chimeric recombinant mammalian paraoxonase-1 enzymes. Biochemistry 2009, 48, 10416–10422. [Google Scholar] [CrossRef] [Green Version]
- Brushia, R.J.; Forte, T.M.; Oda, M.N.; La Du, B.N.; Bielicki, J.K. Baculovirus-mediated expression and purification of human serum paraoxonase 1A. J. Lipid Res. 2001, 42, 951–958. [Google Scholar]
- Zhu, J.; Ze, Y.; Zhang, C.; Zang, Y.; Lu, H.; Chu, P.; Sun, M.; Qin, J. High-level expression of recombinant human paraoxonase 1 Q in silkworm larvae (Bombyx mori). Appl. Microbiol. Biotechnol. 2006, 72, 103–108. [Google Scholar] [CrossRef]
- Telford, G.; Wheeler, D.; Williams, P.; Tomkins, P.T.; Appleby, P.; Sewell, H.; Stewart, G.S.A.B.; Bycroft, B.W.; Pritchard, D.I. The Pseudomonas aeruginosa Quorum-Sensing Signal Molecule N-(3-Oxododecanoyl)-l-Homoserine Lactone Has Immunomodulatory Activity. Infect. Immun. 1998, 66, 36–42. [Google Scholar]
- Aharoni, A.; Gaidukov, L.; Khersonsky, O.; Mc, Q.G.S.; Roodveldt, C.; Tawfik, D.S. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 2005, 37, 73–76. [Google Scholar] [CrossRef]
- Ozer, E.A.; Pezzulo, A.; Shih, D.M.; Chun, C.; Furlong, C.; Lusis, A.J.; Greenberg, E.P.; Zabner, J. Human and murine paraoxonase 1 are host modulators of Pseudomonas aeruginosa quorum-sensing. FEMS Microbiol. Lett. 2005, 253, 29–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordi, C.; Isabel, P.; Frederic, B.; Jorge, J. Paraoxonases as Potential Antibiofilm Agents: Their Relationship with Quorum-Sensing Signals in Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2011, 55, 1325–1331. [Google Scholar]
- Available online: http://www.ncbi.nlm.nih.gov.and uniprot.org/uniprot/096561 (accessed on 3 October 2019).
- Bar-Rogovsky, H.; Hugenmatter, A.; Tawfik, D.S. Tawfik,. The Evolutionary Origins of Detoxifying Enzymes; The mammalian serum paraoxonases (PONs) relate to bacterial homoserine lactonases. J. Biol. Chem. 2013, 288, 23914–23927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddavattam, D.; Khajamohiddin, S.; Manavathi, B.; Pakala, S.B.; Merrick, M. Transposon-like organization of the plasmid-borne organophosphate degradation (opd) gene cluster found in Flavobacterium sp. Appl. Environ. Microbiol. 2003, 2003. 69, 2533–2539. [Google Scholar] [CrossRef] [Green Version]
- Sethunathan, N.; Yoshida, T. A Flavobacterium sp. that degrades diazinon and parathion. Can. J. Microbiol. 1973, 19, 873. [Google Scholar] [CrossRef]
- Draganov, D.I. Lactonases with organophosphataseactivity: Structural and evolutionary perspectives. Chem. Biol. Interact. 2010, 187, 370–372. [Google Scholar] [CrossRef]
- Ozols, J. Isolation and complete covalent structure of liver microsomal paraoxonase. Biochem. J. 1999, 338, 265–272. [Google Scholar]
- Rodrigo, L.; Gil, F.; Hernandez, A.F.; López, O.; Pla, A. Identification of paraoxonase 3 in rat liver microsomes: Purification and biochemical properties. Biochem. J. 2003, 376, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Draganov, D.I.; Teiber, J.F.; Speelman, A.; Osawa, Y.; Sunahara, R.; La Du, B.N. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 2005, 46, 1239–1247. [Google Scholar] [CrossRef] [Green Version]
- La Du, B.N.; Aviram, M.; Billecke, S.; Navab, M.; Primo-Parmo, S.; Sorenson, R.C.; Standiford, T.J. On the physiological role(s) of the paraoxonases. Chem. Interact. 1999, 119, 379–388. [Google Scholar] [CrossRef]
- Getz, G.S.; Reardon, C.A. Paraoxonase, a cardioprotective enzyme: Continuing issues. Curr. Opin. Lipidol. 2004, 15, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Furlong, C.E.; Shih, D.M.; Lusis, A.J.; Richter, R.J.; Costa, L.G. Genetic factors in Susceptibility: Serum PON1 variation between individuals and Species. Int. J. 2004, 8, 31–43. [Google Scholar] [CrossRef]
- Geldmacher von Mallinckrodt, M.; Diepgen, T.L. The human paraoxonase polymorphism and specificity. Toxicol. Environ. Chem. 1988, 18, 179–196. [Google Scholar] [CrossRef]
- Hegele, R.A.; Connelly, P.W.; Scherer, S.W.; Hanley, A.J.G.; Harris, S.B.; Tsui, L.-C.; Zinman, B. Paraoxonase-2 Gene (PON2) G148 Variant Associated with Elevated Fasting Plasma Glucose in Noninsulin-Dependent Diabetes Mellitus 1. J. Clin. Endocrinol. Metab. 1997, 82, 3373–3377. [Google Scholar] [CrossRef] [PubMed]
- Hegele, R.A. Paraoxonase genes and disease. Ann. Med. 1999, 31, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Campo, S.; Sardo, A.M.; Campo, G.M.; Avenoso, A.; Castaldo, M.; D’Ascola, A.; Giunta, E.; Calatroni, A.S. Identification of paraoxonase 3 gene (PON3) missense mutations in a population of southern Italy. Mutat. Res. 2004, 546, 75–80. [Google Scholar] [CrossRef]
- Sanchez, R.; Levy, E.; Seidman, E.; Amre, D.; Costea, F.; Sinnett, D. Paraoxonase 1, 2 and 3 DNA variants and susceptibility to childhood inflammatory bowel disease. Gut 2006, 55, 1820–1821. [Google Scholar] [CrossRef] [Green Version]
- Robertson, K.S.; Hawe, E.; Miller, G.J.; Talmud, P.J.; Humphries, S.E. Human paraoxonase gene cluster polymorphisms as predictors of coronary heart disease risk in the prospective Northwick Park Heart Study II. Biochim. Biophys. Acta 2003, 1639, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Sanghera, D.K.; Manzi, S.; Minster, R.L.; Shaw, P.; Kao, A.; Bontempo, F.; Kamboh, M.I. Genetic variation in the paraoxonase-3 (PON3) gene is associated with serum PON1 activity. Ann. Hum. Genet. 2008, 72, 72–81. [Google Scholar] [CrossRef]
- Aragonès, G.; Guardiola, M.; Barreda, M.; Marsillach, J.; Beltrán-Debón, R.; Rull, A.; Mackness, B.; Mackness, M.; Joven, J.; Simó, J.M.; et al. Measurement of serum PON-3 concentration: Method evaluation, reference values, and influence of genotypes in a population-based study. J. Lipid Res. 2011, 52, 1055–1061. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Fan, Z.; Huang, J.; Su, S.Y.; Yu, Q.; Zhao, J.; Hui, R.; Yao, Z.; Shen, Y.; Qiang, B.; et al. Extensive Association Analysis Between Polymorphisms of PON Gene Cluster With Coronary Heart Disease In Chinese Han Population. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 328–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramji, D.P.; Tadros, M.H.; Hardon, E.M.; Cortese, R. The transcription factor LF-A1 interacts with a bipartite recognition sequence in the promoter regions of several liver-specific genes. Nucleic Acids Res. 1991, 19, 1139–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, O. Hynes. Integrins: Versatility, modulation, and signaling in cell adhesion. Cell 1992, 691, 11–25. [Google Scholar]
- Van Kooyk, Y.; van de Wiel-van Kemenade, P.; Weder, P.; Kuijper, T.W.; Fifdor, C.G. Enhancement of LFA-1 mediated cell adhesion by triggering through CD3orCD3 on TLymphocytes. Nature 1989, 342, 811–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marlin, S.D.; Springer, T.A. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1). Cell 1987, 51, 813–819. [Google Scholar] [CrossRef]
- Marsillach, J.; Becker, J.O.; Vaisar, T.; Hahn, B.H.; Brunzell, J.D.; Furlong, C.E.; De Boer, I.H.; McMahon, M.A.; Hoofnagle, A.N. DCCT/EDIC Research Group Paraoxonase-3 Is Depleted from the High-Density Lipoproteins of Autoimmune Disease Patients with Subclinical Atherosclerosis. J. Proteome Res. 2015, 14, 2046–2054. [Google Scholar] [CrossRef] [Green Version]
- Kumari, P.; Kiran, D.G.; Yash, P.S. Low Paraoxonase 3 activity, circulatory concentration and A99A variants in North West Indian Punjabis; a predictive risk for angiographically proven CAD. Int. J. Non-Commun. Dis. 2017, 2, s-75-AB000R568. Available online: http://www.ijncd.org (accessed on 23 November 2019).
- Suchocka, Z.; Swatowska, J.; Pachecka, J.; Suchocki, P. RP-HPLC determination of paraoxonase 3 activity in human blood serum. J. Pharm. Biomed. Anal. 2006, 42, 113–119. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. Structure−Reactivity Studies of Serum Paraoxonase PON1 Suggest that Its Native Activity Is Lactonase†. Biochemistry 2005, 44, 6371–6382. [Google Scholar] [CrossRef]
- Gaidukov, L.; Tawfik, D.S. High Affinity, Stability, and Lactonase Activity of Serum Paraoxonase PON1 Anchored on HDL with ApoA-I. Biochemistry 2005, 44, 11843–11854. [Google Scholar] [CrossRef]
- Harel, M.; Aharoni, A.; Gaidukov, L.; Brumshtein, B.; Khersonsky, O.; Meged, R.; Dvir, H.; Ravelli, R.B.G.; McCarthy, A.; Toker, L.; et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Boil. 2004, 11, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Adams, T.B.; Greer, D.B.; Doull, J.; Munro, I.C.; Newberne, P.; Portoghese, P.S.; Smith, R.L.; Wagner, B.M.; Weil, C.S.; Woods, L.A.; et al. The FEMA GRAS assessment of lactones used as a flavour ingredients. The Flavor and Extract Manufacturers’ Association. Generally recognized as safe. Food Chem. 1998, 36, 249–278. [Google Scholar] [CrossRef]
- Teiber, J.F.; Xiao, J.; Kramer, G.L.; Ogawa, S.; Ebner, C.; Wolleb, H.; Carreira, E.M.; Shih, D.M.; Haley, R.W. Identification of biologically active d-lactone eicosanoids as paraoxonase substrates. Biochem. Biophys. Res. Commun. 2018, 505, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Erdal, B.; Ahmet, B.; Tulin, B.; Tevfik, N. Comparison of serum acetyl hydrolase (PAF-AH) and paraoxonase1(PON1) values between prostate cancer patients and a control group. Kaohsiung J. Med. Sci. 2017, 11, 572–577. [Google Scholar]
- Carr, R.L.; Dail, M.B.; Chambers, H.W.; Chambers, J.E. Species Differences in Paraoxonase Mediated Hydrolysis of Several Organophosphorus Insecticide Metabolites. J. Toxicol. 2015, 2015, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, G.; Prajapati, R.; Tripathy, R.K.; Bajaj, P.; Iyengar, A.R.S.; Sangamwar, A.T.; Pande, A.H. Towards understanding the catalytic mechanism of human Paraoxonase 1: Site-Specific Mutagenesis at Position 192. PLoS ONE 2016, 11, e0147999. [Google Scholar] [CrossRef]
- Le, Q.A.T.; Kim, S.; Chang, R.; Kim, Y.H. Insights into the Lactonase Mechanism of Serum Paraoxonase 1 (PON1): Experimental and Quantum Mechanics/Molecular Mechanics (QM/MM) Studies. J. Phys. Chem. B 2015, 119, 9571–9585. [Google Scholar] [CrossRef]
- Rothem, L.; Hartman, C.; Dahan, A.; Lachter, J.; Eliakim, R.; Shamir, R. Paraoxonases are associated with intestinal inflammatory diseases and intracellularly localized to the endoplasmic reticulum. Free Radic. Boil. Med. 2007, 43, 730–739. [Google Scholar] [CrossRef]
- Rozenberg, O.; Shih, D.M.; Aviram, M. Paraoxonase 1(PON1) attenuates macrophage oxidative status; studies in PON1 transfected cells and in PON1 transgenic mice. Atherosclerosis 2005, 18, 9–18. [Google Scholar] [CrossRef]
- Shih, D.M.; Xia, Y.-R.; Wang, X.-P.; Wang, S.S.; Bourquard, N.; Fogelman, A.M.; Lusis, A.J.; Reddy, S.T. Decreased obesity and atherosclerosis in human paraoxonase 3 transgenic mice. Circ. Res. 2007, 100, 1200–1207. [Google Scholar] [CrossRef] [Green Version]
- Shih, D.M.; Gu, L.; Xia, Y.-R.; Navab, M.; Li, W.-F.; Hama, S.; Castellani, L.W.; Furlong, C.E.; Costa, L.G.; Fogelman, A.M.; et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 1998, 394, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.J.; Bourquard, N.; Grijalva, V.; Hama, S.; Shih, D.M.; Navab, M.; Fogelman, A.M.; Lusis, A.J.; Young, S.; Reddy, S.T. Paraoxonase-2deficiency aggravates atherosclerosis in mice despite lower apolipoprotein-B-containing lipoproteins: Anti-atherogenic role for paraoxonase-2. J. Biol. Chem. 2006, 281, 29491–29500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- She, Z.G.; Chen, H.Z.; Yan, Y.; Li, H.; Liu, D.P. The Human Paraoxonase Gene Cluster as a Target in the Treatment of Atherosclerosis. Antioxid. Redox Signal. 2012, 16, 597–632. [Google Scholar] [CrossRef]
- Marsillach, J.; Mackness, B.; Mackness, M.; Riu, F.; Beltrán, R.; Joven, J.; Camps, J. Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free Radic. Boil. Med. 2008, 45, 146–157. [Google Scholar] [CrossRef]
- Shih, D.M.; Xia, Y.-R.; Yu, J.M.; Lusis, A.J. Temporal and tissue-specific patterns of Pon3 expression in mouse: In situ hybridization analysis. Single Mol. Single Cell Seq. 2010, 660, 73–87. [Google Scholar]
- Michael, M.; Kulkarni, R.N.; Postic, C.; Previs, S.F.; Shulman, G.I.; Magnuson, M.A.; Kahn, C. Loss of Insulin Signaling in Hepatocytes Leads to Severe Insulin Resistance and Progressive Hepatic Dysfunction. Mol. Cell 2000, 6, 87–97. [Google Scholar] [CrossRef]
- Lu, H.; Zhu, J.; Zang, Y.; Ze, Y.; Qin, J. Cloning, high level expression of human paraoxonase-3 in Sf9 cells and pharmacological characterization of its product. Biochem. Pharmacol. 2005, 70, 1019–1025. [Google Scholar] [CrossRef]
- Reddy, S.T.; Wadleigh, D.J.; Grijalva, V.; Ng, C.; Hama, S.; Gangopadhyay, A.; Shih, D.M.; Lusis, A.J.; Navab, M.; Fogelman, A.M. Human paraoxonase-3 is an HDL-associated enzyme with biological activity similar to paraoxonase-1 protein but is not regulated by oxidized lipids. Arterioscler. Thromb. Vasc. Boil. 2001, 21, 542–547. [Google Scholar] [CrossRef] [Green Version]
- Rosenblat, M.; Draganov, D.; Watson, C.E.; Bisgaier, C.L.; La Du, B.N.; Aviram, M. Mouse Macrophage Paraoxonase 2 Activity Is Increased Whereas Cellular Paraoxonase 3 Activity Is Decreased Under Oxidative Stress. Arterioscler. Thromb. Vasc. Boil. 2003, 23, 468–474. [Google Scholar] [CrossRef] [Green Version]
- Précourt, L.-P.; Amre, D.; Denis, M.-C.; Lavoie, J.-C.; Delvin, E.; Seidman, E.; Levy, E. The three-gene paraoxonase family: Physiologic roles, actions and regulation. Atherosclerosis 2011, 214, 20–36. [Google Scholar] [CrossRef]
- Draganov, D.I. Human PON3, effects beyond the HDL: clues from human PON3 transgenic mice. Circ. Res. 2007, 100, 1104–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frishberg, Y.; Toledano, H.; Becker-Cohen, R.; Feigin, E.; Halle, D. Genetic polymorphism in paraoxonase is a risk factor for childhood focal segmental glomerulosclerosis. Am. J. Kidney Dis. 2000, 36, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Schweikert, E.M.; Devarajan, A.; Witte, I.; Wilgenbus, P.; Amort, J.; Förstermann, U.; Shabazian, A.; Grijalva, V.; Shih, D.M.; Farias-Eisner, R.; et al. PON3 is upregulated in cancer tissues and protects against mitochondrial superoxide-mediated cell death. Cell Death Differ. 2012, 19, 1549–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shamir, R.; Hartman, C.; Karry, R.; Pavlotzky, E.; Eliakim, R.; Lachter, J.; Suissa, A.; Aviram, M. Paraoxonases (PONs) 1, 2, and 3 are expressed in human and mouse gastrointestinal tract and in Caco-2 cell line: Selective secretion of PON1 and PON2. Free Radic. Boil. Med. 2005, 39, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, O.; Howell, A.; Aviram, M. Pomegranate juice sugar fraction reduces macrophage oxidative state, whereas white grape juice sugar fraction increases it. Atherosclerosis 2006, 188, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Jarvik, G.P.; Tsai, N.T.; Mc Kinstry, L.A.; Wani, R.; Brophy, V.H.; Richter, R.J.; Schellenberg, G.D.; Hatsukami, T.S.; Furlong, C.E. Vitamin C and E intake is associated with increased paraoxonase activity. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 1329–1333. [Google Scholar] [CrossRef] [Green Version]
- Romani, R.; De Medio, G.E.; Di Tullio, S.; Lapalombella, R.; Pirisinu, I.; Margonato, V.; Veicsteinas, A.; Marini, M.; Rosi, G. Modulation of paraoxonase 1 and 3 expression after moderate exercise training in the rat. J. Lipid Res. 2009, 50, 2036–2045. [Google Scholar] [CrossRef] [Green Version]
- Ng, C.J.; Bourquard, N.; Hama, S.Y.; Shih, D.; Grijalva, V.R.; Navab, M.; Fogelman, A.M.; Reddy, S.T. Adenovirus-Mediated Expression of Human Paraoxonase 3 Protects Against the Progression of Atherosclerosis in Apolipoprotein E–Deficient Mice. Arterioscler. Thromb. Vasc. Boil. 2007, 27, 1368–1374. [Google Scholar] [CrossRef] [Green Version]
- Belteki, G.; Kempster, S.L.; Forhead, A.J.; Giussani, D.A.; Fowden, A.L.; Curley, A.; Charnock-Jones, D.S.; Smith, G.C.S. Paraoxonase-3, a Putative Circulating Antioxidant, Is Systemically Up-Regulated in Late Gestation in the Fetal Rat, Sheep, and Human. J. Clin. Endocrinol. Metab. 2010, 95, 3798–3805. [Google Scholar] [CrossRef] [Green Version]
- Boehm, D.; Krzystek-Korpacka, M.; Neubauer, K.; Matusiewicz, M.; Berdowska, I.; Zielinski, B.; Paradowski, L.; Andrzej, G. Paraoxonase-1 status in Crohn’s disease and ulcerative colitis. Inflamm. Bowel. Dis. 2009, 15, 93–99. [Google Scholar] [CrossRef]
- Baskol, G.; Baskol, M.; Yurci, A.; Ozbakir, O.; Yucesoy, M. Serum paraoxonase-1 activity and malondialdehyde levels in patients with ulcerative colitis. Cell Biochem. Funct. 2006, 24, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Abbott, C.A.; Mackness, M.I.; Kumar, S.; Boulton, A.J.; Durrington, P.N. Serum Paraoxonase Activity, Concentration, and Phenotype Distribution in Diabetes Mellitus and Its Relationship to Serum Lipids and Lipoproteins. Arterioscler. Thromb. Vasc. Boil. 1995, 15, 1812–1818. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.J.; Shih, D.M.; Hama, S.Y.; Villa, N.; Navab, M.; Reddy, S.T. The paraoxonase gene family and atherosclerosis. Free Radic. Boil. Med. 2005, 38, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Tward, A.; Xia, Y.-R.; Wang, X.-P.; Shi, Y.-S.; Park, C.; Castellani, L.W.; Lusis, A.J.; Shih, D.M.; Fisher, C.; MacLean, M.; et al. Decreased Atherosclerotic Lesion Formation in Human Serum Paraoxonase Transgenic Mice. Circulation 2002, 106, 484–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozenberg, O. Paraoxonase (PON1) deficiency is associated with increased macrophage oxidative stress: Studies in PON1-knockout mice. Free Radic. Boil. Med. 2003, 34, 774–784. [Google Scholar] [CrossRef]
- Deakin, S.; Leviev, I.; Guernier, S.; James, R.W. Simvastatin modulates expression of the PON1 gene and increases serum paraoxonase: A role for sterol regulatory element-binding protein-2. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 2083–2089. [Google Scholar] [CrossRef] [Green Version]
- Shih, D.M.; Yu, J.M.; Vergnes, L.; Dali-Youcef, N.; Champion, M.D.; Devarajan, A.; Zhang, P.; Castellani, L.W.; Brindley, D.N.; Jamey, C.; et al. PON3 knockout mice are susceptible to obesity, gallstone formation, and atherosclerosis. FASEB J. 2015, 29, 1185–1197. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Peng, W.; Wang, M.; Zhu, J.; Zang, Y.; Shi, W.; Zhang, J.; Qin, J. Studies on protective effects of human paraoxonases 1 and 3 on atherosclerosis in apolipoprotein E knockout mice. Gene Ther. 2010, 17, 626–633. [Google Scholar] [CrossRef]
- Peng, W.; Zhang, C.; Lv, H.; Zhu, J.; Zang, Y.; Pang, X.; Zhang, J.; Qin, J. Comparative evaluation of the protective potentials of human paraoxonase 1 and 3 against CCl4-induced liver injury. Toxicol. Lett. 2010, 193, 159–166. [Google Scholar] [CrossRef]
- Peng, W.; Jiang, X.; Haiqin, L.; Zhang, C.; Zhu, J.; Zhang, J.; Zang, Y.; Qin, J. Protective effects of transgene expressed human PON3 against CCl4-induced subacute liver injury in mice. Biomed. Pharmacother. 2009, 63, 592–598. [Google Scholar] [CrossRef]
- Leaf, D.A. The effect of physical exercise on reverse cholesterol transport. Metabolism 2003, 52, 950–957. [Google Scholar] [CrossRef]
- Kurl, S.; Laukkanen, J.; Niskanen, L.; Rauramaa, R.; Tuomainen, T.; Sivenius, J.; Salonen, J. Cardiac Power During Exercise and the Risk of Stroke in Men. Stroke 2005, 36, 820–824. [Google Scholar] [CrossRef] [Green Version]
- Bradamante, S.; Barenghi, L.; Villa, A. Cardiovascular protective effects of resveratrol. Cardiovasc. Drug Rev. 2004, 22, 169–188. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Furlong, C.E. Pharmacological and dietary modulators of paraoxonase 1 (PON1) activity and expression: The hunt goes on. Biochem. Pharmacol. 2011, 81, 337–344. [Google Scholar] [CrossRef] [Green Version]
- Noll, C.; Dairou, J.; Ripoll, C.; Paul, J.-L.; Dupret, J.-M.; Delabar, J.-M.; Rodrigues-Lima, F.; Janel, N. Effect of red wine polyphenol dietary supplementation on two phase II enzymes in liver of hyperhomocysteinemic mice. Food Chem. Toxicol. 2011, 49, 1764–1769. [Google Scholar] [CrossRef]
- Sebai, H.; Sani, M.; Yacoubi, M.T.; Aouani, E.; Ghanem-Boughanmi, N.; Ben-Attia, M. Resveratrol, a red wine polyphenol, attenuates lipopolysaccharide-induced oxidative stress in rat liver. Ecotoxicol. Environ. Saf. 2010, 73, 1078–1083. [Google Scholar] [CrossRef]
- Pais, P.; Pogue, S.; Gerstein, H. Risk factors for acute myocardial infarction in Indians: A case-control study. Lancet 1998, 348, 358–363. [Google Scholar] [CrossRef]
- Gupta, N.; Kandimalla, R.; Priyanka, K.; Singh, G.; Gill, K.D.; Singh, S. Effect of Resveratrol and Nicotine on PON1 gene expression:in vitro study. Ind. J. Clin. Biochem. 2014, 29, 69–73. [Google Scholar] [CrossRef] [Green Version]
- She, Z.-G.; Zheng, W.; Wei, Y.-S.; Chen, H.-Z.; Wang, A.-B.; Li, H.-L.; Liu, G.; Zhang, R.; Liu, J.-J.; Stallcup, W.B.; et al. Human Paraoxonase Gene Cluster Transgenic Overexpression Represses Atherogenesis and Promotes Atherosclerotic Plaque Stability in ApoE-Null Mice. Circ. Res. 2009, 104, 1160–1168. [Google Scholar] [CrossRef] [Green Version]
- Devarajan, A.; Shih, D.; Reddy, S.T. Inflammation, Infection, Cancer and All That…The Role of Paraoxonases. Adv. Exp. Med. Biol. 2014, 824, 33–41. [Google Scholar]
- Witte, I.; Foerstermann, U.; Devarajan, A.; Reddy, S.T.; Horke, S. Protectors or Traitors: The Roles of PON2 and PON3 in Atherosclerosis and Cancer. J. Lipids 2012, 2012, 342806. [Google Scholar] [CrossRef] [Green Version]
- Carlson, C.S.; Heagerty, P.J.; Hatsukami, T.S.; Richter, R.J.; Ranchalis, J.; Lewis, J.; Bacus, T.J.; McKinstry, L.A.; Schellenberg, G.D.; Rieder, M.; et al. TagSNP analyses of the PON gene cluster: Effects on PON1 activity, LDL oxidative susceptibility, and vascular disease. J. Lipid Res. 2006, 47, 1014–1024. [Google Scholar] [CrossRef] [Green Version]
- Marsillach, J.; Aragonès, G.; Beltrán, R.; Caballería, J.; Pedro-Botet, J.; Morcillo-Suárez, C.; Navarro, A.; Joven, J.; Camps, J. The measurement of the lactonase activity of paraoxonase-1 in the clinical evaluation of patients with chronic liver impairment. Clin. Biochem. 2009, 42, 91–98. [Google Scholar] [CrossRef]
- Riedmaier, S.; Klein, K.; Winter, S.; Hofmann, U.; Schwab, M.; Zanger, U.M. Paraoxonase (PON1 and PON3) Polymorphisms: Impact on Liver Expression and Atorvastatin-Lactone Hydrolysis. Front. Pharmacol. 2011, 2, 41. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Burt, A.A.; Ranchalis, J.E.; Richter, R.J.; Marshall, J.K.; Eintracht, J.F.; Rosenthal, E.A.; Furlong, C.E.; Jarvik, G.P. Additional Common Polymorphisms in the PON Gene Cluster Predict PON1 Activity but Not Vascular Disease. J. Lipids 2012, 2012, 476316. [Google Scholar] [CrossRef] [Green Version]
| Study | Subjects Enrolled | PON3 SNPs Studied | Outcome | Author, Year |
|---|---|---|---|---|
| Prospective Northwick Park Heart study II; to evaluate the effect of SNPs on CHD risk | 3052 healthy men | A99A (GCG to GCA) D107N (GAC) to (AAC) | A99A SNP, did not revealed much information about its association with CAD but D107N was absent in the population. | Robertson et al. [50] (2003) |
| Polymorphisms screening of PON cluster in a Chinese Han population | 949 subjects in the 474 cases, 475 controls | −133 C>A | Not found significant effect on CHD risk but detected -133 C >A SNP in PON3 located at a potential binding site for transcription factor LFA-1(Integrin Lymphocyte Function-associated Antigen) | Wang et al. [53] (2003) |
| Identification of PON3 mutations in a population of Southern Italy | 1143 blood donors | G51G G73G A99A S311T G324D | G51G, G73G, A99A were silent and S311T, G324D were missense mutations with no clarity on function in CAD development | Campo et al. [48] (2004) |
| PON gene cluster TagSNPs analysis—on illumina platform | 500 Caucasian males | PON1, PON2 and PON3 | No significant association with CAD disease | Carlson et al. [114] (2006) |
| Association study; PON3 with serum PON1 activity, risk of atherosclerosis in SLE cases | 377 cases and 482 controls (US whites and blacks) | PON3 (A10340C, A2115T), PON1 (L55M, Q192R) | All four SNPs explained 2%, 1%, 8%, and 19% of the variation in PON1 activity, respectively. PON3 SNPs described only 3% of variation in PON1 activity | Sanghera et al. [51] (2008) |
| Influence of genetic polymorphisms of PON on lactonase activity | Healthy population | PON3−567, PON3−665, PON3−746, PON3−4105, PON3−4970, PON3−4984 | Lactonase activity was lower than Paraoxonase activity which indicated evaluation of liver function in clinics | Marsillach et al. [115] (2009) |
| PON3; concentration determination and its association analysis with promoter polymorphisms | n = 356; 156 women, 200 men; mean age: 47 years, of Caucasian from the Mediterranean region of Catalonia | (-567 C/T, -665 A/G, -746 C/T, -4105 G/A, -4970 T/G, -4984 A/G) | Promoter SNPs associated with PON3 serum concentration. TGTAGG, TGTGTA, CACGTA haplotypes were significantly associated with changes in serum PON3 concentration when adjusted for gender, age, BMI | Aragones et al. [52] (2011) |
| PON1 and PON3; atorvastatin hydrolysis | Blood and liver tissues of patients undergoing surgery(n = 150) | -4984A/G, -4105G/A, -1091A/G, -746C/T and F21F | 40 SNPs identified within the PON-locus associated with changes in atorvastatin δ-lactone hydrolysis and expression of PON1 but not PON3. Non genetic factors only were associated with PON3 expression | Riedmaier et al. [116] (2011) |
| Study of 51 common polymorphisms in the PON cluster | 1328 Caucasian males | PON3 (rs17884000, rs9640632, rs468, rs11768074 rs10487132 rs740264) | Predicted PON1 activity but not vascular disease | Daniel S. Kim et al. [117] (2012) |
| Case control study in North West Indian Punjabis | n = 300 cases, n = 300 proven CAD patients | C-133A, A99A, D107N, G324D | Low Paraoxonase 3 activity, circulatory concentration and A99A variants were predictive risks for angiographically proven CAD | K. Priyanka et al. [59] (2017) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Priyanka, K.; Singh, S.; Gill, K. Paraoxonase 3: Structure and Its Role in Pathophysiology of Coronary Artery Disease. Biomolecules 2019, 9, 817. https://doi.org/10.3390/biom9120817
Priyanka K, Singh S, Gill K. Paraoxonase 3: Structure and Its Role in Pathophysiology of Coronary Artery Disease. Biomolecules. 2019; 9(12):817. https://doi.org/10.3390/biom9120817
Chicago/Turabian StylePriyanka, Kumari, Surjit Singh, and Kirandip Gill. 2019. "Paraoxonase 3: Structure and Its Role in Pathophysiology of Coronary Artery Disease" Biomolecules 9, no. 12: 817. https://doi.org/10.3390/biom9120817
APA StylePriyanka, K., Singh, S., & Gill, K. (2019). Paraoxonase 3: Structure and Its Role in Pathophysiology of Coronary Artery Disease. Biomolecules, 9(12), 817. https://doi.org/10.3390/biom9120817




