Genomic, Transcriptomic and Enzymatic Insight into Lignocellulolytic System of a Plant Pathogen Dickeya sp. WS52 to Digest Sweet Pepper and Tomato Stalk
Abstract
1. Introduction
2. Materials and Methods
2.1. Raw Materials and Chemicals
2.2. Bacteria Strain and Culture Conditions
2.3. Bacterial Identification by 16S rRNA and Draft Genome Sequencing of Dickeya sp. WS52
2.4. Transcriptome Sequencing
2.4.1. Sample Collection and RNA Preparation
2.4.2. Library Preparation for Transcriptome Sequencing
2.4.3. Reads Mapping to the Reference Genome
2.4.4. Differential Expression Analysis and Functional Enrichment
2.5. Enzymatic Hydrolysis
2.6. Analytical Method of Monosaccharide Composition
2.7. Accession Numbers of Draft Genome Sequencing and Transcription Sequencing, and Statistical Analyses
3. Results and Discussion
3.1. Isolation and Identification of Plant Pathogen Dickeya sp. WS52
3.2. Genomic analysis of Dickeya sp. WS52
3.3. Transcriptal Profiling of Dickeya sp. WS52
3.3.1. Identification of Expressed Transcripts and Expression Level Analysis in the WS52 Transcriptome
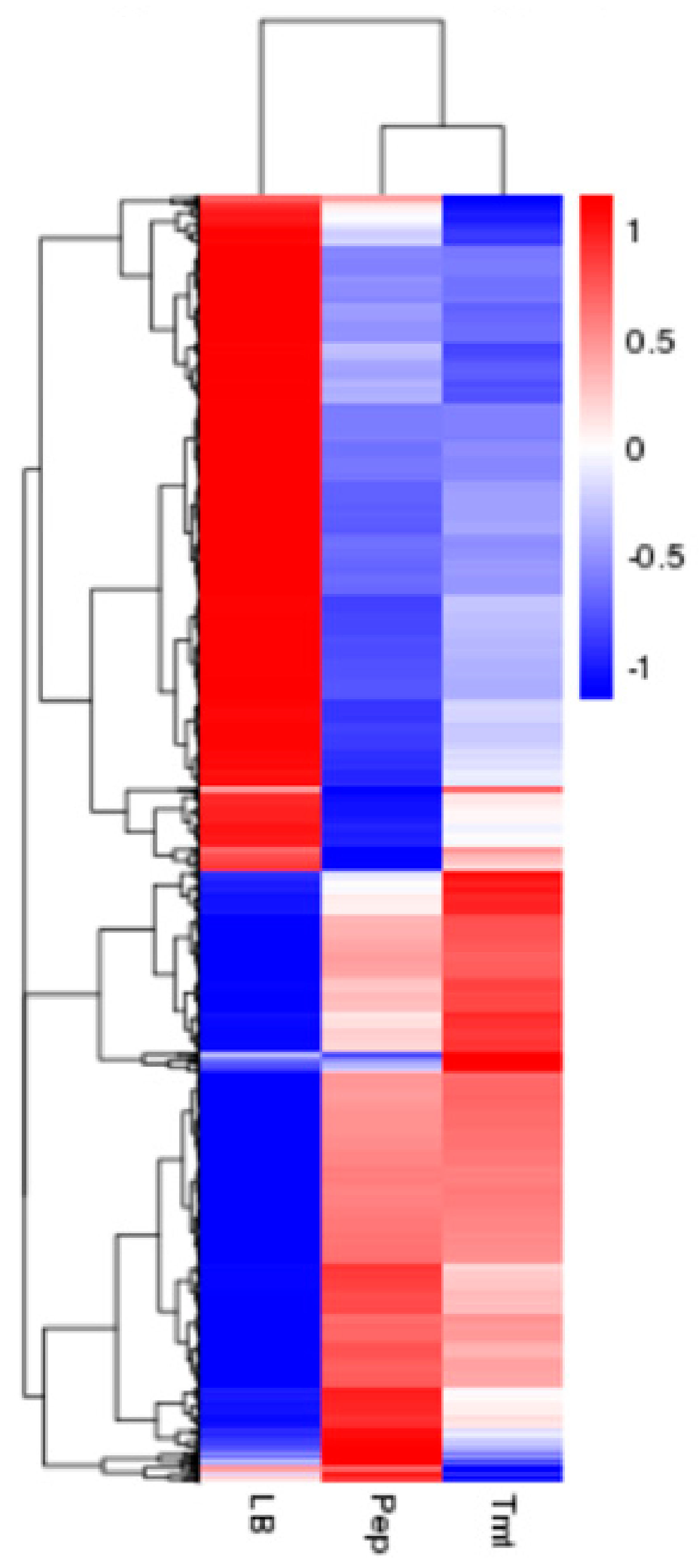
3.3.2. Identification of Differentially Expressed Genes Between Different Samples
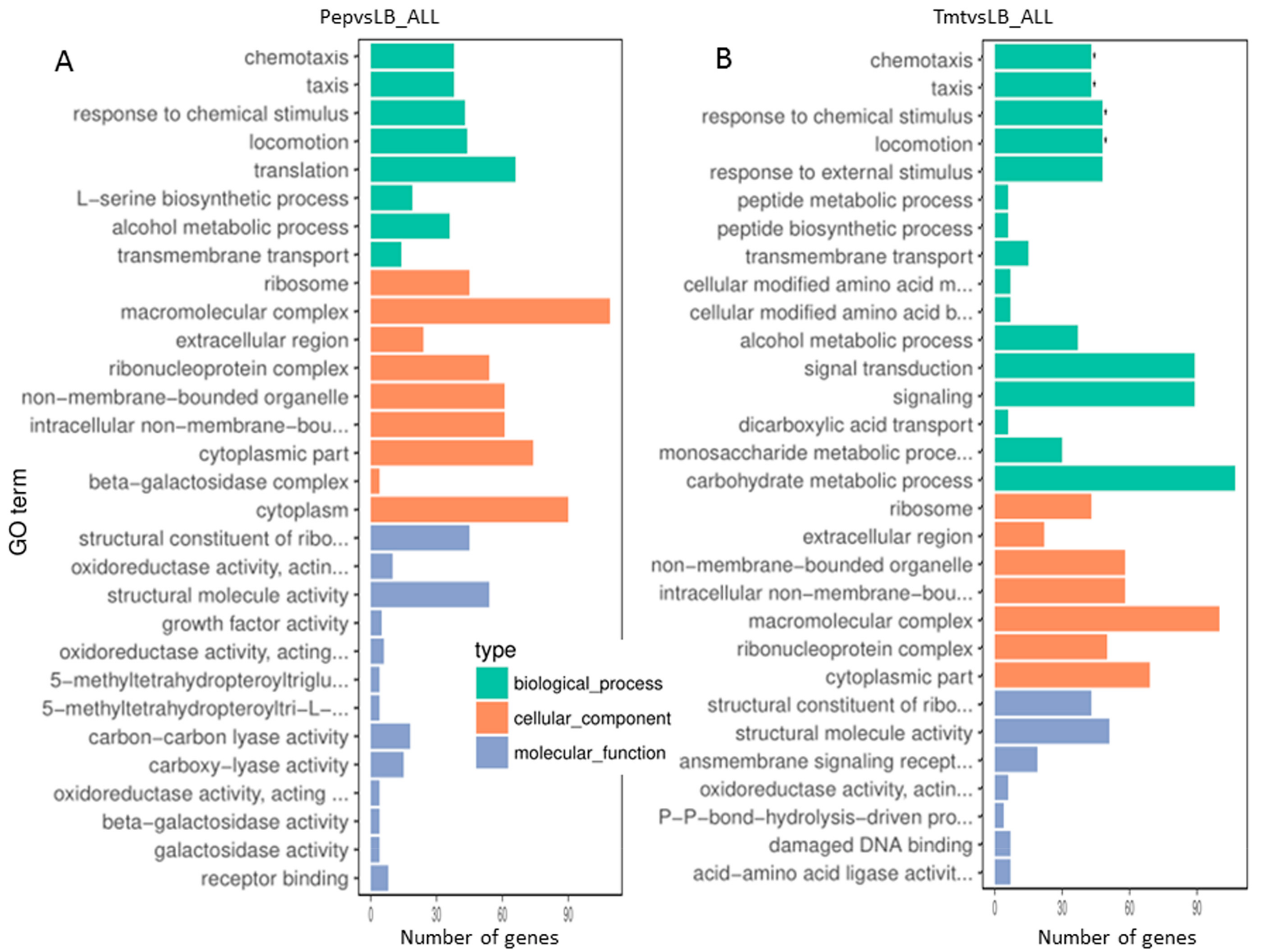
3.3.3. Functional Distribution of Differentially Expressed Genes
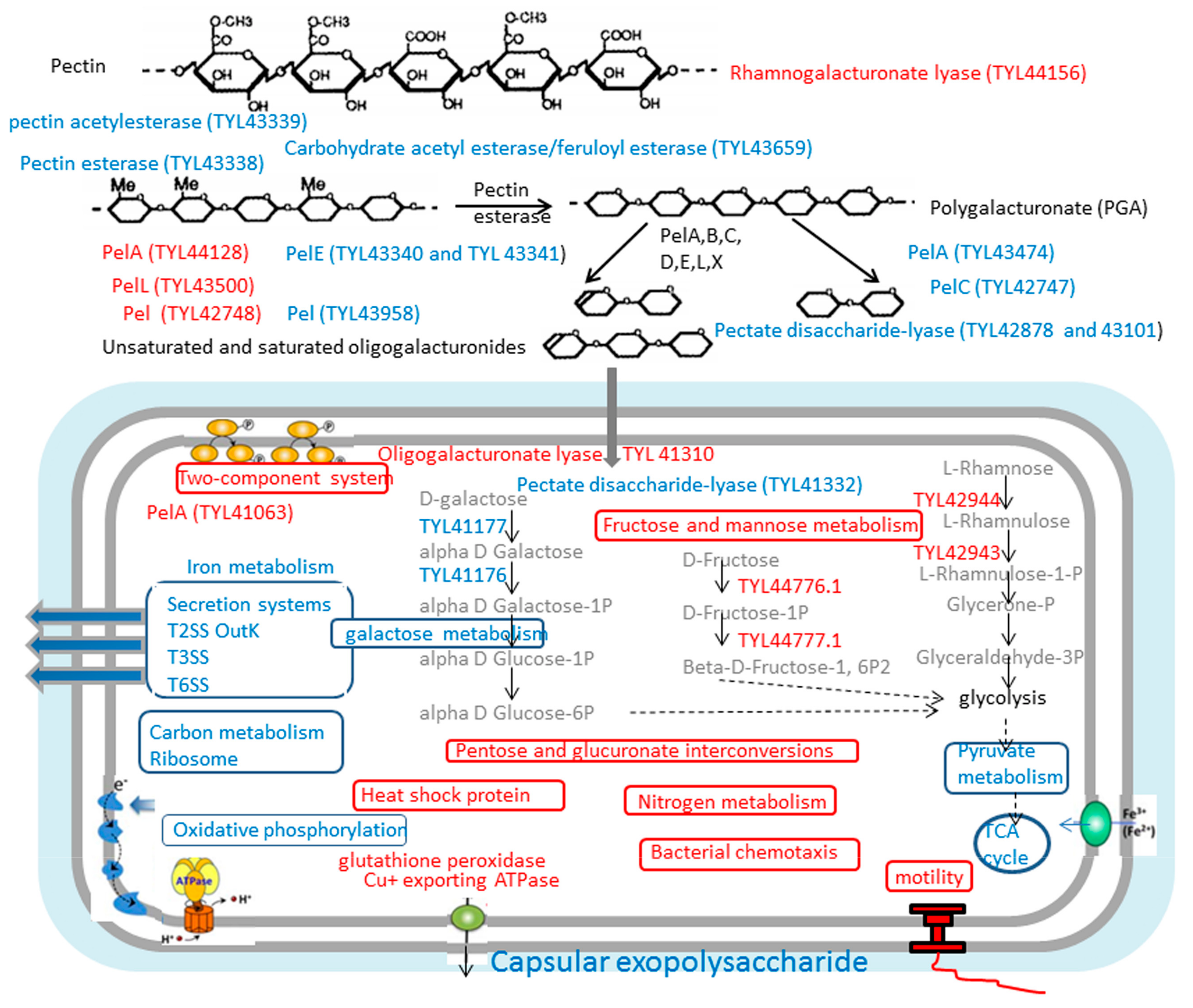
3.3.4. Most Enzymes Related to Carbohydrate Metabolism Were Downregulated under the Vegetable Stalk
3.4. Growth and Enzymatic Profiling of Dickeya sp. WS52 in Sweet Pepper and Tomato Stalk Medium
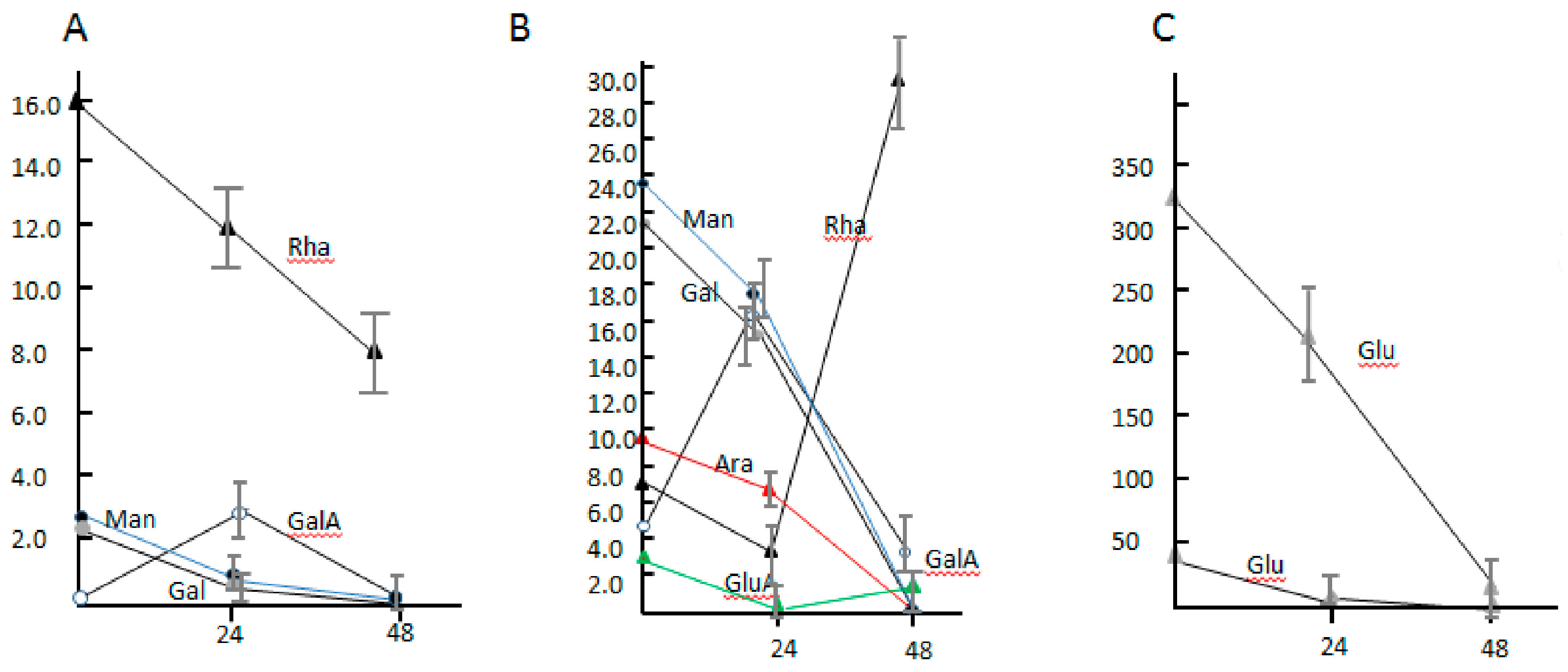
3.5. The Component Analysis of Monosaccharide and the Transcriptional Level of Sugar Glycolysis of Dickeya sp. WS52
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Badhan, A.; Ribeiro, G.O., Jr.; Jones, D.R.; Wang, Y.; Abbott, D.W.; Di Falco, M.; Tsang, A.; McAllister, T.A. Identification of novel enzymes to enhance the ruminal digestion of barley straw. Bioresour. Technol. 2018, 260, 76–84. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, R.; Su, W.; Lu, Q.; Dong, C. Study on pyrolysis characteristics of red pepper stalks to analyze the changes of pyrolytic behaviors from xylophyta to herbage. J. Anal. Appl. Pyrolysis 2016, 120, 330–333. [Google Scholar] [CrossRef]
- Kim, S.; Dale, B.E. Global potential bioethanol production from wasted crops and crop residues. Biomass Bioenergy 2004, 26, 361–375. [Google Scholar] [CrossRef]
- Xiong, X.Q.; Liao, H.D.; Ma, J.S.; Liu, X.M.; Zhang, L.Y.; Shi, X.W.; Yang, X.L.; Lu, X.N.; Zhu, Y.H. Isolation of a rice endophytic bacterium, Pantoea sp. Sd-1, with ligninolytic activity and characterization of its rice straw degradation ability. Lett. Appl. Microbiol. 2014, 58, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Shutao, W.; Jin, Z.; Tong, X. Biochar influences the microbial community structure during tomato stalk composting with chicken manure. Bioresour. Technol. 2014, 154, 148–154. [Google Scholar] [CrossRef]
- Zhang, G.; Li, S.; Xu, Y.; Wang, J.; Wang, F.; Xin, Y.; Shen, Z.; Zhang, H.; Ma, M.; Liu, H. Production of alkaline pectinase: A case study investigating the use of tobacco stalk with the newly isolated strain Bacillus tequilensis CAS-MEI-2-33. BMC Biotechnol. 2019, 19, 45. [Google Scholar] [CrossRef]
- Farias, N.; Almeida, I.; Meneses, C. New Bacterial Phytase through Metagenomic Prospection. Molecules 2018, 23, 448. [Google Scholar] [CrossRef]
- Meneses, C.; Silva, B.; Medeiros, B.; Serrato, R.; Johnston-Monje, D. A Metagenomic Advance for the Cloning and Characterization of a Cellulase from Red Rice Crop Residues. Molecules 2016, 21. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Nakkeeran, E.; Umesh-Kumar, S.; Subramanian, R. Aspergillus carbonarius polygalacturonases purified by integrated membrane process and affinity precipitation for apple juice production. Bioresour. Technol. 2011, 102, 3293–3297. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Vincken, J.P.; Schols, H.A.; Oomen, R.J.; McCann, M.C.; Ulvskov, P.; Voragen, A.G.; Visser, R.G. If homogalacturonan were a side chain of rhamnogalacturonan I. Implications for cell wall architecture. Plant Physiol. 2003, 132, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Broxterman, S.E.; Schols, H.A. Characterisation of pectin-xylan complexes in tomato primary plant cell walls. Carbohydr. Polym. 2018, 197, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Latarullo, M.B.; Tavares, E.Q.; Maldonado, G.P.; Leite, D.C.; Buckeridge, M.S. Pectins, Endopolygalacturonases, and Bioenergy. Front. Plant Sci. 2016, 7, 1401. [Google Scholar] [CrossRef]
- Amin, F.; Bhatti, H.N.; Bilal, M. Recent advances in the production strategies of microbial pectinases—A review. Int. J. Biol. Macromol. 2019, 122, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Christgau, S.; Kofod, L.V.; Halkier, T.; Andersen, L.N.; Hockauf, M.; Dorreich, K.; Dalboge, H.; Kauppinen, S. Pectin methyl esterase from Aspergillus aculeatus: Expression cloning in yeast and characterization of the recombinant enzyme. Biochem. J. 1996, 319, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Coffman, A.M.; Ju, L.K. Development of reproducible assays for polygalacturonase and pectinase. Enzym. Microb. Technol. 2015, 72, 42–48. [Google Scholar] [CrossRef]
- Hoondal, G.S.; Tiwari, R.P.; Tewari, R.; Dahiya, N.; Beg, Q.K. Microbial alkaline pectinases and their industrial applications: A review. Appl. Microbiol. Biotechnol. 2002, 59, 409–418. [Google Scholar] [CrossRef]
- Yim, S.S.; Choi, J.W.; Lee, S.H.; Jeong, K.J. Modular Optimization of a Hemicellulose-Utilizing Pathway in Corynebacterium glutamicum for Consolidated Bioprocessing of Hemicellulosic Biomass. ACS Synth. Biol. 2016, 5, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.R.; Vohra, P.K.; Chopra, S.; Tewari, R. Applications of pectinases in the commercial sector: A review. Bioresour. Technol. 2001, 77, 215–227. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Bernard, T.; Rancurel, C.; Brumer, H.; Coutinho, P.M.; Henrissat, B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 2010, 432, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Garron, M.L.; Cygler, M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 2010, 20, 1547–1573. [Google Scholar] [CrossRef] [PubMed]
- Nakkeeran, E.; Subramanian, R.; Umesh Kumar, S. Improving specific activity of Aspergillus carbonarius polygalacturonase using polymeric membranes. Appl. Biochem. Biotechnol. 2008, 151, 233–243. [Google Scholar] [CrossRef]
- Busto, M.D.; Garcia-Tramontin, K.E.; Ortega, N.; Perez-Mateos, M. Preparation and properties of an immobilized pectinlyase for the treatment of fruit juices. Bioresour. Technol. 2006, 97, 1477–1483. [Google Scholar] [CrossRef]
- Bajpai, P. Application of enzymes in the pulp and paper industry. Biotechnol. Prog. 1999, 15, 147–157. [Google Scholar] [CrossRef]
- Omogbenigun, F.O.; Nyachoti, C.M.; Slominski, B.A. Dietary supplementation with multienzyme preparations improves nutrient utilization and growth performance in weaned pigs. J. Anim. Sci. 2004, 82, 1053–1061. [Google Scholar] [CrossRef]
- Tang, Y.; Wu, P.; Jiang, S.; Selvaraj, J.N.; Yang, S.; Zhang, G. A new cold-active and alkaline pectate lyase from Antarctic bacterium with high catalytic efficiency. Appl. Microbiol. Biotechnol. 2019, 103, 5231–5241. [Google Scholar] [CrossRef]
- Bonavita, A.; Carratore, V.; Ciardiello, M.A.; Giovane, A.; Servillo, L.; D’Avino, R. Influence of pH on the Structure and Function of Kiwi Pectin Methylesterase Inhibitor. J. Agric. Food Chem. 2016, 64, 5866–5876. [Google Scholar] [CrossRef]
- Cheirsilp, B.; Umsakul, K. Processing of banana-based wine product using pectinase and alpha-amylase. J. Food Process. Eng. 2008, 31, 78–90. [Google Scholar] [CrossRef]
- Alencar, B.R.A.; Dutra, E.D.; Sampaio, E.; Menezes, R.S.C.; Morais, M.A. Enzymatic hydrolysis of cactus pear varieties with high solids loading for bioethanol production. Bioresour. Technol. 2018, 250, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Shevchik, V.E. Bacterial pectate lyases, structural and functional diversity. Environ. Microbiol. Rep. 2014, 6, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Laothanachareon, T.; Bunterngsook, B.; Suwannarangsee, S.; Eurwilaichitr, L.; Champreda, V. Synergistic action of recombinant accessory hemicellulolytic and pectinolytic enzymes to Trichoderma reesei cellulase on rice straw degradation. Bioresour. Technol. 2015, 198, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chio, C.; Chen, X.; Su, E.; Cao, F.; Jin, Y.; Qin, W. Efficient saccharification of agave biomass using Aspergillus niger produced low-cost enzyme cocktail with hyperactive pectinase activity. Bioresour. Technol. 2019, 272, 26–33. [Google Scholar] [CrossRef]
- Kobayashi, T.; Hatada, Y.; Higaki, N.; Lusterio, D.D.; Ozawa, T.; Koike, K.; Kawai, S.; Ito, S. Enzymatic properties and deduced amino acid sequence of a high-alkaline pectate lyase from an alkaliphilic Bacillus isolate. Biochim. Biophys. Acta 1999, 1427, 145–154. [Google Scholar] [CrossRef]
- Blanco, P.; Sieiro, C.; Villa, T.G. Production of pectic enzymes in yeasts. FEMS Microbiol. Lett. 1999, 175, 1–9. [Google Scholar] [CrossRef]
- Blandino, A.; Iqbalsyah, T.; Pandiella, S.S.; Cantero, D.; Webb, C. Polygalacturonase production by Aspergillus awamori on wheat in solid-state fermentation. Appl. Microbiol. Biotechnol. 2002, 58, 164–169. [Google Scholar] [CrossRef]
- Bruhlmann, F. Purification and characterization of an extracellular pectate lyase from an Amycolata sp. Appl. Environ. Microbiol. 1995, 61, 3580–3585. [Google Scholar]
- Reverchon, S.; Nasser, W. Dickeya ecology, environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 2013, 5, 622–636. [Google Scholar] [CrossRef]
- Wormit, A.; Usadel, B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. [Google Scholar] [CrossRef]
- Kazemi-Pour, N.; Condemine, G.; Hugouvieux-Cotte-Pattat, N. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics 2004, 4, 3177–3186. [Google Scholar] [CrossRef] [PubMed]
- Nivaskumar, M.; Francetic, O. Type II secretion system: A magic beanstalk or a protein escalator. Biochim. Biophys. Acta 2014, 1843, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Zghidi-Abouzid, O.; Oger-Desfeux, C.; Hommais, F.; Greliche, N.; Muskhelishvili, G.; Nasser, W.; Reverchon, S. Global transcriptional response of Dickeya dadantii to environmental stimuli relevant to the plant infection. Environ. Microbiol. 2016, 18, 3651–3672. [Google Scholar] [CrossRef] [PubMed]
- Reverchon, S.; Robert-Baudouy, J. Regulation of expression of pectate lyase genes pelA, pelD, and pelE in Erwinia chrysanthemi. J. Bacteriol. 1987, 169, 2417–2423. [Google Scholar] [CrossRef]
- Tardy, F.; Nasser, W.; Robert-Baudouy, J.; Hugouvieux-Cotte-Pattat, N. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: Enzyme characteristics and potential inhibitors. J. Bacteriol. 1997, 179, 2503–2511. [Google Scholar] [CrossRef]
- Lowe-Power, T.M.; Hendrich, C.G.; von Roepenack-Lahaye, E.; Li, B.; Wu, D.; Mitra, R.; Dalsing, B.L.; Ricca, P.; Naidoo, J.; Cook, D.; et al. Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Environ. Microbiol. 2018, 20, 1330–1349. [Google Scholar] [CrossRef]
- Raoul des Essarts, Y.; Pedron, J.; Blin, P.; Van Dijk, E.; Faure, D.; Van Gijsegem, F. Common and distinctive adaptive traits expressed in Dickeya dianthicola and Dickeya solani pathogens when exploiting potato plant host. Environ. Microbiol. 2019, 21, 1004–1018. [Google Scholar] [CrossRef]
- Fahy, A.; McGenity, T.J.; Timmis, K.N.; Ball, A.S. Heterogeneous aerobic benzene-degrading communities in oxygen-depleted groundwaters. FEMS Microbiol. Ecol. 2006, 58, 260–270. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Yang, Y.J.; Singh, R.P.; Lan, X.; Zhang, C.S.; Sheng, D.H.; Li, Y.Q. Whole transcriptome analysis and gene deletion to understand the chloramphenicol resistance mechanism and develop a screening method for homologous recombination in Myxococcus xanthus. Microb. Cell Factories 2019, 18, 123. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Yang, Y.J.; Singh, R.P.; Lan, X.; Zhang, C.S.; Li, Y.Z.; Li, Y.Q.; Sheng, D.H. Genome Editing in Model Strain Myxococcus xanthus DK1622 by a Site-Specific Cre/loxP Recombination System. Biomolecules 2018, 8. [Google Scholar] [CrossRef]
- Guo, H.; Wu, Y.; Hong, C.; Chen, H.; Chen, X.; Zheng, B.; Jiang, D.; Qin, W. Enhancing digestibility of Miscanthus using lignocellulolytic enzyme produced by Bacillus. Bioresour. Technol. 2017, 245, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zou, P.; Zhou, J.; Geng, Y.; Fan, J.; Clark, J.; Li, Y.; Zhang, C. Microwave-assisted hydrothermal extraction of non-structural carbohydrates and hemicelluloses from tobacco biomass. Carbohydr. Polym. 2019, 223, 115043. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Manchanda, G.; Maurya, I.K.; Maheshwari, N.K.; Tiwari, P.K.; Rai, A.R. Streptomyces from rotten wheat straw endowed the high plant growth potential traits and agro-active compounds. Bio. Agri. Biotechnol. 2019, 17, 507–513. [Google Scholar] [CrossRef]

| Species | Genome Size (bp) | Protein Coding Genes | % CAZy | GHs | CEs | CBMs | PLs |
|---|---|---|---|---|---|---|---|
| Dickeya sp. WS52 | 4,744,455 | 4082 | 2.1 | 45 | 14 | 9 | 18 |
| D. Aquatica 174/2 | 4,501,560 | 4079 | 1.8 | 38 | 6 | 12 | 16 |
| D. chrysanthemi Ech1591 | 4,813,854 | 4112 | 1.8 | 42 | 6 | 8 | 18 |
| D. Dadantii 3937 | 4,922,802 | 4244 | 1.9 | 46 | 6 | 9 | 17 |
| D. fangzhongdai DSM101947 | 503,245 | 4298 | 1.8 | 43 | 6 | 11 | 17 |
| D. fangzhongdai PA1 | 4,979,223 | 4208 | 1.8 | 42 | 6 | 13 | 16 |
| D. solani IFB0223 | 4,937,554 | 4143 | 1.8 | 42 | 6 | 12 | 16 |
| D. Zeae Ech586 | 4,818,394 | 4113 | 1.7 | 41 | 5 | 8 | 14 |
| E. Amylovora ATCC49946 | 3,905,604 | 3488 | 1.4 | 36 | 3 | 8 | 2 |
| P. carotovorum SCC1 | 4,980,322 | 4294 | 1.9 | 47 | 6 | 13 | 15 |
| C. thermocellum ATCC27405 | 3,843,300 | 3263 | 5.6 | 74 | 16 | 90 | 4 |
| C. bescii DSM6725 | 2,931,660 | 2662 | 2.9 | 49 | 6 | 18 | 3 |
| Sphingobium sp. SYK-6 | 4,348,130 | 3939 | 0.7 | 21 | 1 | 3 | 0 |
| E. lignolyticus SCF1 | 4,814,050 | 4350 | 1.2 | 42 | 5 | 7 | 0 |
| P. ananatis Sd-1 | 4,927,500 | 4332 | 2.2 | 59 | 25 | 11 | 2 |
| ORF Number (TYL) | Annotation | GH Family | AA Length | Pepper | Tomato | SignalP |
|---|---|---|---|---|---|---|
| 43657.1 | β-glucosidase | 1 | 429 | −6.76 | −6.38 | N |
| 43004.1 | β-glucosidase | 1 | 429 | −1.57 | n.d. | N |
| 42629.1 | GH 1 protein | 1 | 429 | −3.28 | −3.24 | N |
| 42630.1 | GH 1 protein | 1 | 429 | −2.91 | −3.63 | N |
| 42639.1 | GH 1 protein | 1 | 429 | n.d. | n.d. | N |
| 42214.1 | β-galactosidase | 2 | 752 | −1.22 | −1.39 | N |
| 43595.1 | beta-N-acehexosaminidase | 3 | 216 | n.d. | n.d. | N |
| 41537.1 | β-glucosidase | 3 | 216 | −2.50 | −1.80 | Y |
| 41574.1 | GH 3 protein | 3 | 216 | n.d. | n.d. | Y |
| 43501.1 | cellulase family glycosylhydrolase | 5 | 275 | n.d. | −1.44 | N |
| 41080.1 | endoglucanase | 8 | 320 | n.d. | 1.08 | Y |
| 42790.1 | glycogen debranching protein | 13 | 299 | n.d. | n.d. | N |
| 42791.1 | 1,4-alpha-glucan branching protein | 13 | 299 | n.d. | n.d. | N |
| 42742.1 | GH 19 protein | 19 | 231 | 3.87 | 5.04 | N |
| 44181.1 | lytic murein transglycosylase | 23 | 135 | −1.04 | n.d. | Y |
| 43265.1 | murein transglycosylase D | 23 | 135 | 1.22 | 1.53 | N |
| 43333.1 | lytic murein transglycosylase | 23 | 135 | n.d. | n.d. | N |
| 42383.1 | murein transglycosylase | 23 | 135 | 1.01 | 1.03 | Y |
| 43901.1 | lysozyme | 24 | 137 | n.d. | n.d. | N |
| 44351.1 | GH 28 protein | 28 | 325 | n.d. | n.d. | Y |
| 42856.1 | GH 28 protein | 28 | 325 | n.d. | 1.18 | Y |
| 42857.1 | GH 28 protein | 28 | 325 | 1.10 | n.d. | Y |
| 42558.1 | GH 28 protein | 28 | 325 | n.d. | n.d. | N |
| 43601.1 | GH 31 protein | 31 | 427 | 1.58 | 1.73 | N |
| 43027.1 | sucrose-6-phosphate hydrolase | 32 | 293 | n.d. | n.d. | N |
| 42500.1 | glycosyl hydrolase | 33 | 342 | n.d. | n.d. | N |
| 42149.1 | alpha-galactosidase | 36 | 688 | −4.26 | −4.57 | N |
| 44116.1 | β-galactosidase | 42 | 371 | −1.23 | −1.57 | N |
| 44137.1 | Xylan 1,3-β-xylosidase | 43 | 248 | n.d. | −1.05 | Y |
| 41284.1 | GH43 protein | 43 | 248 | n.d. | n.d. | N |
| 44117.1 | galactosidase | 53 | 342 | −2.27 | −2.16 | Y |
| 43528.1 | peptidoglycan hydrolase | 73 | 128 | n.d. | n.d. | N |
| 41480.1 | hypothetical protein | 73 | 128 | n.d. | n.d. | N |
| 42781.1 | 4-alpha-glucanotransferase | 77 | 494 | n.d. | n.d. | N |
| 43305.1 | alpha-L-rhamnosidase | 78 | 504 | n.d. | n.d. | N |
| 44201.1 | murein transglycosylase A | 102 | 157 | n.d. | n.d. | N |
| 41622.1 | lytic murein transglycosylase | 103 | 295 | 1.68 | 1.74 | Y |
| 41246.1 | lytic murein transglycosylase B | 103 | 295 | −1.88 | −1.67 | N |
| 43711.1 | GH 104 protein | 104 | 145 | n.d. | n.d. | N |
| 43498.1 | GH 104 protein | 104 | 145 | n.d. | n.d. | N |
| 42562.1 | GH 105 protein | 105 | 332 | n.d. | n.d. | N |
| 42513.1 | GH 105 protein | 105 | 332 | −4.96 | −3.73 | N |
| 44519.1 | Oxidoreductase | 109 | 126 | n.d. | n.d. | N |
| 44637.1 | Gfo/Idh/MocA family-oxidoreductase | 109 | 126 | −1.33 | −1.20 | N |
| ORF Number (TYL) | Annotation | PL | AA Length | Log2FC in Pep | Log2FC in Tmt | SignalP |
|---|---|---|---|---|---|---|
| 43957.1 | hypothetical protein | 1 | 202 | 2.14 | 1.94 | N |
| 43340.1 | Pectate lyase E | 1 | 202 | n.d. | −1.85 | Y |
| 43341.1 | Pectate lyase E | 1 | 202 | −4.21 | −4.24 | Y |
| 43474.1 | Pectate lyase A | 1 | 202 | n.d. | −1.83 | Y |
| 42747.1 | Pectate lyase C | 1 | 202 | n.d. | −1.33 | Y |
| 42748.1 | Pectate lyase C | 1 | 202 | 1.11 | n.d. | Y |
| 42878.1 | Pectate disaccharide-lyase | 1 | 202 | −1.68 | −2.31 | Y |
| 41268.1 | Pectin lyase | 1 | 202 | n.d. | n.d. | N |
| 41332.1 | Pectate disaccharide-lyase | 2 | 530 | −1.04 | −1.81 | N |
| 44128.1 | Pectate lyase A | 3 | 197 | 1.75 | n.d. | Y |
| 41063.1 | Pectate lyase A | 3 | 197 | n.d. | 1.73 | N |
| 44527.1 | Rhamnogalacturonate lyase | 4 | 567 | n.d. | n.d. | Y |
| 44156.1 | Rhamnogalacturonate lyase | 4 | 567 | 2.57 | 2.08 | Y |
| 43101.1 | Pectate disaccharide-lyase | 9 | 374 | −1.33 | −2.13 | Y |
| 43500.1 | Pectate lyase L | 9 | 374 | 1.30 | n.d. | Y |
| 44668.1 | Pectate lyase L | 9 | 374 | n.d. | n.d. | Y |
| 43958.1 | pectate lyase | 10 | 287 | −3.92 | −4.85 | Y |
| 41310.1 | Oligogalacturonate lyase | 22 | 265 | 3.05 | 1.50 | N |
| ORF Number (TYL) | Annotation | CE | AA Length | Log2FC in Pep | Log2FC in Tmt | SignalP |
|---|---|---|---|---|---|---|
| 44765.1 | S-formylglutathione hydrolase | 1 | 227 | −2.63 | −2.26 | N |
| 44076.1 | enterochelin esterase | 1 | 227 | n.d. | n.d. | N |
| 43659.1 | Carbohydrate acetyl esterase/feruloyl esterase | 1 | 227 | −2.28 | −2.07 | Y |
| 43338.1 | Pectin esterase A | 8 | 288 | −1.02 | −2.24 | Y |
| 41030.1 | Pectin esterase B | 8 | 288 | n.d. | n.d. | Y |
| 44436.1 | N-acetylglucosamine-6-phosphate deacetylase | 9 | 373 | n.d. | n.d. | N |
| 43959.1 | alpha/beta hydrolase | 10 | 341 | n.d. | 2.15 | N |
| 43308.1 | alpha/beta hydrolase | 10 | 341 | −2.60 | −1.67 | Y |
| 40812.1 | alpha/beta hydrolase | 10 | 341 | n.d. | n.d. | Y |
| 40810.1 | alpha/beta hydrolase | 10 | 341 | n.d. | −1.28 | N |
| 41844.1 | alpha/beta hydrolase | 10 | 341 | n.d. | n.d. | Y |
| 41326.1 | alpha/beta hydrolase | 10 | 341 | −1.62 | −1.87 | N |
| 42473.1 | N-acetylglucosamine deacetylase | 11 | 271 | 1.09 | n.d. | N |
| 43339.1 | pectin acetylesterase | 12 | 210 | −1.86 | −1.47 | Y |
| ORF Number (TYL) | Log2FC in Pep | Log2FC in Tmt | Annotation |
|---|---|---|---|
| 44776.1 | 2.26 | 2.38 | PTS fructose transporter subunit IIBC |
| 44777.1 | 2.90 | 3.02 | 1-phosphofructokinase |
| 44778.1 | 1.47 | 1.27 | fused PTS fructose transporter subunit IIA |
| 42943.1 | 3.30 | 3.67 | rhamnulokinase |
| 42944.1 | 4.04 | 4.80 | L-rhamnose isomerase |
| 42946.1 | 4.02 | 3.74 | L-rhamnose mutarotase |
| 44512.1 | n.d. | n.d. | mannose-6-phosphate isomerase |
| 44525.1 | n.d. | n.d. | L-arabinose isomerase |
| 43854.1 | n.d. | n.d. | mannose-6-phosphate isomerase |
| 41342.1 | 1.17 | 1.68 | PTS mannose transporter subunit IID |
| 41344.1 | n.d. | n.d. | PTS mannose transporter subunit IIAB |
| 44504.1 | 1.20 | 1.48 | mannosyl-3-phosphoglycerate |
| 42336.1 | n.d. | n.d. | glucuronate isomerase |
| 41873.1 | n.d. | n.d. | glucose-6-phosphate isomerase |
| 41752.1 | −1.84 | n.d. | arabinose-5-phosphate isomerase KdsD |
| 43476.1 | −1.28 | −1.30 | xylose isomerase |
| 43187.1 | −1.72 | −2.11 | xylose isomerase |
| 41176.1 | −2.21 | −2.45 | galactokinase |
| 41177.1 | −1.39 | −1.63 | galactose mutarotase |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.-J.; Lin, W.; Singh, R.P.; Xu, Q.; Chen, Z.; Yuan, Y.; Zou, P.; Li, Y.; Zhang, C. Genomic, Transcriptomic and Enzymatic Insight into Lignocellulolytic System of a Plant Pathogen Dickeya sp. WS52 to Digest Sweet Pepper and Tomato Stalk. Biomolecules 2019, 9, 753. https://doi.org/10.3390/biom9120753
Yang Y-J, Lin W, Singh RP, Xu Q, Chen Z, Yuan Y, Zou P, Li Y, Zhang C. Genomic, Transcriptomic and Enzymatic Insight into Lignocellulolytic System of a Plant Pathogen Dickeya sp. WS52 to Digest Sweet Pepper and Tomato Stalk. Biomolecules. 2019; 9(12):753. https://doi.org/10.3390/biom9120753
Chicago/Turabian StyleYang, Ying-Jie, Wei Lin, Raghvendra Pratap Singh, Qian Xu, Zhihou Chen, Yuan Yuan, Ping Zou, Yiqiang Li, and Chengsheng Zhang. 2019. "Genomic, Transcriptomic and Enzymatic Insight into Lignocellulolytic System of a Plant Pathogen Dickeya sp. WS52 to Digest Sweet Pepper and Tomato Stalk" Biomolecules 9, no. 12: 753. https://doi.org/10.3390/biom9120753
APA StyleYang, Y.-J., Lin, W., Singh, R. P., Xu, Q., Chen, Z., Yuan, Y., Zou, P., Li, Y., & Zhang, C. (2019). Genomic, Transcriptomic and Enzymatic Insight into Lignocellulolytic System of a Plant Pathogen Dickeya sp. WS52 to Digest Sweet Pepper and Tomato Stalk. Biomolecules, 9(12), 753. https://doi.org/10.3390/biom9120753






