Shaping the Nascent Ribosome: AAA-ATPases in Eukaryotic Ribosome Biogenesis
Abstract
1. Introduction to Eukaryotic Ribosome Biogenesis
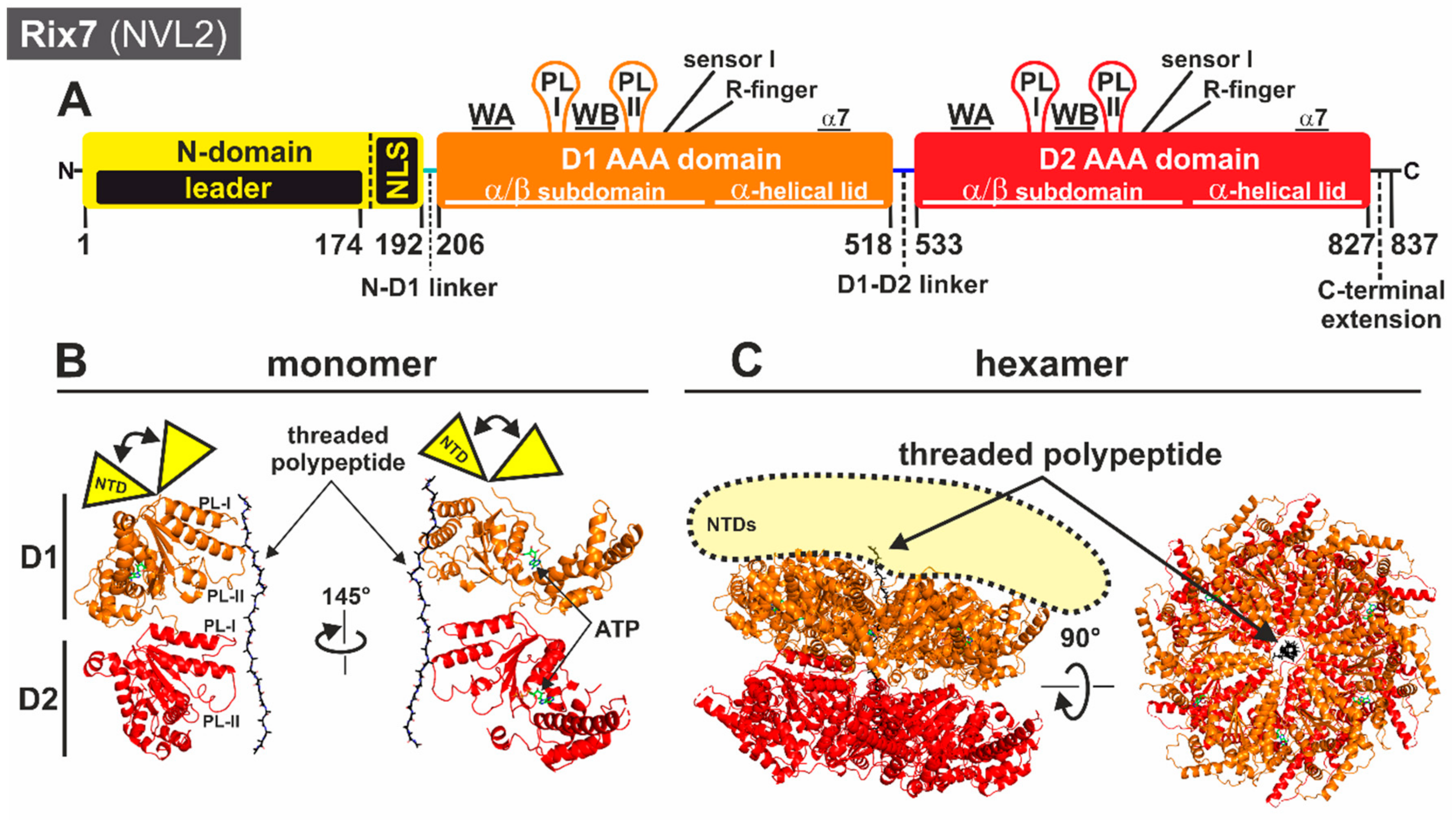
2. AAA-ATPases in Ribosome Biogenesis
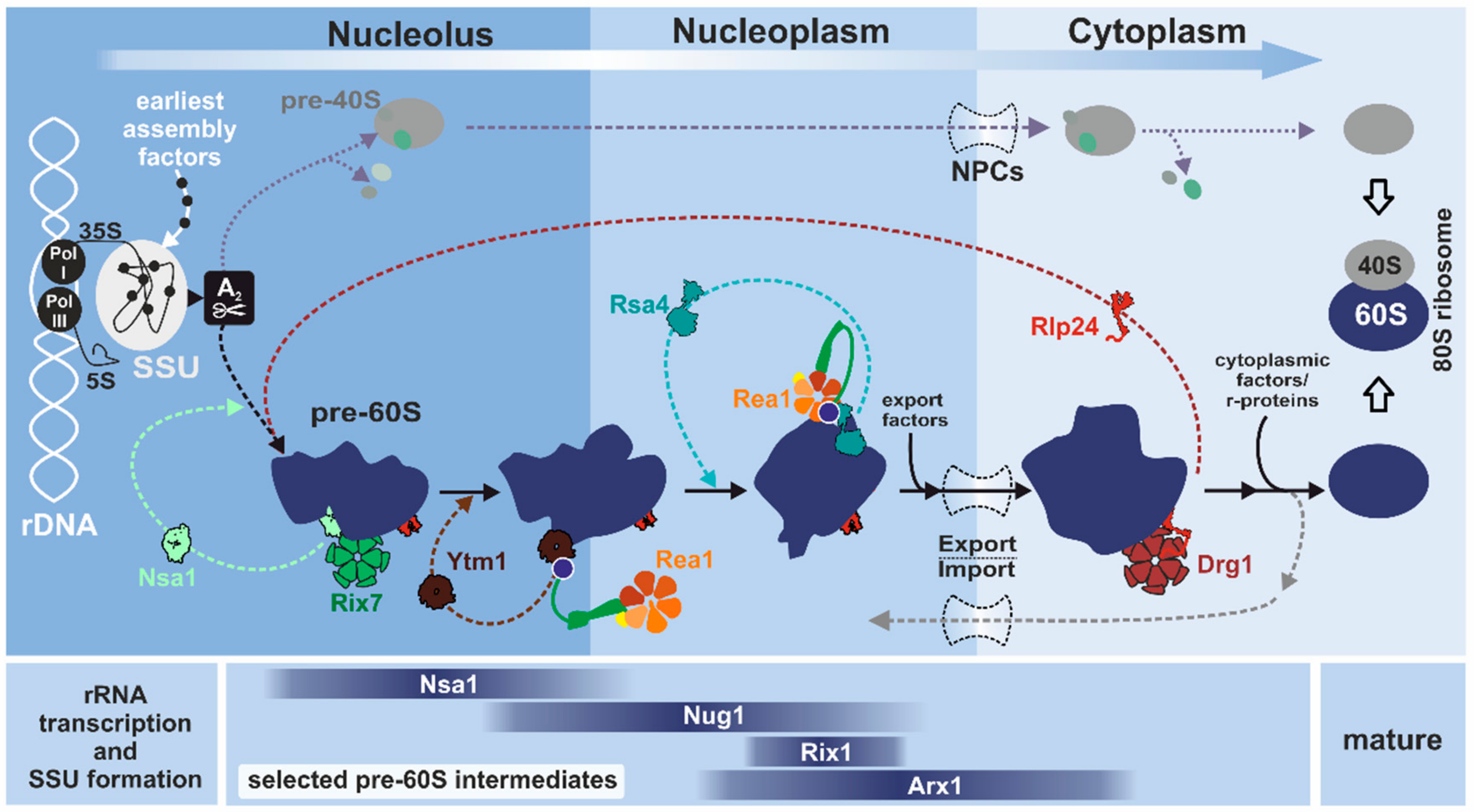
2.1. How to Sculpt a Eukaryotic Ribosome Step by Step
2.2. Order is Key: Molecular Machines Keep in Line
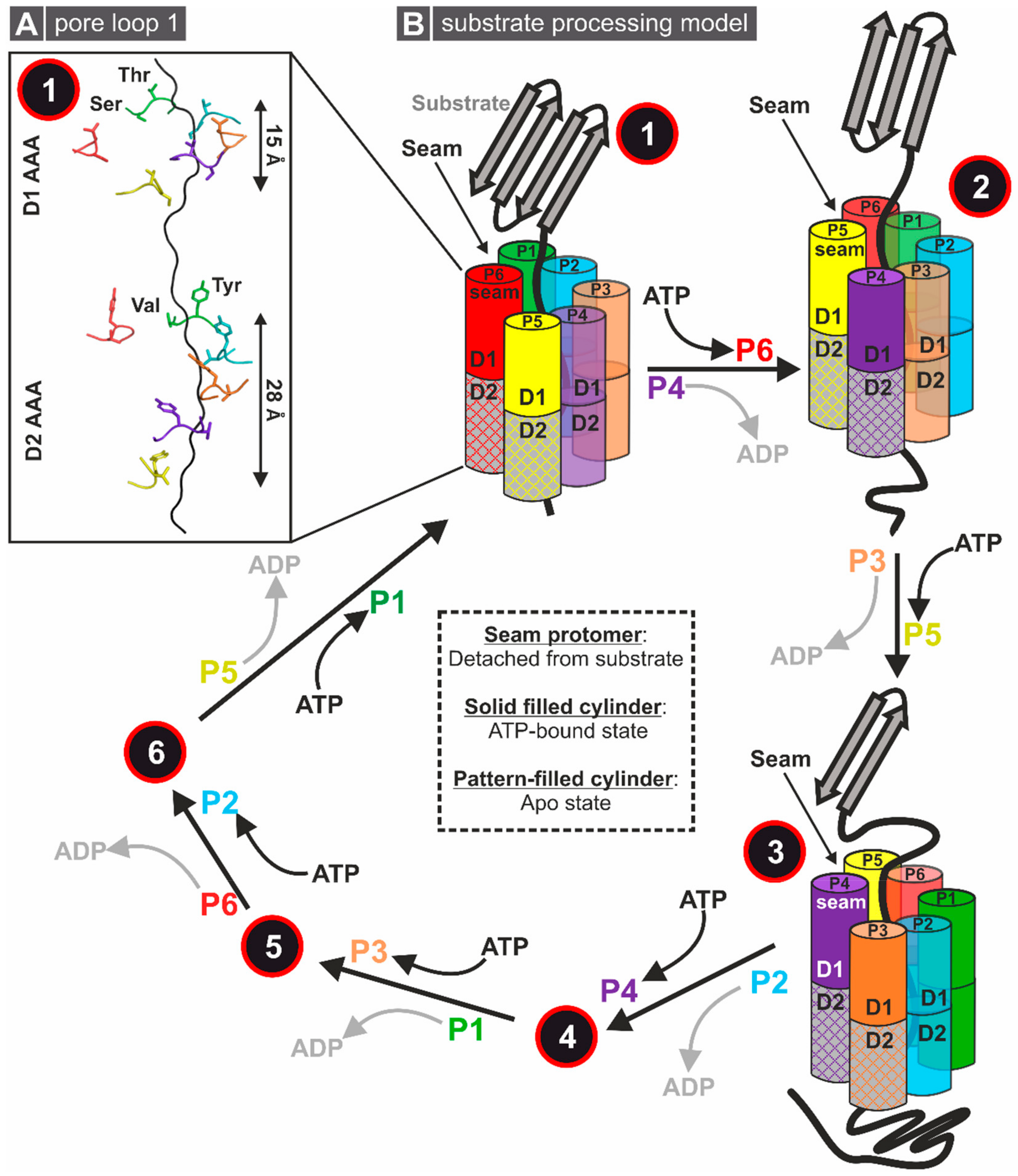
2.3. General Modes of Substrate Processing
3. From the Nucleolus to the Cytoplasm: Mechanistic Insights and Cellular Functions of Rix7, Rea1, and Drg1
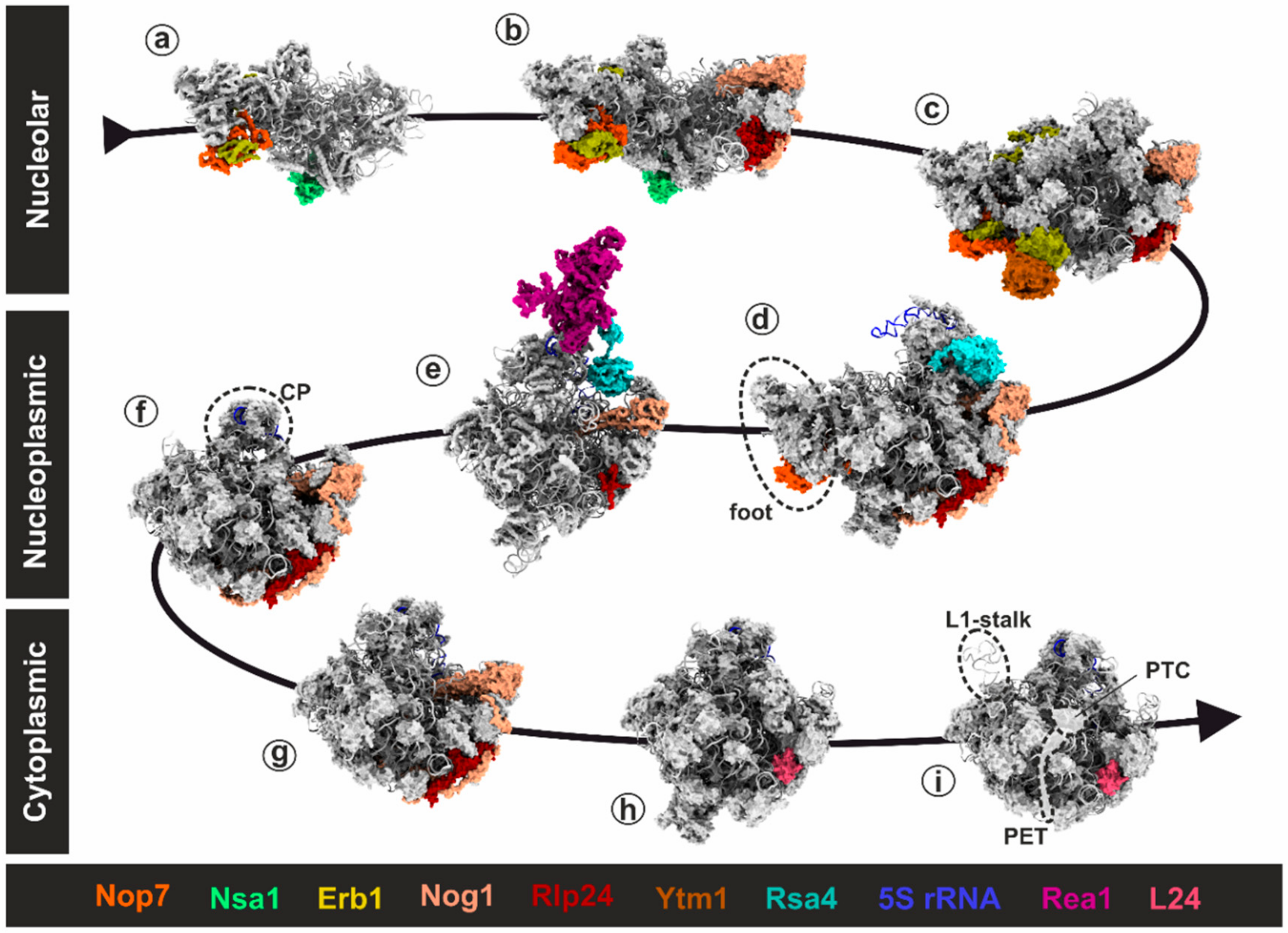
3.1. Rix7: Remodeling of Nucleolar Pre-60S Particles
3.1.1. Structural Insights into the Molecular Mechanism of Rix7
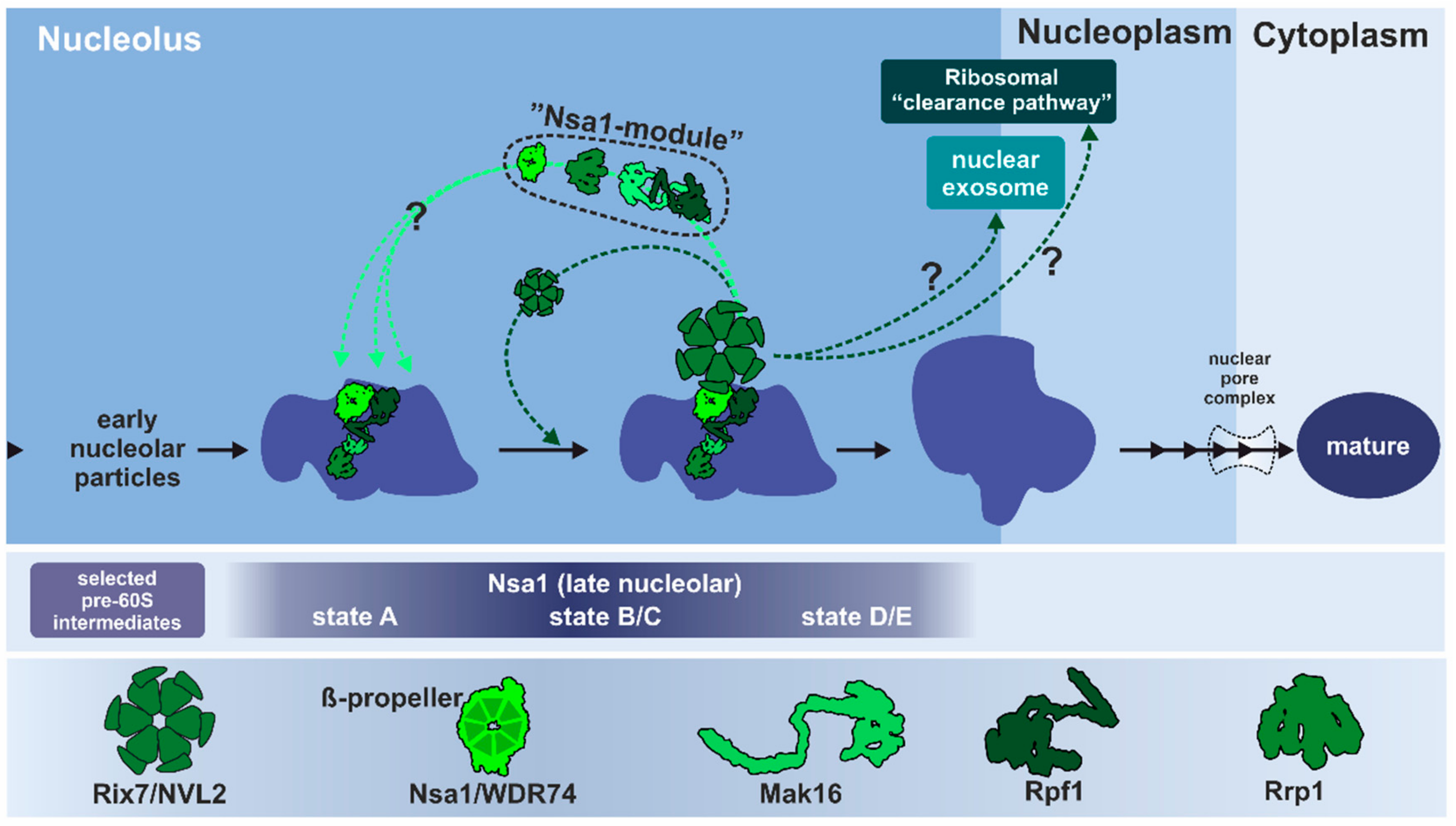
3.1.2. Remodeling of the Nucleolar Nsa1-Particle in Yeast
3.1.3. Conservation of Rix7 Function in Eukaryotes
3.2. Rea1: A Colossus among Giants
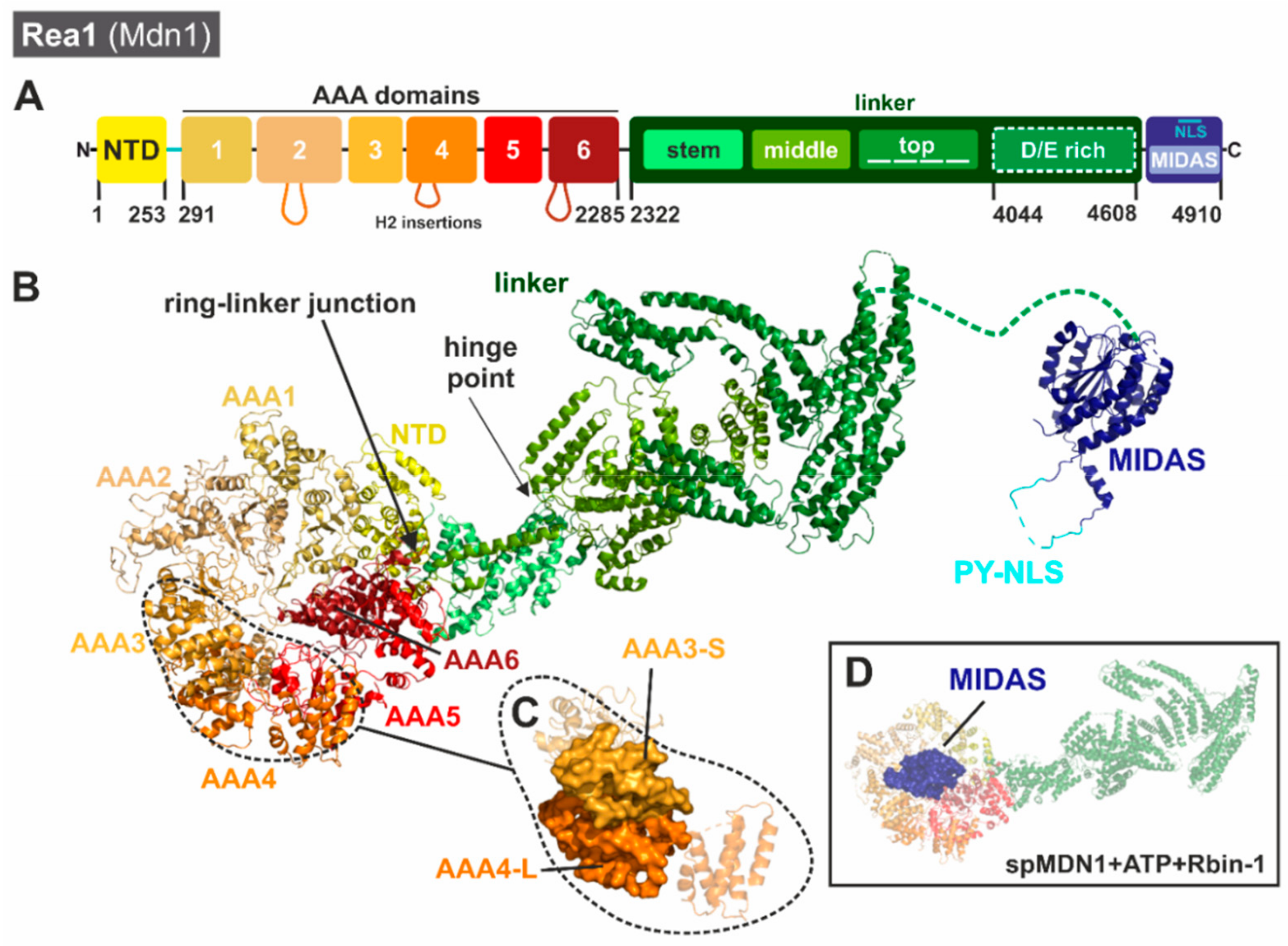
3.2.1. Unique Structural Features of Rea1/Mdn1
3.2.2. One Giant Ratchet for Two Jobs: Pre-60S Remodeling by Rea1 at Multiple Stages
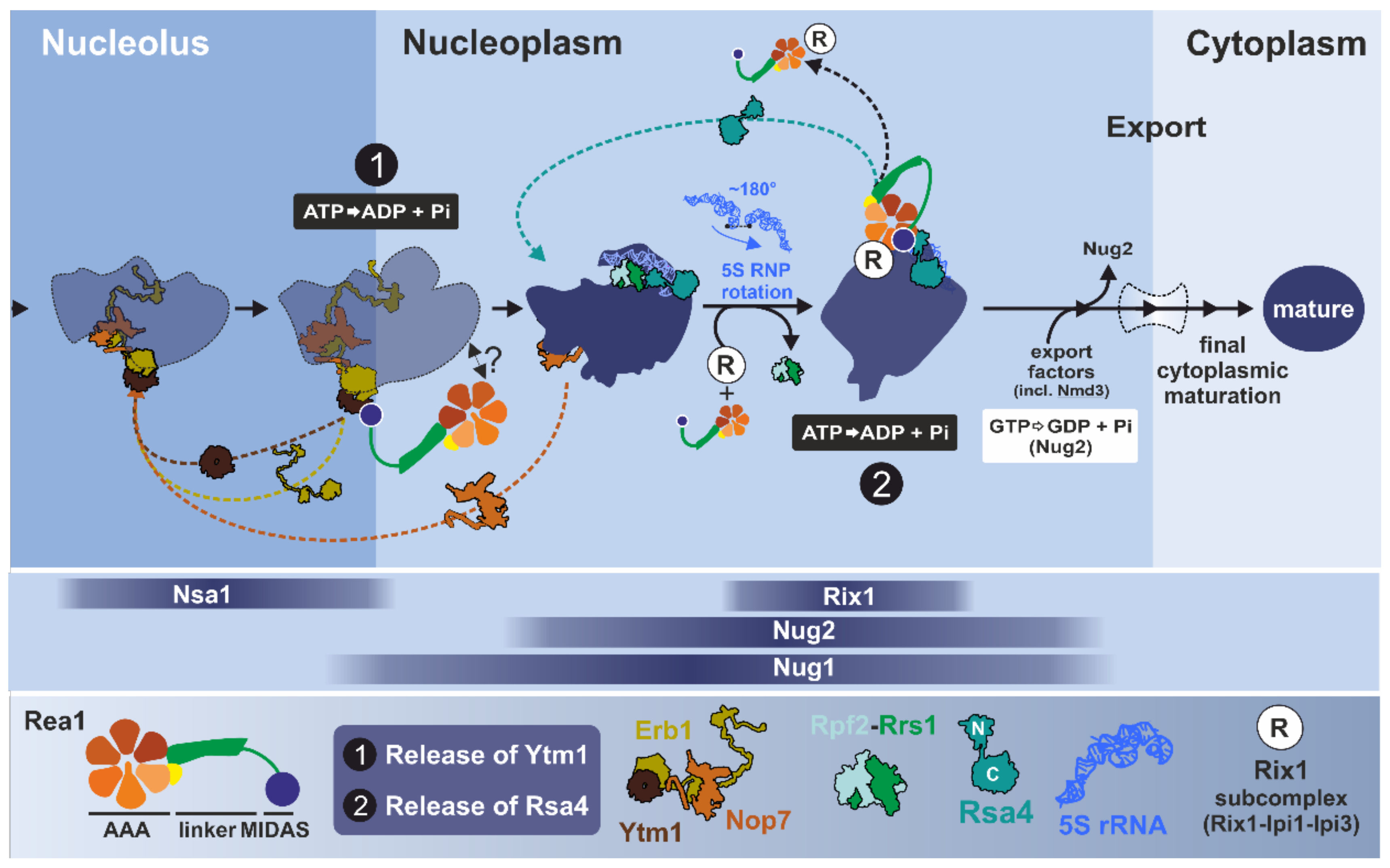
3.2.3. Step 1: Remodeling of the Transiting Pre-60S Particle
3.2.4. Step 2: Remodeling of the Nucleoplasmic Rix1-Associated Pre-60S Particle
3.2.5. Inhibitor-Based Analysis of Mdn1
3.3. Drg1: AAA-ATPase Ante Portas
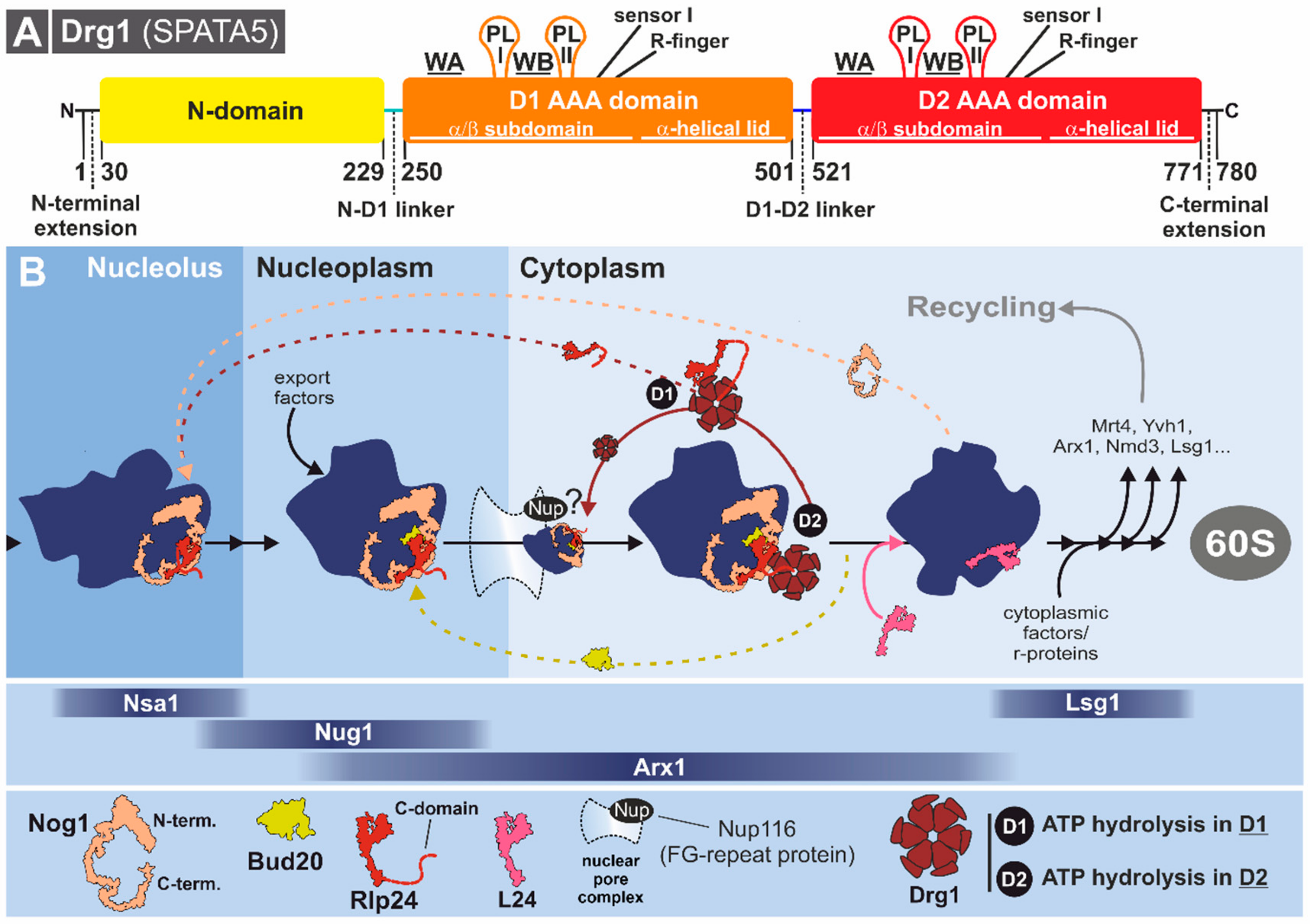
3.3.1. Structural Characteristics of Drg1
3.3.2. Drg1 Initiates the Cytoplasmic Pre-60S Maturation Cascade
3.3.3. Substrate Recognition and Processing by Drg1
3.3.4. Capturing Ribosome Biogenesis Dynamics with the Drg1-Inhibitor Diazaborine
4. Concluding Remarks and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bohnsack, K.E.; Bohnsack, M.T. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 2019, 38, e100278. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef]
- Baßler, J.; Hurt, E. Eukaryotic Ribosome Assembly. Annu. Rev. Biochem. 2019, 88, 281–306. [Google Scholar] [CrossRef] [PubMed]
- Klinge, S.; Woolford, J.L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef]
- Konikkat, S.; Woolford, J.L. Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem. J. 2017, 474, 195–214. [Google Scholar] [CrossRef]
- Kressler, D.; Hurt, E.; Baßler, J. A Puzzle of Life: Crafting Ribosomal Subunits. Trends Biochem. Sci. 2017, 42, 640–654. [Google Scholar] [CrossRef]
- Tafforeau, L.; Zorbas, C.; Langhendries, J.-L.; Mullineux, S.-T.; Stamatopoulou, V.; Mullier, R.; Wacheul, L.; Lafontaine, D.L.J. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell 2013, 51, 539–551. [Google Scholar] [CrossRef]
- Brighenti, E.; Treré, D.; Derenzini, M. Targeted cancer therapy with ribosome biogenesis inhibitors: A real possibility? Oncotarget 2015, 6, 38617–38627. [Google Scholar] [CrossRef]
- Burger, K.; Mühl, B.; Harasim, T.; Rohrmoser, M.; Malamoussi, A.; Orban, M.; Kellner, M.; Gruber-Eber, A.; Kremmer, E.; Hölzel, M.; et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J. Biol. Chem. 2010, 285, 12416–12425. [Google Scholar] [CrossRef]
- Catez, F.; Dalla Venezia, N.; Marcel, V.; Zorbas, C.; Lafontaine, D.L.J.; Diaz, J.-J. Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 2019, 159, 74–81. [Google Scholar] [CrossRef]
- Awad, D.; Prattes, M.; Kofler, L.; Rössler, I.; Loibl, M.; Pertl, M.; Zisser, G.; Wolinski, H.; Pertschy, B.; Bergler, H. Inhibiting eukaryotic ribosome biogenesis. BMC Biol. 2019, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, M.; Montanaro, L.; Trerè, D. Ribosome biogenesis and cancer. Acta Histochem. 2017, 119, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Montanaro, L.; Treré, D.; Derenzini, M. The Ribosome Biogenesis—Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Aspesi, A.; Ellis, S.R. Rare ribosomopathies: Insights into mechanisms of cancer. Nat. Rev. Cancer 2019, 19, 228. [Google Scholar] [CrossRef]
- Sulima, S.O.; Kampen, K.R.; De Keersmaecker, K. Cancer Biogenesis in Ribosomopathies. Cells 2019, 8, 229. [Google Scholar] [CrossRef]
- Danilova, N.; Gazda, H.T. Ribosomopathies: How a common root can cause a tree of pathologies. Dis. Model. Mech. 2015, 8, 1013–1026. [Google Scholar] [CrossRef]
- Farley-Barnes, K.I.; Ogawa, L.M.; Baserga, S.J. Ribosomopathies: Old Concepts, New Controversies. Trends Genet. 2019, 35, 754–767. [Google Scholar] [CrossRef]
- Kampen, K.R.; Sulima, S.O.; Vereecke, S.; De Keersmaecker, K. Hallmarks of ribosomopathies. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Cerezo, E.; Plisson-Chastang, C.; Henras, A.K.; Lebaron, S.; Gleizes, P.-E.; O’Donohue, M.-F.; Romeo, Y.; Henry, Y. Maturation of pre-40S particles in yeast and humans. Wiley Interdiscip. Rev. RNA 2019, 10, e1516. [Google Scholar] [CrossRef]
- Fernández-Pevida, A.; Kressler, D.; de la Cruz, J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip. Rev. RNA 2015, 6, 191–209. [Google Scholar] [CrossRef]
- Dragon, F.; Gallagher, J.E.G.; Compagnone-Post, P.A.; Mitchell, B.M.; Porwancher, K.A.; Wehner, K.A.; Wormsley, S.; Settlage, R.E.; Shabanowitz, J.; Osheim, Y.; et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002, 417, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Grandi, P.; Rybin, V.; Baßler, J.; Petfalski, E.; Strauß, D.; Marzioch, M.; Schäfer, T.; Kuster, B.; Tschochner, H.; Tollervey, D.; et al. 90S Pre-Ribosomes Include the 35S Pre-rRNA, the U3 snoRNP, and 40S Subunit Processing Factors but Predominantly Lack 60S Synthesis Factors. Mol. Cell 2002, 10, 105–115. [Google Scholar] [CrossRef]
- Osheim, Y.N.; French, S.L.; Keck, K.M.; Champion, E.A.; Spasov, K.; Dragon, F.; Baserga, S.J.; Beyer, A.L. Pre-18S Ribosomal RNA Is Structurally Compacted into the SSU Processome Prior to Being Cleaved from Nascent Transcripts in Saccharomyces cerevisiae. Mol. Cell 2004, 16, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Barandun, J.; Hunziker, M.; Klinge, S. Assembly and structure of the SSU processome—A nucleolar precursor of the small ribosomal subunit. Curr. Opin. Struct. Biol. 2018, 49, 85–93. [Google Scholar] [CrossRef]
- Peña, C.; Hurt, E.; Panse, V.G. Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 2017, 24, 689–699. [Google Scholar] [CrossRef]
- Kressler, D.; Hurt, E.; Bergler, H.; Baßler, J. The power of AAA-ATPases on the road of pre-60S ribosome maturation—Molecular machines that strip pre-ribosomal particles. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 92–100. [Google Scholar] [CrossRef]
- Barrio-Garcia, C.; Thoms, M.; Flemming, D.; Kater, L.; Berninghausen, O.; Baßler, J.; Beckmann, R.; Hurt, E. Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat. Struct. Mol. Biol. 2016, 23, 37–44. [Google Scholar] [CrossRef]
- Greber, B.J. Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo-electron microscopy. RNA 2016, 22, 1643–1662. [Google Scholar] [CrossRef]
- Greber, B.J.; Boehringer, D.; Montellese, C.; Ban, N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2012, 19, 1228–1233. [Google Scholar] [CrossRef]
- Kargas, V.; Castro-Hartmann, P.; Escudero-Urquijo, N.; Dent, K.; Hilcenko, C.; Sailer, C.; Zisser, G.; Marques-Carvalho, M.J.; Pellegrino, S.; Wawiórka, L.; et al. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. eLife 2019, 8, e44904. [Google Scholar] [CrossRef]
- Kater, L.; Thoms, M.; Barrio-Garcia, C.; Cheng, J.; Ismail, S.; Ahmed, Y.L.; Bange, G.; Kressler, D.; Berninghausen, O.; Sinning, I.; et al. Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes. Cell 2017, 171, 1599–1610. [Google Scholar] [CrossRef] [PubMed]
- Leidig, C.; Thoms, M.; Holdermann, I.; Bradatsch, B.; Berninghausen, O.; Bange, G.; Sinning, I.; Hurt, E.; Beckmann, R. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 2014, 5, 3491. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Tutuncuoglu, B.; Yan, K.; Brown, H.; Zhang, Y.; Tan, D.; Gamalinda, M.; Yuan, Y.; Li, Z.; Jakovljevic, J.; et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 2016, 534, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Zhu, X.; Zheng, S.; Tan, D.; Dong, M.-Q.; Ye, K. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell 2019, 10, 120–130. [Google Scholar] [CrossRef]
- Zhou, Y.; Musalgaonkar, S.; Johnson, A.W.; Taylor, D.W. Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome. Nat. Commun. 2019, 10, 958. [Google Scholar] [CrossRef]
- Biedka, S.; Wu, S.; LaPeruta, A.J.; Gao, N.; Woolford, J.L., Jr. Insights into remodeling events during eukaryotic large ribosomal subunit assembly provided by high resolution cryo-EM structures. RNA Biol. 2017, 14, 1306–1313. [Google Scholar] [CrossRef][Green Version]
- Sanghai, Z.A.; Miller, L.; Molloy, K.R.; Barandun, J.; Hunziker, M.; Chaker-Margot, M.; Wang, J.; Chait, B.T.; Klinge, S. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 2018, 556, 126–129. [Google Scholar] [CrossRef]
- Kressler, D.; Hurt, E.; Baβler, J. Driving ribosome assembly. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 673–683. [Google Scholar] [CrossRef]
- Strunk, B.S.; Karbstein, K. Powering through ribosome assembly. RNA 2009, 15, 2083–2104. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Karbstein, K. Quality Control Mechanisms during Ribosome Maturation. Trends Cell Biol. 2013, 23, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.J. A ‘garbage can’ for ribosomes: How eukaryotes degrade their ribosomes. Trends Biochem. Sci. 2010, 35, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Thoms, M.; Barrio-Garcia, C.; Thomson, E.; Flemming, D.; Beckmann, R.; Hurt, E. Preribosomes escaping from the nucleus are caught during translation by cytoplasmic quality control. Nat. Struct. Mol. Biol. 2017, 24, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Gamalinda, M.; Ohmayer, U.; Jakovljevic, J.; Kumcuoglu, B.; Woolford, J.; Mbom, B.; Lin, L.; Woolford, J.L. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014, 28, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Biedka, S.; Micic, J.; Wilson, D.; Brown, H.; Diorio-Toth, L.; Woolford, J.L. Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J. Cell Biol. 2018, 217, 2503–2518. [Google Scholar] [CrossRef]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2016, 14, 680–692. [Google Scholar] [CrossRef]
- Ammelburg, M.; Frickey, T.; Lupas, A.N. Classification of AAA+ proteins. J. Struct. Biol. 2006, 156, 2–11. [Google Scholar] [CrossRef]
- Erzberger, J.P.; Berger, J.M. Evolutionary Relationships and Structural Mechanisms of Aaa+ Proteins. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 93–114. [Google Scholar] [CrossRef]
- Frickey, T.; Lupas, A.N. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 2004, 146, 2–10. [Google Scholar] [CrossRef]
- Fröhlich, K.U. An AAA family tree. J. Cell Sci. 2001, 114, 1601–1602. [Google Scholar]
- Iyer, L.M.; Leipe, D.D.; Koonin, E.V.; Aravind, L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004, 146, 11–31. [Google Scholar] [CrossRef] [PubMed]
- Hanson, P.I.; Whiteheart, S.W. AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 2005, 6, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Latterich, M.; Patel, S. The AAA team: Related ATPases with diverse functions. Trends Cell Biol. 1998, 8, 65–71. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A Class of Chaperone-Like ATPases Associated with the Assembly, Operation, and Disassembly of Protein Complexes. Genome Res. 1999, 9, 27–43. [Google Scholar]
- Snider, J.; Thibault, G.; Houry, W.A. The AAA+ superfamily of functionally diverse proteins. Genome Biol. 2008, 9, 216. [Google Scholar] [CrossRef]
- Xia, D.; Tang, W.K.; Ye, Y. Structure and function of the AAA + ATPase p97/Cdc48p. Gene 2016, 583, 64–77. [Google Scholar] [CrossRef]
- Walker, J.E.; Saraste, M.; Runswick, M.J.; Gay, N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982, 1, 945–951. [Google Scholar] [CrossRef]
- Wendler, P.; Ciniawsky, S.; Kock, M.; Kube, S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 2–14. [Google Scholar] [CrossRef]
- Bell, T.A.; Baker, T.A.; Sauer, R.T. Hinge–Linker Elements in the AAA+ Protein Unfoldase ClpX Mediate Intersubunit Communication, Assembly, and Mechanical Activity. Biochemistry 2018, 57, 6787–6796. [Google Scholar] [CrossRef]
- Hänzelmann, P.; Schindelin, H. Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97. Structure 2016, 24, 127–139. [Google Scholar] [CrossRef]
- Huang, C.; Li, G.; Lennarz, W.J. Dynamic flexibility of the ATPase p97 is important for its interprotomer motion transmission. Proc. Natl. Acad. Sci. USA 2012, 109, 9792–9797. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Huang, C.; Zhao, G.; Lennarz, W.J. Interprotomer motion-transmission mechanism for the hexameric AAA ATPase p97. Proc. Natl. Acad. Sci. USA 2012, 109, 3737–3741. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; Sun, S.; Sui, S.-F. The Role of the N-D1 Linker of the N-Ethylmaleimide-Sensitive Factor in the SNARE Disassembly. PLoS ONE 2013, 8, e64346. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.K.; Xia, D. Role of the D1-D2 Linker of Human VCP/p97 in the Asymmetry and ATPase Activity of the D1-domain. Sci. Rep. 2016, 6, 20037. [Google Scholar] [CrossRef] [PubMed]
- Gates, S.N.; Yokom, A.L.; Lin, J.; Jackrel, M.E.; Rizo, A.N.; Kendsersky, N.M.; Buell, C.E.; Sweeny, E.A.; Mack, K.L.; Chuang, E.; et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 2017, 357, 273–279. [Google Scholar] [CrossRef]
- Ripstein, Z.A.; Huang, R.; Augustyniak, R.; Kay, L.E.; Rubinstein, J.L. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife 2017, 6, e25754. [Google Scholar] [CrossRef]
- Cooney, I.; Han, H.; Stewart, M.G.; Carson, R.H.; Hansen, D.T.; Iwasa, J.H.; Price, J.C.; Hill, C.P.; Shen, P.S. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science 2019, 365, 502–505. [Google Scholar] [CrossRef]
- Lo, Y.-H.; Sobhany, M.; Hsu, A.L.; Ford, B.L.; Krahn, J.M.; Borgnia, M.J.; Stanley, R.E. Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7. Nat. Commun. 2019, 10, 513. [Google Scholar] [CrossRef]
- Twomey, E.C.; Ji, Z.; Wales, T.E.; Bodnar, N.O.; Ficarro, S.B.; Marto, J.A.; Engen, J.R.; Rapoport, T.A. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 2019, 365. [Google Scholar] [CrossRef]
- Han, H.; Fulcher, J.M.; Dandey, V.P.; Iwasa, J.H.; Sundquist, W.I.; Kay, M.S.; Shen, P.S.; Hill, C.P. Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases. eLife 2019, 8, e44071. [Google Scholar] [CrossRef]
- White, K.I.; Zhao, M.; Choi, U.B.; Pfuetzner, R.A.; Brunger, A.T. Structural principles of SNARE complex recognition by the AAA+ protein NSF. eLife 2018, 7, e38888. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, N.O.; Rapoport, T.A. Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 2017, 169, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, N.; Rapoport, T. Toward an understanding of the Cdc48/p97 ATPase. F1000Research 2017, 6, 1318. [Google Scholar] [CrossRef] [PubMed]
- Olivares, A.O.; Baker, T.A.; Sauer, R.T. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat. Rev. Microbiol. 2015, 14, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Kudriaeva, A.A.; Belogurov, A.A. Proteasome: A Nanomachinery of Creative Destruction. Biochemistry 2019, 84, 159–192. [Google Scholar] [CrossRef] [PubMed]
- Sauer, R.T.; Baker, T.A. AAA+ Proteases: ATP-Fueled Machines of Protein Destruction. Annu. Rev. Biochem. 2011, 80, 587–612. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.B.; Zhao, M.; White, K.I.; Pfuetzner, R.A.; Esquivies, L.; Zhou, Q.; Brunger, A.T. NSF-mediated disassembly of on- and off-pathway SNARE complexes and inhibition by complexin. eLife 2018, 7, e36497. [Google Scholar] [CrossRef]
- Cipriano, D.J.; Jung, J.; Vivona, S.; Fenn, T.D.; Brunger, A.T.; Bryant, Z. Processive ATP-driven Substrate Disassembly by the N-Ethylmaleimide-sensitive Factor (NSF) Molecular Machine. J. Biol. Chem. 2013, 288, 23436–23445. [Google Scholar] [CrossRef]
- Zhao, M.; Brunger, A.T. Recent Advances in Deciphering the Structure and Molecular Mechanism of the AAA + ATPase N-Ethylmaleimide-Sensitive Factor (NSF). J. Mol. Biol. 2016, 428, 1912–1926. [Google Scholar] [CrossRef]
- Gadal, O.; Strauß, D.; Braspenning, J.; Hoepfner, D.; Petfalski, E.; Philippsen, P.; Tollervey, D.; Hurt, E. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 2001, 20, 3695–3704. [Google Scholar] [CrossRef]
- Kressler, D.; Roser, D.; Pertschy, B.; Hurt, E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol. 2008, 181, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Germain-Lee, E.L.; Obie, C.; Valle, D. NVL: A New Member of the AAA Family of ATPases Localized to the Nucleus. Genomics 1997, 44, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Galani, K.; Nissan, T.A.; Petfalski, E.; Tollervey, D.; Hurt, E. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 2004, 279, 55411–55418. [Google Scholar] [CrossRef] [PubMed]
- Buchberger, A.; Schindelin, H.; Hänzelmann, P. Control of p97 function by cofactor binding. FEBS Lett. 2015, 589, 2578–2589. [Google Scholar] [CrossRef] [PubMed]
- Kappel, L.; Loibl, M.; Zisser, G.; Klein, I.; Fruhmann, G.; Gruber, C.; Unterweger, S.; Rechberger, G.; Pertschy, B.; Bergler, H. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 2012, 199, 771–782. [Google Scholar] [CrossRef]
- Niwa, H.; Ewens, C.A.; Tsang, C.; Yeung, H.O.; Zhang, X.; Freemont, P.S. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J. Biol. Chem. 2012, 287, 8561–8570. [Google Scholar] [CrossRef]
- Prattes, M.; Loibl, M.; Zisser, G.; Luschnig, D.; Kappel, L.; Rössler, I.; Grassegger, M.; Hromic, A.; Krieger, E.; Gruber, K.; et al. A conserved inter-domain communication mechanism regulates the ATPase activity of the AAA-protein Drg1. Sci. Rep. 2017, 7, srep44751. [Google Scholar] [CrossRef]
- Shiozawa, K.; Maita, N.; Tomii, K.; Seto, A.; Goda, N.; Akiyama, Y.; Shimizu, T.; Shirakawa, M.; Hiroaki, H. Structure of the N-terminal domain of PEX1 AAA-ATPase. Characterization of a putative adaptor-binding domain. J. Biol. Chem. 2004, 279, 50060–50068. [Google Scholar] [CrossRef]
- Lo, Y.-H.; Romes, E.M.; Pillon, M.C.; Sobhany, M.; Stanley, R.E. Structural Analysis Reveals Features of Ribosome Assembly Factor Nsa1/WDR74 Important for Localization and Interaction with Rix7/NVL2. Structure 2017, 25, 762–772. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Fujiwara, K.; Goda, N.; Iwaya, N.; Tenno, T.; Shirakawa, M.; Hiroaki, H. Structure and Function of the N-terminal Nucleolin Binding Domain of Nuclear Valosin-containing Protein-like 2 (NVL2) Harboring a Nucleolar Localization Signal. J. Biol. Chem. 2011, 286, 21732–21741. [Google Scholar] [CrossRef]
- Nagahama, M.; Hara, Y.; Seki, A.; Yamazoe, T.; Kawate, Y.; Shinohara, T.; Hatsuzawa, K.; Tani, K.; Tagaya, M. NVL2 Is a Nucleolar AAA-ATPase that Interacts with Ribosomal Protein L5 through Its Nucleolar Localization Sequence. Mol. Biol. Cell 2004, 15, 5712–5723. [Google Scholar] [CrossRef] [PubMed]
- Kondo, K.; Inouye, M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J. Biol. Chem. 1992, 267, 16252–16258. [Google Scholar] [PubMed]
- Lee, W.C.; Xue, Z.X.; Mélèse, T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J. Cell Biol. 1991, 113, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Zabetakis, D.; Mélèse, T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol. Cell. Biol. 1992, 12, 3865–3871. [Google Scholar] [CrossRef]
- Yan, C.; Mélèse, T. Multiple regions of NSR1 are sufficient for accumulation of a fusion protein within the nucleolus. J. Cell Biol. 1993, 123, 1081–1091. [Google Scholar] [CrossRef]
- Deville, C.; Carroni, M.; Franke, K.B.; Topf, M.; Bukau, B.; Mogk, A.; Saibil, H.R. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv. 2017, 3, e1701726. [Google Scholar] [CrossRef]
- Glynn, S.E.; Martin, A.; Nager, A.R.; Baker, T.A.; Sauer, R.T. Structures of Asymmetric ClpX Hexamers Reveal Nucleotide-Dependent Motions in a AAA+ Protein-Unfolding Machine. Cell 2009, 139, 744–756. [Google Scholar] [CrossRef]
- de la Peña, A.H.; Goodall, E.A.; Gates, S.N.; Lander, G.C.; Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 2018, 362. [Google Scholar] [CrossRef]
- Schuller, J.M.; Falk, S.; Fromm, L.; Hurt, E.; Conti, E. Structure of the nuclear exosome captured on a maturing preribosome. Science 2018, 360, 219–222. [Google Scholar] [CrossRef]
- Pellett, S.; Tracy, J.W. Mak16p is required for the maturation of 25S and 5.8S rRNAs in the yeast Saccharomyces cerevisiae. Yeast 2006, 23, 495–506. [Google Scholar] [CrossRef]
- Pratte, D.; Singh, U.; Murat, G.; Kressler, D. Mak5 and Ebp2 act together on early pre-60S particles and their reduced functionality bypasses the requirement for the essential pre-60S factor Nsa1. PLoS ONE 2013, 8, e82741. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hiraishi, N.; Ishida, Y.; Nagahama, M. AAA-ATPase NVL2 acts on MTR4-exosome complex to dissociate the nucleolar protein WDR74. Biochem. Biophys. Res. Commun. 2015, 467, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, N.; Ishida, Y.; Sudo, H.; Nagahama, M. WDR74 participates in an early cleavage of the pre-rRNA processing pathway in cooperation with the nucleolar AAA-ATPase NVL2. Biochem. Biophys. Res. Commun. 2018, 495, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, M.; Yamazoe, T.; Hara, Y.; Tani, K.; Tsuji, A.; Tagaya, M. The AAA-ATPase NVL2 is a component of pre-ribosomal particles that interacts with the DExD/H-box RNA helicase DOB1. Biochem. Biophys. Res. Commun. 2006, 346, 1075–1082. [Google Scholar] [CrossRef]
- Yoshikatsu, Y.; Ishida, Y.; Sudo, H.; Yuasa, K.; Tsuji, A.; Nagahama, M. NVL2, a nucleolar AAA-ATPase, is associated with the nuclear exosome and is involved in pre-rRNA processing. Biochem. Biophys. Res. Commun. 2015, 464, 780–786. [Google Scholar] [CrossRef]
- Wang, M.; Chen, J.; He, K.; Wang, Q.; Li, Z.; Shen, J.; Wen, Z.; Song, Z.; Xu, Y.; Shi, Y. The NVL gene confers risk for both major depressive disorder and schizophrenia in the Han Chinese population. Prog. Neuropsychopharmacol. Biol. Psychiatry 2015, 62, 7–13. [Google Scholar] [CrossRef]
- Zhao, S.G.; Evans, J.R.; Kothari, V.; Sun, G.; Larm, A.; Mondine, V.; Schaeffer, E.M.; Ross, A.E.; Klein, E.A.; Den, R.B.; et al. The Landscape of Prognostic Outlier Genes in High-Risk Prostate Cancer. Clin. Cancer Res. 2016, 22, 1777–1786. [Google Scholar] [CrossRef]
- Büttner, K.; Wenig, K.; Hopfner, K.-P. The exosome: A macromolecular cage for controlled RNA degradation. Mol. Microbiol. 2006, 61, 1372–1379. [Google Scholar] [CrossRef]
- Kilchert, C.; Wittmann, S.; Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016, 17, 227–239. [Google Scholar] [CrossRef]
- Schilders, G.; van Dijk, E.; Pruijn, G.J.M. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007, 35, 2564–2572. [Google Scholar] [CrossRef]
- Thoms, M.; Thomson, E.; Baßler, J.; Gnädig, M.; Griesel, S.; Hurt, E. The Exosome Is Recruited to RNA Substrates through Specific Adaptor Proteins. Cell 2015, 162, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Zinder, J.C.; Lima, C.D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017, 31, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Fromm, L.; Falk, S.; Flemming, D.; Schuller, J.M.; Thoms, M.; Conti, E.; Hurt, E. Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat. Commun. 2017, 8, 1787. [Google Scholar] [CrossRef] [PubMed]
- Lingaraju, M.; Johnsen, D.; Schlundt, A.; Langer, L.M.; Basquin, J.; Sattler, M.; Jensen, T.H.; Falk, S.; Conti, E. The MTR4 helicase recruits nuclear adaptors of the human RNA exosome using distinct arch-interacting motifs. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Suvorova, E.S.; Radke, J.B.; Ting, L.-M.; Vinayak, S.; Alvarez, C.A.; Kratzer, S.; Kim, K.; Striepen, B.; White, M.W. A nucleolar AAA-NTPase is required for parasite division. Mol. Microbiol. 2013, 90, 338–355. [Google Scholar] [CrossRef]
- Wu, D.; Chen, P.J.; Chen, S.; Hu, Y.; Nunez, G.; Ellis, R.E. C. elegans MAC-1, an essential member of the AAA family of ATPases, can bind CED-4 and prevent cell death. Development 1999, 126, 2021–2031. [Google Scholar]
- Nissan, T.A.; Galani, K.; Maco, B.; Tollervey, D.; Aebi, U.; Hurt, E. A Pre-Ribosome with a Tadpole-like Structure Functions in ATP-Dependent Maturation of 60S Subunits. Mol. Cell 2004, 15, 295–301. [Google Scholar] [CrossRef]
- Ulbrich, C.; Diepholz, M.; Baßler, J.; Kressler, D.; Pertschy, B.; Galani, K.; Böttcher, B.; Hurt, E. Mechanochemical Removal of Ribosome Biogenesis Factors from Nascent 60S Ribosomal Subunits. Cell 2009, 138, 911–922. [Google Scholar] [CrossRef]
- Garbarino, J.E.; Gibbons, I.R. Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genom. 2002, 3, 18. [Google Scholar] [CrossRef]
- Li, P.-C.; Ma, J.-J.; Zhou, X.-M.; Li, G.-H.; Zhao, C.-Z.; Xia, H.; Fan, S.-J.; Wang, X.-J. Arabidopsis MDN1 Is Involved in the Establishment of a Normal Seed Proteome and Seed Germination. Front. Plant Sci. 2019, 10, 1118. [Google Scholar] [CrossRef]
- Li, P.-C.; Li, K.; Wang, J.; Zhao, C.-Z.; Zhao, S.-Z.; Hou, L.; Xia, H.; Ma, C.-L.; Wang, X.-J. The AAA-ATPase MIDASIN 1 Functions in Ribosome Biogenesis and Is Essential for Embryo and Root Development. Plant Physiol. 2019, 180, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Raman, N.; Weir, E.; Müller, S. The AAA ATPase MDN1 Acts as a SUMO-Targeted Regulator in Mammalian Pre-ribosome Remodeling. Mol. Cell 2016, 64, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Hook, P.; Vallee, R.B. The dynein family at a glance. J. Cell Sci. 2006, 119, 4369–4371. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.J.; Kon, T.; Knight, P.J.; Sutoh, K.; Burgess, S.A. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Carter, A.P. Review: Structure and mechanism of the dynein motor ATPase. Biopolymers 2016, 105, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Y.L.; Thoms, M.; Mitterer, V.; Sinning, I.; Hurt, E. Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin–ligand-type complexes. Nat. Commun. 2019, 10, 3050. [Google Scholar] [CrossRef] [PubMed]
- Sosnowski, P.; Urnavicius, L.; Boland, A.; Fagiewicz, R.; Busselez, J.; Papai, G.; Schmidt, H. The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1. eLife 2018, 7, e39163. [Google Scholar] [CrossRef]
- Chen, Z.; Suzuki, H.; Kobayashi, Y.; Wang, A.C.; DiMaio, F.; Kawashima, S.A.; Walz, T.; Kapoor, T.M. Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis. Cell 2018, 175, 822–834. [Google Scholar] [CrossRef]
- Matsuo, Y.; Granneman, S.; Thoms, M.; Manikas, R.-G.; Tollervey, D.; Hurt, E. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 2014, 505, 112–116. [Google Scholar] [CrossRef]
- Kawashima, S.A.; Chen, Z.; Aoi, Y.; Patgiri, A.; Kobayashi, Y.; Nurse, P.; Kapoor, T.M. Potent, Reversible, and Specific Chemical Inhibitors of Eukaryotic Ribosome Biogenesis. Cell 2016, 167, 512–524. [Google Scholar] [CrossRef]
- Baßler, J.; Kallas, M.; Pertschy, B.; Ulbrich, C.; Thoms, M.; Hurt, E. The AAA-ATPase Rea1 Drives Removal of Biogenesis Factors during Multiple Stages of 60S Ribosome Assembly. Mol. Cell 2010, 38, 712–721. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thoms, M.; Ahmed, Y.L.; Maddi, K.; Hurt, E.; Sinning, I. Concerted removal of the Erb1–Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res. 2016, 44, 926–939. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Harnpicharnchai, P.; Jakovljevic, J.; Horsey, E.; Miles, T.; Roman, J.; Rout, M.; Meagher, D.; Imai, B.; Guo, Y.; Brame, C.J.; et al. Composition and Functional Characterization of Yeast 66S Ribosome Assembly Intermediates. Mol. Cell 2001, 8, 505–515. [Google Scholar] [CrossRef]
- Miles, T.D.; Jakovljevic, J.; Horsey, E.W.; Harnpicharnchai, P.; Tang, L.; Woolford, J.L. Ytm1, Nop7, and Erb1 Form a Complex Necessary for Maturation of Yeast 66S Preribosomes. Mol. Cell. Biol. 2005, 25, 10419–10432. [Google Scholar] [CrossRef] [PubMed]
- Sahasranaman, A.; Dembowski, J.; Strahler, J.; Andrews, P.; Maddock, J.; Woolford, J.L. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: Role of factors required for 27S pre-rRNA processing. EMBO J. 2011, 30, 4020–4032. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Sahasranaman, A.; Jakovljevic, J.; Schleifman, E.; Woolford, J.L. Interactions among Ytm1, Erb1, and Nop7 Required for Assembly of the Nop7-Subcomplex in Yeast Preribosomes. Mol. Biol. Cell 2008, 19, 2844–2856. [Google Scholar] [CrossRef] [PubMed]
- Romes, E.M.; Sobhany, M.; Stanley, R.E. The Crystal Structure of the Ubiquitin-like Domain of Ribosome Assembly Factor Ytm1 and Characterization of Its Interaction with the AAA-ATPase Midasin. J. Biol. Chem. 2016, 291, 882–893. [Google Scholar] [CrossRef]
- Konikkat, S.; Biedka, S.; Woolford, J.L. The assembly factor Erb1 functions in multiple remodeling events during 60S ribosomal subunit assembly in S. cerevisiae. Nucleic Acids Res. 2017, 45, 4853–4865. [Google Scholar] [CrossRef][Green Version]
- Marcin, W.; Neira, J.L.; Bravo, J. The Carboxy-Terminal Domain of Erb1 Is a Seven-Bladed ß-Propeller that Binds RNA. PLoS ONE 2015, 10, e0123463. [Google Scholar] [CrossRef]
- Wegrecki, M.; Rodríguez-Galán, O.; de la Cruz, J.; Bravo, J. The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two β-propellers during the assembly of large ribosomal subunits in eukaryotes. Nucleic Acids Res. 2015, 43, 11017–11030. [Google Scholar] [CrossRef]
- Granneman, S.; Petfalski, E.; Tollervey, D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011, 30, 4006–4019. [Google Scholar] [CrossRef] [PubMed]
- Bradatsch, B.; Leidig, C.; Granneman, S.; Gnädig, M.; Tollervey, D.; Böttcher, B.; Beckmann, R.; Hurt, E. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat. Struct. Mol. Biol. 2012, 19, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Zisser, G.; Ohmayer, U.; Mauerhofer, C.; Mitterer, V.; Klein, I.; Rechberger, G.N.; Wolinski, H.; Prattes, M.; Pertschy, B.; Milkereit, P.; et al. Viewing pre-60S maturation at a minute’s timescale. Nucleic Acids Res. 2018, 46, 3140–3151. [Google Scholar] [CrossRef] [PubMed]
- Grimm, T.; Hölzel, M.; Rohrmoser, M.; Harasim, T.; Malamoussi, A.; Gruber-Eber, A.; Kremmer, E.; Eick, D. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006, 34, 3030–3043. [Google Scholar] [CrossRef]
- Hölzel, M.; Rohrmoser, M.; Schlee, M.; Grimm, T.; Harasim, T.; Malamoussi, A.; Gruber-Eber, A.; Kremmer, E.; Hiddemann, W.; Bornkamm, G.W.; et al. Mammalian WDR12 is a novel member of the Pes1–Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 2005, 170, 367–378. [Google Scholar] [CrossRef]
- Hölzel, M.; Grimm, T.; Rohrmoser, M.; Malamoussi, A.; Harasim, T.; Gruber-Eber, A.; Kremmer, E.; Eick, D. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res. 2007, 35, 789–800. [Google Scholar] [CrossRef]
- Kellner, M.; Rohrmoser, M.; Forné, I.; Voss, K.; Burger, K.; Mühl, B.; Gruber-Eber, A.; Kremmer, E.; Imhof, A.; Eick, D. DEAD-box helicase DDX27 regulates 3′ end formation of ribosomal 47S RNA and stably associates with the PeBoW-complex. Exp. Cell Res. 2015, 334, 146–159. [Google Scholar] [CrossRef]
- Lapik, Y.R.; Fernandes, C.J.; Lau, L.F.; Pestov, D.G. Physical and Functional Interaction between Pes1 and Bop1 in Mammalian Ribosome Biogenesis. Mol. Cell 2004, 15, 17–29. [Google Scholar] [CrossRef]
- Rohrmoser, M.; Hölzel, M.; Grimm, T.; Malamoussi, A.; Harasim, T.; Orban, M.; Pfisterer, I.; Gruber-Eber, A.; Kremmer, E.; Eick, D. Interdependence of Pes1, Bop1, and WDR12 Controls Nucleolar Localization and Assembly of the PeBoW Complex Required for Maturation of the 60S Ribosomal Subunit. Mol. Cell. Biol. 2007, 27, 3682–3694. [Google Scholar] [CrossRef]
- Strezoska, Ž.; Pestov, D.G.; Lau, L.F. Functional Inactivation of the Mouse Nucleolar Protein Bop1 Inhibits Multiple Steps in Pre-rRNA Processing and Blocks Cell Cycle Progression. J. Biol. Chem. 2002, 277, 29617–29625. [Google Scholar] [CrossRef]
- Castle, C.D.; Cassimere, E.K.; Denicourt, C. LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol. Biol. Cell 2012, 23, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, E.; Haindl, M.; Muller, S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011, 30, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Chantha, S.-C.; Matton, D.P. Underexpression of the plant NOTCHLESS gene, encoding a WD-repeat protein, causes pleitropic phenotype during plant development. Planta 2007, 225, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Chantha, S.-C.; Emerald, B.S.; Matton, D.P. Characterization of the plant Notchless homolog, a WD repeat protein involved in seed development. Plant Mol. Biol. 2006, 62, 897–912. [Google Scholar] [CrossRef] [PubMed]
- Chantha, S.-C.; Gray-Mitsumune, M.; Houde, J.; Matton, D.P. The MIDASIN and NOTCHLESS genes are essential for female gametophyte development in Arabidopsis thaliana. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2010, 16, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Beckmann, R.; Cate, J.H.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014, 24, 165–169. [Google Scholar] [CrossRef]
- Dinman, J.D. 5S rRNA: Structure and Function from Head to Toe. Int. J. Biomed. Sci. 2005, 1, 2–7. [Google Scholar]
- Kressler, D.; Bange, G.; Ogawa, Y.; Stjepanovic, G.; Bradatsch, B.; Pratte, D.; Amlacher, S.; Strauß, D.; Yoneda, Y.; Katahira, J.; et al. Synchronizing Nuclear Import of Ribosomal Proteins with Ribosome Assembly. Science 2012, 338, 666–671. [Google Scholar] [CrossRef]
- Pelava, A.; Schneider, C.; Watkins, N.J. The importance of ribosome production, and the 5S RNP–MDM2 pathway, in health and disease. Biochem. Soc. Trans. 2016, 44, 1086–1090. [Google Scholar] [CrossRef]
- Sloan, K.E.; Bohnsack, M.T.; Watkins, N.J. The 5S RNP Couples p53 Homeostasis to Ribosome Biogenesis and Nucleolar Stress. Cell Rep. 2013, 5, 237–247. [Google Scholar] [CrossRef]
- Asano, N.; Kato, K.; Nakamura, A.; Komoda, K.; Tanaka, I.; Yao, M. Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis. Nucleic Acids Res. 2015, 43, 4746–4757. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kharde, S.; Calviño, F.R.; Gumiero, A.; Wild, K.; Sinning, I. The structure of Rpf2–Rrs1 explains its role in ribosome biogenesis. Nucleic Acids Res. 2015, 43, 7083–7095. [Google Scholar] [CrossRef] [PubMed]
- Madru, C.; Lebaron, S.; Blaud, M.; Delbos, L.; Pipoli, J.; Pasmant, E.; Réty, S.; Leulliot, N. Chaperoning 5S RNA assembly. Genes Dev. 2015, 29, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Harnpicharnchai, P.; Jakovljevic, J.; Tang, L.; Guo, Y.; Oeffinger, M.; Rout, M.P.; Hiley, S.L.; Hughes, T.; Woolford, J.L. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007, 21, 2580–2592. [Google Scholar] [CrossRef] [PubMed]
- Thoms, M.; Mitterer, V.; Kater, L.; Falquet, L.; Beckmann, R.; Kressler, D.; Hurt, E. Suppressor mutations in Rpf2–Rrs1 or Rpl5 bypass the Cgr1 function for pre-ribosomal 5S RNP-rotation. Nat. Commun. 2018, 9, 4094. [Google Scholar] [CrossRef] [PubMed]
- Loibl, M.; Klein, I.; Prattes, M.; Schmidt, C.; Kappel, L.; Zisser, G.; Gungl, A.; Krieger, E.; Pertschy, B.; Bergler, H. The drug diazaborine blocks ribosome biogenesis by inhibiting the AAA-ATPase Drg1. J. Biol. Chem. 2014, 289, 3913–3922. [Google Scholar] [CrossRef] [PubMed]
- Pertschy, B.; Zisser, G.; Schein, H.; Köffel, R.; Rauch, G.; Grillitsch, K.; Morgenstern, C.; Durchschlag, M.; Högenauer, G.; Bergler, H. Diazaborine Treatment of Yeast Cells Inhibits Maturation of the 60S Ribosomal Subunit. Mol. Cell. Biol. 2004, 24, 6476–6487. [Google Scholar] [CrossRef]
- Wendler, F.; Bergler, H.; Prutej, K.; Jungwirth, H.; Zisser, G.; Kuchler, K.; Högenauer, G. Diazaborine Resistance in the Yeast Saccharomyces cerevisiae Reveals a Link between YAP1 and the Pleiotropic Drug Resistance Genes PDR1 andPDR3. J. Biol. Chem. 1997, 272, 27091–27098. [Google Scholar] [CrossRef]
- Steinman, J.B.; Kapoor, T.M. Using chemical inhibitors to probe AAA protein conformational dynamics and cellular functions. Curr. Opin. Chem. Biol. 2019, 50, 45–54. [Google Scholar] [CrossRef]
- Pertschy, B.; Saveanu, C.; Zisser, G.; Lebreton, A.; Tengg, M.; Jacquier, A.; Liebminger, E.; Nobis, B.; Kappel, L.; van der Klei, I.; et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 2007, 27, 6581–6592. [Google Scholar] [CrossRef]
- Jungwirth, H.; Bergler, H.; Högenauer, G. Diazaborine Treatment of Baker’s Yeast Results in Stabilization of Aberrant mRNAs. J. Biol. Chem. 2001, 276, 36419–36424. [Google Scholar] [CrossRef] [PubMed]
- Zakalskiy, A.; Högenauer, G.; Ishikawa, T.; Wehrschütz-Sigl, E.; Wendler, F.; Teis, D.; Zisser, G.; Steven, A.C.; Bergler, H. Structural and enzymatic properties of the AAA protein Drg1p from Saccharomyces cerevisiae. Decoupling of intracellular function from ATPase activity and hexamerization. J. Biol. Chem. 2002, 277, 26788–26795. [Google Scholar] [CrossRef] [PubMed]
- Baek, G.H.; Cheng, H.; Choe, V.; Bao, X.; Shao, J.; Luo, S.; Rao, H. Cdc48: A swiss army knife of cell biology. J. Amino Acids 2013, 2013, 183421. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, P.; Schindelin, H. The Interplay of Cofactor Interactions and Post-translational Modifications in the Regulation of the AAA+ ATPase p97. Front. Mol. Biosci. 2017, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Black, J.; Kisiel, N.; Kulesz-Martin, M.F. SPAF, a new AAA-protein specific to early spermatogenesis and malignant conversion. Oncogene 2000, 19, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Heallen, T.R.; Adams, H.P.; Furuta, T.; Verbrugghe, K.J.; Schumacher, J.M. An Afg2/Spaf-Related Cdc48-like AAA ATPase Regulates the Stability and Activity of the C. elegans Aurora B Kinase AIR-2. Dev. Cell 2008, 15, 603–616. [Google Scholar] [CrossRef]
- Puusepp, S.; Kovacs-Nagy, R.; Alhaddad, B.; Braunisch, M.; Hoffmann, G.F.; Kotzaeridou, U.; Lichvarova, L.; Liiv, M.; Makowski, C.; Mandel, M.; et al. Compound heterozygous SPATA5 variants in four families and functional studies of SPATA5 deficiency. Eur. J. Hum. Genet. 2018, 26, 407–419. [Google Scholar] [CrossRef]
- Tanaka, A.J.; Cho, M.T.; Millan, F.; Juusola, J.; Retterer, K.; Joshi, C.; Niyazov, D.; Garnica, A.; Gratz, E.; Deardorff, M.; et al. Mutations in SPATA5 Are Associated with Microcephaly, Intellectual Disability, Seizures, and Hearing Loss. Am. J. Hum. Genet. 2015, 97, 457–464. [Google Scholar] [CrossRef]
- Buchert, R.; Nesbitt, A.I.; Tawamie, H.; Krantz, I.D.; Medne, L.; Helbig, I.; Matalon, D.R.; Reis, A.; Santani, A.; Sticht, H.; et al. SPATA5 mutations cause a distinct autosomal recessive phenotype of intellectual disability, hypotonia and hearing loss. Orphanet J. Rare Dis. 2016, 11, 130. [Google Scholar] [CrossRef]
- Kurata, H.; Terashima, H.; Nakashima, M.; Okazaki, T.; Matsumura, W.; Ohno, K.; Saito, Y.; Maegaki, Y.; Kubota, M.; Nanba, E.; et al. Characterization of SPATA5-related encephalopathy in early childhood. Clin. Genet. 2016, 90, 437–444. [Google Scholar] [CrossRef]
- Szczałuba, K.; Szymańska, K.; Kosińska, J.; Pollak, A.; Murcia, V.; Kędra, A.; Stawiński, P.; Rydzanicz, M.; Demkow, U.; Płoski, R. Isolated Hearing Impairment Caused by SPATA5 Mutations in a Family with Variable Phenotypic Expression. Adv. Exp. Med. Biol. 2017, 980, 59–66. [Google Scholar] [PubMed]
- Hetman, M.; Slomnicki, L.P. Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies. J. Neurochem. 2019, 148, 325–347. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.-Y.; Li, Z.; Bussiere, C.; Bresson, S.; Marcotte, E.M.; Johnson, A.W. Defining the Pathway of Cytoplasmic Maturation of the 60S Ribosomal Subunit. Mol. Cell 2010, 39, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Espinar-Marchena, F.J.; Babiano, R.; Cruz, J. Placeholder factors in ribosome biogenesis: Please, pave my way. Microb. Cell 2017, 4, 144–168. [Google Scholar] [CrossRef] [PubMed]
- Saveanu, C.; Namane, A.; Gleizes, P.-E.; Lebreton, A.; Rousselle, J.-C.; Noaillac-Depeyre, J.; Gas, N.; Jacquier, A.; Fromont-Racine, M. Sequential Protein Association with Nascent 60S Ribosomal Particles. Mol. Cell. Biol. 2003, 23, 4449–4460. [Google Scholar] [CrossRef]
- Altvater, M.; Chang, Y.; Melnik, A.; Occhipinti, L.; Schütz, S.; Rothenbusch, U.; Picotti, P.; Panse, V.G. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol. Syst. Biol. 2012, 8, 628. [Google Scholar] [CrossRef]
- Bassler, J.; Klein, I.; Schmidt, C.; Kallas, M.; Thomson, E.; Wagner, M.A.; Bradatsch, B.; Rechberger, G.; Strohmaier, H.; Hurt, E.; et al. The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol. Cell. Biol. 2012, 32, 4898–4912. [Google Scholar] [CrossRef]
- Greber, B.J.; Gerhardy, S.; Leitner, A.; Leibundgut, M.; Salem, M.; Boehringer, D.; Leulliot, N.; Aebersold, R.; Panse, V.G.; Ban, N. Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation. Cell 2016, 164, 91–102. [Google Scholar] [CrossRef]
- Nerurkar, P.; Gillet, L.; Pena, C.; Schubert, O.T.; Altvater, M.; Chang, Y.; Aebersold, R.; Panse, V. The GTPase Nog1 couples polypeptide exit tunnel quality control with ribosomal stalk assembly. BioRxiv 2018. [Google Scholar] [CrossRef]
- Fernández-Pevida, A.; Rodríguez-Galán, O.; Díaz-Quintana, A.; Kressler, D.; Cruz, J. de la Yeast Ribosomal Protein L40 Assembles Late into Precursor 60 S Ribosomes and Is Required for Their Cytoplasmic Maturation. J. Biol. Chem. 2012, 287, 38390–38407. [Google Scholar] [CrossRef]
- Hedges, J.; West, M.; Johnson, A.W. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005, 24, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Kemmler, S.; Occhipinti, L.; Veisu, M.; Panse, V.G. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 2009, 186, 863–880. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.E.; Hoover, L.A.; Craig, E.A. The Cytosolic J-protein, Jjj1, and Rei1 Function in the Removal of the Pre-60 S Subunit Factor Arx1. J. Biol. Chem. 2010, 285, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Sulima, S.O.; Gülay, S.P.; Anjos, M.; Patchett, S.; Meskauskas, A.; Johnson, A.W.; Dinman, J.D. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res. 2014, 42, 2049–2063. [Google Scholar] [CrossRef]
- Weis, F.; Giudice, E.; Churcher, M.; Jin, L.; Hilcenko, C.; Wong, C.C.; Traynor, D.; Kay, R.R.; Warren, A.J. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015, 22, 914–919. [Google Scholar] [CrossRef]
- Brown, R.S.H.; Zhao, C.; Chase, A.R.; Wang, J.; Schlieker, C. The mechanism of Torsin ATPase activation. Proc. Natl. Acad. Sci. USA 2014, 111, E4822–E4831. [Google Scholar] [CrossRef]
- Horsnell, W.G.C.; Steel, G.J.; Morgan, A. Analysis of NSF Mutants Reveals Residues Involved in SNAP Binding and ATPase Stimulation. Biochemistry 2002, 41, 5230–5235. [Google Scholar] [CrossRef]
- Morgan, A.; Dimaline, R.; Burgoyne, R.D. The ATPase activity of N-ethylmaleimide-sensitive fusion protein (NSF) is regulated by soluble NSF attachment proteins. J. Biol. Chem. 1994, 269, 29347–29350. [Google Scholar]
- Schlieker, C.; Weibezahn, J.; Patzelt, H.; Tessarz, P.; Strub, C.; Zeth, K.; Erbse, A.; Schneider-Mergener, J.; Chin, J.W.; Schultz, P.G.; et al. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 2004, 11, 607. [Google Scholar] [CrossRef]
- Söllner, T.; Whiteheart, S.W.; Brunner, M.; Erdjument-Bromage, H.; Geromanos, S.; Tempst, P.; Rothman, J.E. SNAP receptors implicated in vesicle targeting and fusion. Nature 1993, 362, 318. [Google Scholar] [CrossRef]
- Steel, G.J.; Morgan, A. Selective stimulation of the D1 ATPase domain of N-ethylmaleimide-sensitive fusion protein (NSF) by soluble NSF attachment proteins. FEBS Lett. 1998, 423, 113–116. [Google Scholar] [CrossRef]
- Ye, Q.; Rosenberg, S.C.; Moeller, A.; Speir, J.A.; Su, T.Y.; Corbett, K.D. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife 2015, 4, e07367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wigley, D.B. The “Glutamate Switch”: A link between ATPase activity and ligand binding in AAA+ proteins. Nat. Struct. Mol. Biol. 2008, 15, 1223–1227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gui, L.; Zhang, X.; Bulfer, S.L.; Sanghez, V.; Wong, D.E.; Lee, Y.; Lehmann, L.; Lee, J.S.; Shih, P.-Y.; et al. Altered cofactor regulation with disease-associated p97/VCP mutations. Proc. Natl. Acad. Sci. USA 2015, 112, E1705–E1714. [Google Scholar] [CrossRef] [PubMed]
- Shchepachev, V.; Bresson, S.; Spanos, C.; Petfalski, E.; Fischer, L.; Rappsilber, J.; Tollervey, D. Defining the RNA interactome by total RNA-associated protein purification. Mol. Syst. Biol. 2019, 15. [Google Scholar] [CrossRef]
- Albert, B.; Kos-Braun, I.C.; Henras, A.K.; Dez, C.; Rueda, M.P.; Zhang, X.; Gadal, O.; Kos, M.; Shore, D. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. eLife 2019, 8, e45002. [Google Scholar] [CrossRef]
- Tye, B.W.; Commins, N.; Ryazanova, L.V.; Wühr, M.; Springer, M.; Pincus, D.; Churchman, L.S. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. eLife 2019, 8, e43002. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prattes, M.; Lo, Y.-H.; Bergler, H.; Stanley, R.E. Shaping the Nascent Ribosome: AAA-ATPases in Eukaryotic Ribosome Biogenesis. Biomolecules 2019, 9, 715. https://doi.org/10.3390/biom9110715
Prattes M, Lo Y-H, Bergler H, Stanley RE. Shaping the Nascent Ribosome: AAA-ATPases in Eukaryotic Ribosome Biogenesis. Biomolecules. 2019; 9(11):715. https://doi.org/10.3390/biom9110715
Chicago/Turabian StylePrattes, Michael, Yu-Hua Lo, Helmut Bergler, and Robin E. Stanley. 2019. "Shaping the Nascent Ribosome: AAA-ATPases in Eukaryotic Ribosome Biogenesis" Biomolecules 9, no. 11: 715. https://doi.org/10.3390/biom9110715
APA StylePrattes, M., Lo, Y.-H., Bergler, H., & Stanley, R. E. (2019). Shaping the Nascent Ribosome: AAA-ATPases in Eukaryotic Ribosome Biogenesis. Biomolecules, 9(11), 715. https://doi.org/10.3390/biom9110715




