Cell Fate Control by Translation: mRNA Translation Initiation as a Therapeutic Target for Cancer Development and Stem Cell Fate Control
Abstract
:1. Introduction
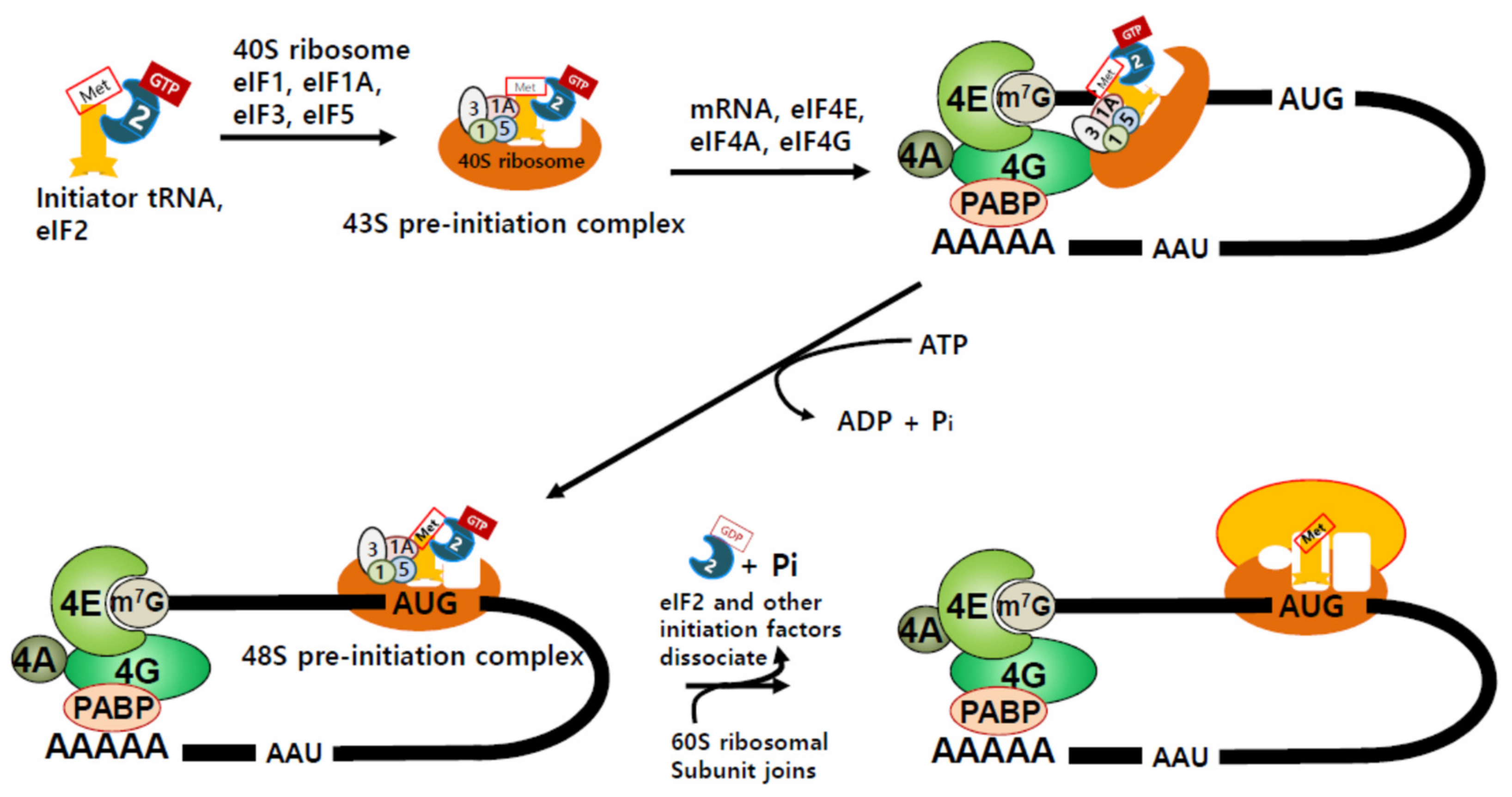
2. Translation Initiation and Regulation of Protein Synthesis
3. Molecular Signals that Regulate Protein Synthesis
3.1. Mitogen-Activated Protein Kinase Kinase (MEK) Signals and Protein Synthesis
3.2. mTOR Signals and Protein Synthesis
4. Protein Synthesis and Cancer
4.1. eIF4E and Cancer
4.2. mTOR, 4EBP, and Cancer
4.3. eIF2 and Cancer
4.4. Cap-Independent Translation and Cancer
5. Translational Control of the Stem Cell Fate
5.1. Differences in the Rate and Amount of Translation in Stem and Differentiated Cells
5.2. eIF4E, 4EBP, and the Control of Stem Cell Fate
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kong, S.Y.; Kim, W.; Lee, H.R.; Kim, H.J. The histone demethylase KDM5A is required for the repression of astrocytogenesis and regulated by the translational machinery in neural progenitor cells. FASEB J. 2018, 32, 1108–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, P.A.; Smith, K.; Poulsom, R.; De Benedetti, A.; Bicknell, R.; Harris, A.L. Differential expression of vascular endothelial growth factor mRNA vs protein isoform expression in human breast cancer and relationship to eIF-4E. Br. J. Cancer 1998, 77, 2120–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenwald, I.B.; Kaspar, R.; Rousseau, D.; Gehrke, L.; Leboulch, P.; Chen, J.J.; Schmidt, E.V.; Sonenberg, N.; London, I.M. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J. Biol. Chem. 1995, 270, 21176–21180. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Hayday, A.C.; Wiman, K.; Hayward, W.S.; Tonegawa, S. Activation of the c-myc gene by translocation: A model for translational control. Proc. Natl. Acad. Sci. USA 1983, 80, 7476–7480. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.P.; Topisirovic, I. Signaling Pathways Involved in the Regulation of mRNA Translation. Mol. Cell. Biol. 2018, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radhakrishnan, A.; Green, R. Connections Underlying Translation and mRNA Stability. J. Mol. Biol. 2016, 428, 3558–3564. [Google Scholar] [CrossRef]
- Buszczak, M.; Signer, R.A.; Morrison, S.J. Cellular differences in protein synthesis regulate tissue homeostasis. Cell 2014, 159, 242–251. [Google Scholar] [CrossRef]
- Truitt, M.L.; Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 2016, 16, 288–304. [Google Scholar] [CrossRef] [Green Version]
- Wilson, D.N.; Doudna Cate, J.H. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef]
- Attardi, G.; Amaldi, F. Structure and synthesis of ribosomal RNA. Annu. Rev. Biochem. 1970, 39, 183–226. [Google Scholar] [CrossRef]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Sonenberg, N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005, 433, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004, 32, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.; Etchison, D.; Hershey, J.W. Protein synthesis eukaryotic initiation factors 4A and 4B are not altered by poliovirus infection of HeLa cells. J. Biol. Chem. 1983, 258, 7236–7239. [Google Scholar] [PubMed]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Pause, A.; Haghighat, A.; Pyronnet, S.; Witherell, G.; Belsham, G.J.; Sonenberg, N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 2001, 7, 382–394. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Robichaud, N.; Sonenberg, N. Translational control and the cancer cell response to stress. Curr. Opin. Cell Biol. 2017, 45, 102–109. [Google Scholar] [CrossRef]
- Proud, C.G. Mnks, eIF4E phosphorylation and cancer. Biochim. Biophys. Acta 2015, 1849, 766–773. [Google Scholar] [CrossRef]
- Morita, M.; Gravel, S.P.; Hulea, L.; Larsson, O.; Pollak, M.; St-Pierre, J.; Topisirovic, I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 2015, 14, 473–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genheden, M.; Kenney, J.W.; Johnston, H.E.; Manousopoulou, A.; Garbis, S.D.; Proud, C.G. BDNF stimulation of protein synthesis in cortical neurons requires the MAP kinase-interacting kinase MNK1. J. Neurosci. 2015, 35, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Waskiewicz, A.J.; Flynn, A.; Proud, C.G.; Cooper, J.A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997, 16, 1909–1920. [Google Scholar] [CrossRef] [PubMed]
- Scheper, G.C.; Morrice, N.A.; Kleijn, M.; Proud, C.G. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 2001, 21, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, R.; Hunter, T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997, 16, 1921–1933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyronnet, S.; Imataka, H.; Gingras, A.C.; Fukunaga, R.; Hunter, T.; Sonenberg, N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999, 18, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Waskiewicz, A.J.; Johnson, J.C.; Penn, B.; Mahalingam, M.; Kimball, S.R.; Cooper, J.A. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 1999, 19, 1871–1880. [Google Scholar] [CrossRef]
- Furic, L.; Rong, L.; Larsson, O.; Koumakpayi, I.H.; Yoshida, K.; Brueschke, A.; Petroulakis, E.; Robichaud, N.; Pollak, M.; Gaboury, L.A.; et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. USA 2010, 107, 14134–14139. [Google Scholar] [CrossRef] [Green Version]
- Duncan, R.; Milburn, S.C.; Hershey, J.W. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem. 1987, 262, 380–388. [Google Scholar]
- Marcotrigiano, J.; Gingras, A.C.; Sonenberg, N.; Burley, S.K. X-ray studies of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Nucleic Acids Symp. Ser. 1997, 36, 8–11. [Google Scholar]
- Scheper, G.C.; Proud, C.G. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 2002, 269, 5350–5359. [Google Scholar] [CrossRef]
- Batool, A.; Aashaq, S.; Andrabi, K.I. Reappraisal to the study of 4E-BP1 as an mTOR substrate—A normative critique. Eur. J. Cell Biol. 2017, 96, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Hay, N.; Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 2004, 18, 1926–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cargnello, M.; Tcherkezian, J.; Roux, P.P. The expanding role of mTOR in cancer cell growth and proliferation. Mutagenesis 2015, 30, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaeschke, A.; Hartkamp, J.; Saitoh, M.; Roworth, W.; Nobukuni, T.; Hodges, A.; Sampson, J.; Thomas, G.; Lamb, R. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J. Cell Biol. 2002, 159, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Pan, D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001, 15, 1383–1392. [Google Scholar] [CrossRef]
- Inoki, K.; Li, Y.; Zhu, T.; Wu, J.; Guan, K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002, 4, 648–657. [Google Scholar] [CrossRef]
- Xiao, G.H.; Shoarinejad, F.; Jin, F.; Golemis, E.A.; Yeung, R.S. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J. Biol. Chem. 1997, 272, 6097–6100. [Google Scholar] [CrossRef]
- Saucedo, L.J.; Gao, X.; Chiarelli, D.A.; Li, L.; Pan, D.; Edgar, B.A. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 2003, 5, 566–571. [Google Scholar] [CrossRef]
- Guertin, D.A.; Sabatini, D.M. Defining the role of mTOR in cancer. Cancer Cell 2007, 12, 9–22. [Google Scholar] [CrossRef]
- Pelletier, J.; Graff, J.; Ruggero, D.; Sonenberg, N. Targeting the eIF4F translation initiation complex: A critical nexus for cancer development. Cancer Res. 2015, 75, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Kaizuka, T.; Hara, T.; Oshiro, N.; Kikkawa, U.; Yonezawa, K.; Takehana, K.; Iemura, S.; Natsume, T.; Mizushima, N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 2010, 285, 20109–20116. [Google Scholar] [CrossRef] [PubMed]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009, 137, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Korets, S.B.; Czok, S.; Blank, S.V.; Curtin, J.P.; Schneider, R.J. Targeting the mTOR/4E-BP pathway in endometrial cancer. Clin. Cancer Res. 2011, 17, 7518–7528. [Google Scholar] [CrossRef] [PubMed]
- You, J.S.; McNally, R.M.; Jacobs, B.L.; Privett, R.E.; Gundermann, D.M.; Lin, K.H.; Steinert, N.D.; Goodman, C.A.; Hornberger, T.A. The role of raptor in the mechanical load-induced regulation of mTOR signaling, protein synthesis, and skeletal muscle hypertrophy. FASEB J. 2019, 33, 4021–4034. [Google Scholar] [CrossRef] [PubMed]
- Gruner, S.; Peter, D.; Weber, R.; Wohlbold, L.; Chung, M.Y.; Weichenrieder, O.; Valkov, E.; Igreja, C.; Izaurralde, E. The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation. Mol. Cell 2016, 64, 467–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mader, S.; Lee, H.; Pause, A.; Sonenberg, N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995, 15, 4990–4997. [Google Scholar] [CrossRef] [Green Version]
- Marcotrigiano, J.; Gingras, A.C.; Sonenberg, N.; Burley, S.K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 1999, 3, 707–716. [Google Scholar] [CrossRef]
- Merrick, W.C. eIF4F: A retrospective. J. Biol. Chem. 2015, 290, 24091–24099. [Google Scholar] [CrossRef]
- Siddiqui, N.; Sonenberg, N. Signalling to eIF4E in cancer. Biochem. Soc. Trans. 2015, 43, 763–772. [Google Scholar] [CrossRef]
- Wang, X.; Li, W.; Williams, M.; Terada, N.; Alessi, D.R.; Proud, C.G. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001, 20, 4370–4379. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Xie, T.; Zhang, S.Y.; Liu, B. Eukaryotic elongation factor-2 kinase (eEF2K): A potential therapeutic target in cancer. Apoptosis 2014, 19, 1527–1531. [Google Scholar] [CrossRef]
- Ryazanov, A.G.; Shestakova, E.A.; Natapov, P.G. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature 1988, 334, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Faller, W.J.; Jackson, T.J.; Knight, J.R.; Ridgway, R.A.; Jamieson, T.; Karim, S.A.; Jones, C.; Radulescu, S.; Huels, D.J.; Myant, K.B.; et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 2015, 517, 497–500. [Google Scholar] [CrossRef] [PubMed]
- D’Abronzo, L.S.; Ghosh, P.M. eIF4E Phosphorylation in Prostate Cancer. Neoplasia 2018, 20, 563–573. [Google Scholar] [CrossRef]
- Coleman, L.J.; Peter, M.B.; Teall, T.J.; Brannan, R.A.; Hanby, A.M.; Honarpisheh, H.; Shaaban, A.M.; Smith, L.; Speirs, V.; Verghese, E.T.; et al. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br. J. Cancer 2009, 100, 1393–1399. [Google Scholar] [CrossRef] [Green Version]
- Uniacke, J.; Perera, J.K.; Lachance, G.; Francisco, C.B.; Lee, S. Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer Res. 2014, 74, 1379–1389. [Google Scholar] [CrossRef]
- Hsieh, A.C.; Costa, M.; Zollo, O.; Davis, C.; Feldman, M.E.; Testa, J.R.; Meyuhas, O.; Shokat, K.M.; Ruggero, D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 2010, 17, 249–261. [Google Scholar] [CrossRef]
- Truitt, M.L.; Conn, C.S.; Shi, Z.; Pang, X.; Tokuyasu, T.; Coady, A.M.; Seo, Y.; Barna, M.; Ruggero, D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 2015, 162, 59–71. [Google Scholar] [CrossRef] [Green Version]
- Graff, J.R.; Konicek, B.W.; Vincent, T.M.; Lynch, R.L.; Monteith, D.; Weir, S.N.; Schwier, P.; Capen, A.; Goode, R.L.; Dowless, M.S.; et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 2007, 117, 2638–2648. [Google Scholar] [CrossRef]
- Galicia-Vazquez, G.; Cencic, R.; Robert, F.; Agenor, A.Q.; Pelletier, J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA 2012, 18, 1373–1384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, R.; Hershey, J.W. Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J. Biol. Chem. 1983, 258, 7228–7235. [Google Scholar] [PubMed]
- Jones, R.M.; Branda, J.; Johnston, K.A.; Polymenis, M.; Gadd, M.; Rustgi, A.; Callanan, L.; Schmidt, E.V. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol. Cell. Biol. 1996, 16, 4754–4764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenwald, I.B.; Rhoads, D.B.; Callanan, L.D.; Isselbacher, K.J.; Schmidt, E.V. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc. Natl. Acad. Sci. USA 1993, 90, 6175–6178. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.J.; Cencic, R.; Mills, J.R.; Robert, F.; Pelletier, J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008, 68, 5326–5334. [Google Scholar] [CrossRef]
- Cunningham, J.T.; Moreno, M.V.; Lodi, A.; Ronen, S.M.; Ruggero, D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell 2014, 157, 1088–1103. [Google Scholar] [CrossRef]
- Hsieh, A.C.; Liu, Y.; Edlind, M.P.; Ingolia, N.T.; Janes, M.R.; Sher, A.; Shi, E.Y.; Stumpf, C.R.; Christensen, C.; Bonham, M.J.; et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012, 485, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Joshi, S.; Platanias, L.C. Mnk Kinases in Cytokine Signaling and Regulation of Cytokine Responses. Biomol. Concepts 2012, 3, 127–139. [Google Scholar] [CrossRef]
- Maimon, A.; Mogilevsky, M.; Shilo, A.; Golan-Gerstl, R.; Obiedat, A.; Ben-Hur, V.; Lebenthal-Loinger, I.; Stein, I.; Reich, R.; Beenstock, J.; et al. Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts Ras-induced transformation. Cell Rep. 2014, 7, 501–513. [Google Scholar] [CrossRef]
- Rousseau, D.; Kaspar, R.; Rosenwald, I.; Gehrke, L.; Sonenberg, N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 1996, 93, 1065–1070. [Google Scholar] [CrossRef]
- Graff, J.R.; Zimmer, S.G. Translational control and metastatic progression: Enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin. Exp. Metastasis 2003, 20, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Kevil, C.; Carter, P.; Hu, B.; DeBenedetti, A. Translational enhancement of FGF-2 by eIF-4 factors, and alternate utilization of CUG and AUG codons for translation initiation. Oncogene 1995, 11, 2339–2348. [Google Scholar] [PubMed]
- Robichaud, N.; del Rincon, S.V.; Huor, B.; Alain, T.; Petruccelli, L.A.; Hearnden, J.; Goncalves, C.; Grotegut, S.; Spruck, C.H.; Furic, L.; et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 2015, 34, 2032–2042. [Google Scholar] [CrossRef] [PubMed]
- Lazaris-Karatzas, A.; Montine, K.S.; Sonenberg, N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5’ cap. Nature 1990, 345, 544–547. [Google Scholar] [CrossRef]
- Diab-Assaf, M.; Abou-Khouzam, R.; Saadallah-Zeidan, N.; Habib, K.; Bitar, N.; Karam, W.; Liagre, B.; Harakeh, S.; Azar, R. Expression of eukaryotic initiation factor 4E and 4E binding protein 1 in colorectal carcinogenesis. Int. J. Clin. Exp. Pathol. 2015, 8, 404–413. [Google Scholar]
- Heikkinen, T.; Korpela, T.; Fagerholm, R.; Khan, S.; Aittomaki, K.; Heikkila, P.; Blomqvist, C.; Carpen, O.; Nevanlinna, H. Eukaryotic translation initiation factor 4E (eIF4E) expression is associated with breast cancer tumor phenotype and predicts survival after anthracycline chemotherapy treatment. Breast Cancer Res. Treat. 2013, 141, 79–88. [Google Scholar] [CrossRef]
- Zismanov, V.; Attar-Schneider, O.; Lishner, M.; Heffez Aizenfeld, R.; Tartakover Matalon, S.; Drucker, L. Multiple myeloma proteostasis can be targeted via translation initiation factor eIF4E. Int. J. Oncol. 2015, 46, 860–870. [Google Scholar] [CrossRef]
- Pettersson, F.; Del Rincon, S.V.; Emond, A.; Huor, B.; Ngan, E.; Ng, J.; Dobocan, M.C.; Siegel, P.M.; Miller, W.H., Jr. Genetic and pharmacologic inhibition of eIF4E reduces breast cancer cell migration, invasion, and metastasis. Cancer Res. 2015, 75, 1102–1112. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, R.Z.; Gu, Y.; Shi, P.F.; Qian, S. Targeting of phospho-eIF4E by homoharringtonine eradicates a distinct subset of human acute myeloid leukemia. Leuk Lymphoma 2018, 1–13. [Google Scholar] [CrossRef]
- Volpon, L.; Culjkovic-Kraljacic, B.; Sohn, H.S.; Blanchet-Cohen, A.; Osborne, M.J.; Borden, K.L.B. A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA 2017, 23, 927–937. [Google Scholar] [CrossRef] [Green Version]
- Pettersson, F.; Yau, C.; Dobocan, M.C.; Culjkovic-Kraljacic, B.; Retrouvey, H.; Puckett, R.; Flores, L.M.; Krop, I.E.; Rousseau, C.; Cocolakis, E.; et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin. Cancer Res. 2011, 17, 2874–2884. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Zhang, X.; Li, D.; He, P.; Tian, W.; Zeng, B. Expression analysis and clinical significance of eIF4E, VEGF-C, E-cadherin and MMP-2 in colorectal adenocarcinoma. Oncotarget 2016, 7, 85502–85514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salehi, Z.; Mashayekhi, F.; Shahosseini, F. Significance of eIF4E expression in skin squamous cell carcinoma. Cell Biol. Int. 2007, 31, 1400–1404. [Google Scholar] [CrossRef] [PubMed]
- Crew, J.P.; Fuggle, S.; Bicknell, R.; Cranston, D.W.; de Benedetti, A.; Harris, A.L. Eukaryotic initiation factor-4E in superficial and muscle invasive bladder cancer and its correlation with vascular endothelial growth factor expression and tumour progression. Br. J. Cancer 2000, 82, 161–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Makala, L.; Wu, D.; Cai, Y. Targeting translation: eIF4E as an emerging anticancer drug target. Expert Rev. Mol. Med. 2016, 18, e2. [Google Scholar] [CrossRef] [PubMed]
- Moerke, N.J.; Aktas, H.; Chen, H.; Cantel, S.; Reibarkh, M.Y.; Fahmy, A.; Gross, J.D.; Degterev, A.; Yuan, J.; Chorev, M.; et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 2007, 128, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Holm, N.; Byrnes, K.; Johnson, L.; Abreo, F.; Sehon, K.; Alley, J.; Meschonat, C.; Md, Q.C.; Li, B.D. A prospective trial on initiation factor 4E (eIF4E) overexpression and cancer recurrence in node-negative breast cancer. Ann. Surg. Oncol. 2008, 15, 3207–3215. [Google Scholar] [CrossRef]
- Kentsis, A.; Topisirovic, I.; Culjkovic, B.; Shao, L.; Borden, K.L. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc. Natl. Acad. Sci. USA 2004, 101, 18105–18110. [Google Scholar] [CrossRef] [Green Version]
- Koromilas, A.E. Roles of the translation initiation factor eIF2alpha serine 51 phosphorylation in cancer formation and treatment. Biochim. Biophys. Acta 2015, 1849, 871–880. [Google Scholar] [CrossRef]
- Rajesh, K.; Krishnamoorthy, J.; Kazimierczak, U.; Tenkerian, C.; Papadakis, A.I.; Wang, S.; Huang, S.; Koromilas, A.E. Phosphorylation of the translation initiation factor eIF2alpha at serine 51 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death Dis. 2015, 6, e1591. [Google Scholar] [CrossRef]
- Wang, M.; Kaufman, R.J. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 2014, 14, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Leprivier, G.; Rotblat, B.; Khan, D.; Jan, E.; Sorensen, P.H. Stress-mediated translational control in cancer cells. Biochim. Biophys. Acta 2015, 1849, 845–860. [Google Scholar] [CrossRef] [PubMed]
- Holcik, M.; Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.J.; Chambers, J.E.; Liniker, E.; Marciniak, S.J. Endoplasmic reticulum stress in malignancy. Cancer Cell 2014, 25, 563–573. [Google Scholar] [CrossRef]
- Proud, C.G. Regulation of eukaryotic initiation factor eIF2B. Prog. Mol. Subcell Biol. 2001, 26, 95–114. [Google Scholar]
- Boye, E.; Grallert, B. eIF2alpha phosphorylation and the regulation of translation. Curr Genet. 2019. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [Green Version]
- B’Chir, W.; Maurin, A.C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013, 41, 7683–7699. [Google Scholar] [CrossRef]
- Rozpedek, W.; Pytel, D.; Mucha, B.; Leszczynska, H.; Diehl, J.A.; Majsterek, I. The Role of the PERK/eIF2alpha/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr. Mol. Med. 2016, 16, 533–544. [Google Scholar] [CrossRef]
- Fernandez, J.; Yaman, I.; Sarnow, P.; Snider, M.D.; Hatzoglou, M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2alpha. J. Biol. Chem. 2002, 277, 19198–19205. [Google Scholar] [CrossRef]
- Guo, L.; Chi, Y.; Xue, J.; Ma, L.; Shao, Z.; Wu, J. Phosphorylated eIF2alpha predicts disease-free survival in triple-negative breast cancer patients. Sci. Rep. 2017, 7, 44674. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, I.B.; Hutzler, M.J.; Wang, S.; Savas, L.; Fraire, A.E. Expression of eukaryotic translation initiation factors 4E and 2alpha is increased frequently in bronchioloalveolar but not in squamous cell carcinomas of the lung. Cancer 2001, 92, 2164–2171. [Google Scholar] [CrossRef]
- Wang, S.; Lloyd, R.V.; Hutzler, M.J.; Rosenwald, I.B.; Safran, M.S.; Patwardhan, N.A.; Khan, A. Expression of eukaryotic translation initiation factors 4E and 2alpha correlates with the progression of thyroid carcinoma. Thyroid 2001, 11, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, I.B.; Koifman, L.; Savas, L.; Chen, J.J.; Woda, B.A.; Kadin, M.E. Expression of the translation initiation factors eIF-4E and eIF-2* is frequently increased in neoplastic cells of Hodgkin lymphoma. Hum. Pathol. 2008, 39, 910–916. [Google Scholar] [CrossRef]
- Rosenwald, I.B.; Wang, S.; Savas, L.; Woda, B.; Pullman, J. Expression of translation initiation factor eIF-2alpha is increased in benign and malignant melanocytic and colonic epithelial neoplasms. Cancer 2003, 98, 1080–1088. [Google Scholar] [CrossRef]
- Lobo, M.V.; Martin, M.E.; Perez, M.I.; Alonso, F.J.; Redondo, C.; Alvarez, M.I.; Salinas, M. Levels, phosphorylation status and cellular localization of translational factor eIF2 in gastrointestinal carcinomas. Histochem. J. 2000, 32, 139–150. [Google Scholar] [CrossRef]
- Pelletier, J.; Thomas, G.; Volarevic, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef]
- Lacerda, R.; Menezes, J.; Romao, L. More than just scanning: The importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell. Mol. Life Sci. 2017, 74, 1659–1680. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef]
- Hellen, C.U.; Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001, 15, 1593–1612. [Google Scholar] [CrossRef] [Green Version]
- Shatsky, I.N.; Dmitriev, S.E.; Terenin, I.M.; Andreev, D.E. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol. Cells 2010, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Morfoisse, F.; Kuchnio, A.; Frainay, C.; Gomez-Brouchet, A.; Delisle, M.B.; Marzi, S.; Helfer, A.C.; Hantelys, F.; Pujol, F.; Guillermet-Guibert, J.; et al. Hypoxia induces VEGF-C expression in metastatic tumor cells via a HIF-1alpha-independent translation-mediated mechanism. Cell Rep. 2014, 6, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.M.; Shih, Y.H.; Tseng, J.T.; Lai, M.C.; Wu, C.H.; Li, Y.H.; Tsai, S.J.; Sun, H.S. Overexpression of FGF9 in colon cancer cells is mediated by hypoxia-induced translational activation. Nucleic Acids Res. 2014, 42, 2932–2944. [Google Scholar] [CrossRef] [PubMed]
- Vagner, S.; Gensac, M.C.; Maret, A.; Bayard, F.; Amalric, F.; Prats, H.; Prats, A.C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 1995, 15, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martineau, Y.; Le Bec, C.; Monbrun, L.; Allo, V.; Chiu, I.M.; Danos, O.; Moine, H.; Prats, H.; Prats, A.C. Internal ribosome entry site structural motifs conserved among mammalian fibroblast growth factor 1 alternatively spliced mRNAs. Mol. Cell. Biol. 2004, 24, 7622–7635. [Google Scholar] [CrossRef]
- Stein, I.; Itin, A.; Einat, P.; Skaliter, R.; Grossman, Z.; Keshet, E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: Implications for translation under hypoxia. Mol. Cell. Biol. 1998, 18, 3112–3119. [Google Scholar] [CrossRef]
- Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; Gritsenko, A.A.; Stern-Ginossar, N.; Yakhini, Z.; Weinberger, A.; Segal, E. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016, 351. [Google Scholar] [CrossRef]
- Hershey, J.W.; Sonenberg, N.; Mathews, M.B. Principles of translational control: An overview. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef]
- Sriram, A.; Bohlen, J.; Teleman, A.A. Translation acrobatics: How cancer cells exploit alternate modes of translational initiation. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Shi, Y.; Yang, Y.; Hoang, B.; Bardeleben, C.; Holmes, B.; Gera, J.; Lichtenstein, A. Therapeutic potential of targeting IRES-dependent c-myc translation in multiple myeloma cells during ER stress. Oncogene 2016, 35, 1015–1024. [Google Scholar] [CrossRef]
- Bert, A.G.; Grepin, R.; Vadas, M.A.; Goodall, G.J. Assessing IRES activity in the HIF-1alpha and other cellular 5′ UTRs. RNA 2006, 12, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Bode, B.; Koromilas, A.; Diehl, J.A.; Krukovets, I.; Snider, M.D.; Hatzoglou, M. Translation mediated by the internal ribosome entry site of the cat-1 mRNA is regulated by glucose availability in a PERK kinase-dependent manner. J. Biol. Chem. 2002, 277, 11780–11787. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Herrera, I.G.; Prado-Lourenco, L.; Teshima-Kondo, S.; Kondo, K.; Cabon, F.; Arnal, J.F.; Bayard, F.; Prats, A.C. IRES-dependent regulation of FGF-2 mRNA translation in pathophysiological conditions in the mouse. Biochem. Soc. Trans. 2006, 34, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Shatsky, I.N.; Terenin, I.M.; Smirnova, V.V.; Andreev, D.E. Cap-Independent Translation: What’s in a Name? Trends Biochem. Sci. 2018, 43, 882–895. [Google Scholar] [CrossRef]
- Blanco-Perez, M.; Perez-Canamas, M.; Ruiz, L.; Hernandez, C. Efficient Translation of Pelargonium line pattern virus RNAs Relies on a TED-Like 3 -Translational Enhancer that Communicates with the Corresponding 5 -Region through a Long-Distance RNA-RNA Interaction. PLoS ONE 2016, 11, e0152593. [Google Scholar] [CrossRef]
- Andreev, D.E.; Dmitriev, S.E.; Zinovkin, R.; Terenin, I.M.; Shatsky, I.N. The 5′ untranslated region of Apaf-1 mRNA directs translation under apoptosis conditions via a 5’ end-dependent scanning mechanism. FEBS Lett. 2012, 586, 4139–4143. [Google Scholar] [CrossRef]
- Signer, R.A.; Magee, J.A.; Salic, A.; Morrison, S.J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 2014, 509, 49–54. [Google Scholar] [CrossRef]
- Signer, R.A.; Qi, L.; Zhao, Z.; Thompson, D.; Sigova, A.A.; Fan, Z.P.; DeMartino, G.N.; Young, R.A.; Sonenberg, N.; Morrison, S.J. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 2016, 30, 1698–1703. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; McMillan, E.; Han, F.; Svendsen, C.N. Regionally specified human neural progenitor cells derived from the mesencephalon and forebrain undergo increased neurogenesis following overexpression of ASCL1. Stem Cells 2009, 27, 390–398. [Google Scholar] [CrossRef]
- Wright, L.S.; Li, J.; Caldwell, M.A.; Wallace, K.; Johnson, J.A.; Svendsen, C.N. Gene expression in human neural stem cells: Effects of leukemia inhibitory factor. J. Neurochem. 2003, 86, 179–195. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Tahmasebi, S.; Jafarnejad, S.M.; Tam, I.S.; Gonatopoulos-Pournatzis, T.; Matta-Camacho, E.; Tsukumo, Y.; Yanagiya, A.; Li, W.; Atlasi, Y.; Caron, M.; et al. Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc. Natl. Acad. Sci. USA 2016, 113, 12360–12367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Barna, M.; Ruggero, D. Tailor made protein synthesis for HSCs. Cell Stem Cell 2014, 14, 423–424. [Google Scholar] [CrossRef]
- Blanco, S.; Bandiera, R.; Popis, M.; Hussain, S.; Lombard, P.; Aleksic, J.; Sajini, A.; Tanna, H.; Cortes-Garrido, R.; Gkatza, N.; et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016, 534, 335–340. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, C.G.; Teixeira, F.K.; Czech, B.; Preall, J.B.; Zamparini, A.L.; Seifert, J.R.; Malone, C.D.; Hannon, G.J.; Lehmann, R. Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell 2016, 18, 276–290. [Google Scholar] [CrossRef]
- Browning, K.S.; Maia, D.M.; Lax, S.R.; Ravel, J.M. Identification of a new protein synthesis initiation factor from wheat germ. J. Biol. Chem. 1987, 262, 538–541. [Google Scholar]
- Rhoads, R.E. eIF4E: New family members, new binding partners, new roles. J. Biol. Chem. 2009, 284, 16711–16715. [Google Scholar] [CrossRef]
- Uniacke, J.; Holterman, C.E.; Lachance, G.; Franovic, A.; Jacob, M.D.; Fabian, M.R.; Payette, J.; Holcik, M.; Pause, A.; Lee, S. An oxygen-regulated switch in the protein synthesis machinery. Nature 2012, 486, 126–129. [Google Scholar] [CrossRef] [Green Version]
- Osborne, M.J.; Volpon, L.; Kornblatt, J.A.; Culjkovic-Kraljacic, B.; Baguet, A.; Borden, K.L. eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proc. Natl. Acad. Sci. USA 2013, 110, 3877–3882. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Smibert, C.A.; Kaplan, D.R.; Miller, F.D. An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron 2014, 84, 723–739. [Google Scholar] [CrossRef] [PubMed]
- Yoffe, Y.; David, M.; Kalaora, R.; Povodovski, L.; Friedlander, G.; Feldmesser, E.; Ainbinder, E.; Saada, A.; Bialik, S.; Kimchi, A. Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev. 2016, 30, 1991–2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imataka, H.; Gradi, A.; Sonenberg, N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998, 17, 7480–7489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamphear, B.J.; Kirchweger, R.; Skern, T.; Rhoads, R.E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995, 270, 21975–21983. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, H.; Takahashi, K.; Yamamoto, T.; Iwasaki, M.; Narita, M.; Nakamura, M.; Rand, T.A.; Nakagawa, M.; Watanabe, A.; Yamanaka, S. Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2017, 114, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S.; Zhang, X.Y.; Maeda, M.; Miura, K.; Wang, S.; Farese, R.V., Jr.; Iwao, H.; Innerarity, T.L. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 2000, 19, 5533–5541. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-J. Cell Fate Control by Translation: mRNA Translation Initiation as a Therapeutic Target for Cancer Development and Stem Cell Fate Control. Biomolecules 2019, 9, 665. https://doi.org/10.3390/biom9110665
Kim H-J. Cell Fate Control by Translation: mRNA Translation Initiation as a Therapeutic Target for Cancer Development and Stem Cell Fate Control. Biomolecules. 2019; 9(11):665. https://doi.org/10.3390/biom9110665
Chicago/Turabian StyleKim, Hyun-Jung. 2019. "Cell Fate Control by Translation: mRNA Translation Initiation as a Therapeutic Target for Cancer Development and Stem Cell Fate Control" Biomolecules 9, no. 11: 665. https://doi.org/10.3390/biom9110665
APA StyleKim, H.-J. (2019). Cell Fate Control by Translation: mRNA Translation Initiation as a Therapeutic Target for Cancer Development and Stem Cell Fate Control. Biomolecules, 9(11), 665. https://doi.org/10.3390/biom9110665



