GLYI4 Plays A Role in Methylglyoxal Detoxification and Jasmonate-Mediated Stress Responses in Arabidopsis thaliana
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Chemical Treatments
2.3. RNA Extraction and RT-qPCR
2.4. Total Protein Extraction and Western Blotting
2.5. Metabolite Extraction and LC-MS Analysis
2.6. Metabolomic Data Processing and Statistical Analysis
2.7. Plant Fitness Parameters
2.8. ROS Detection in Arabidopsis Leaves
2.9. Stomata Staining
2.10. Resistance Assays
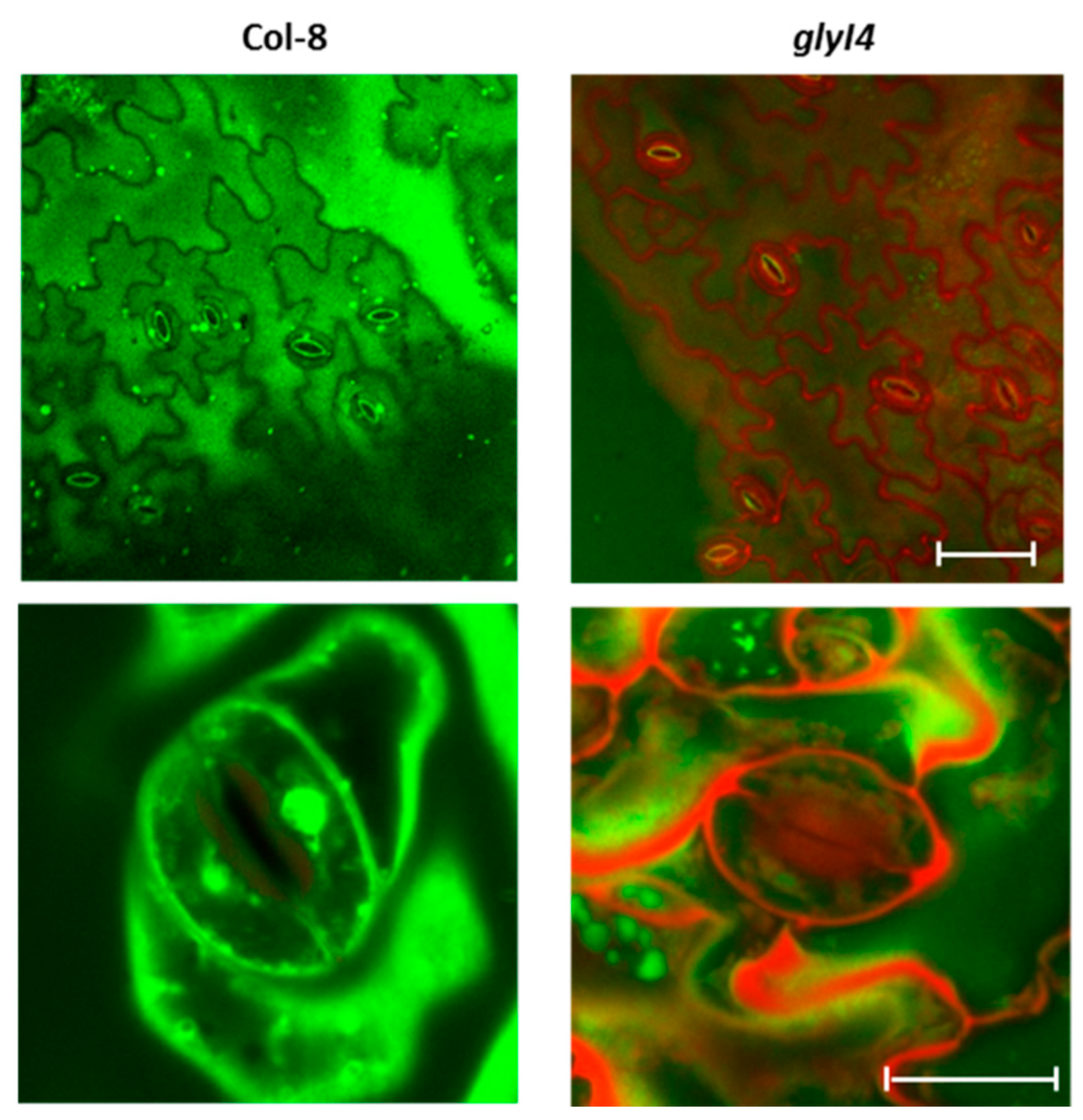
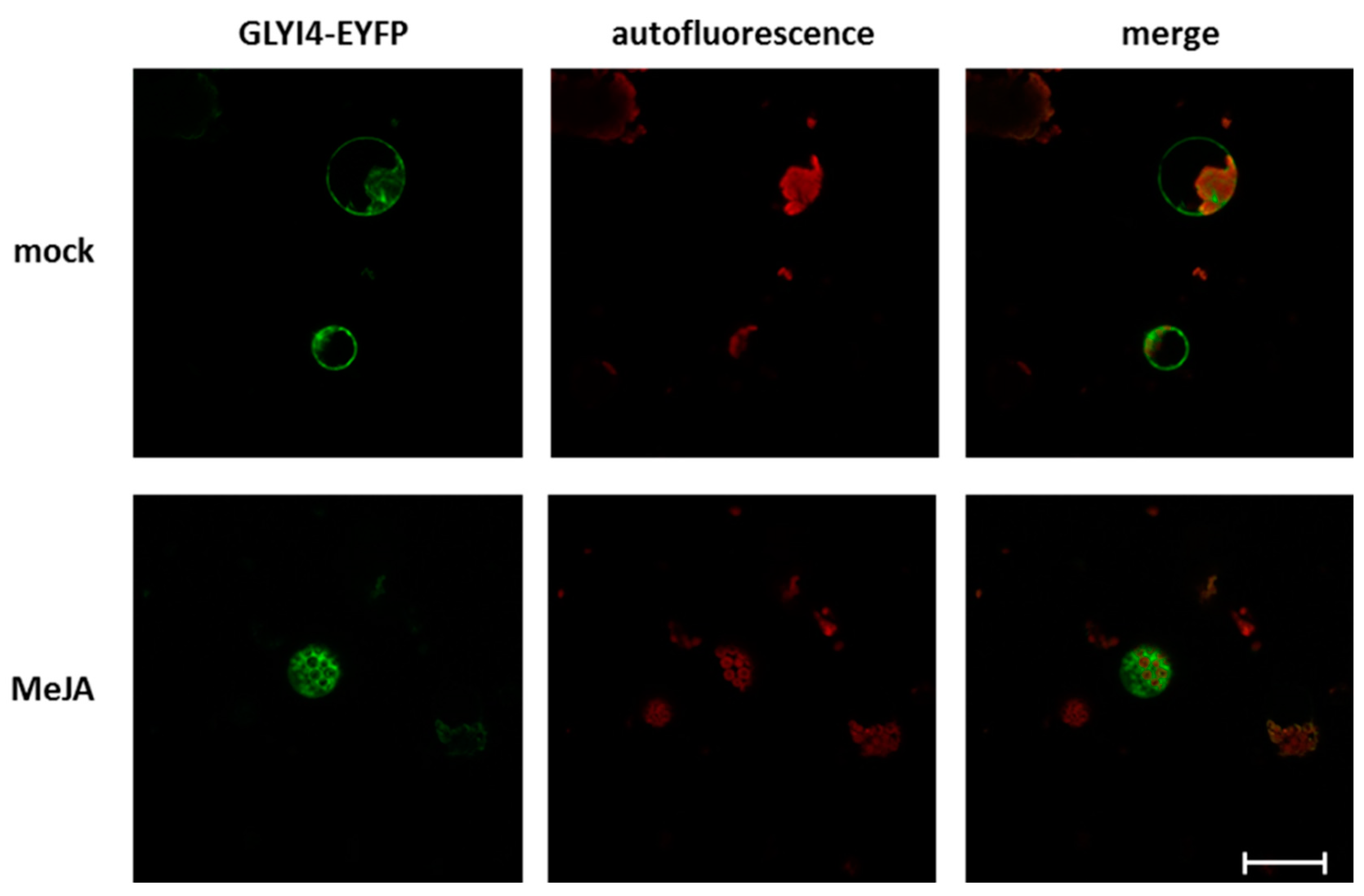
2.11. GLYI4 Subcellular Localization
2.12. Statistical Analyses
3. Results
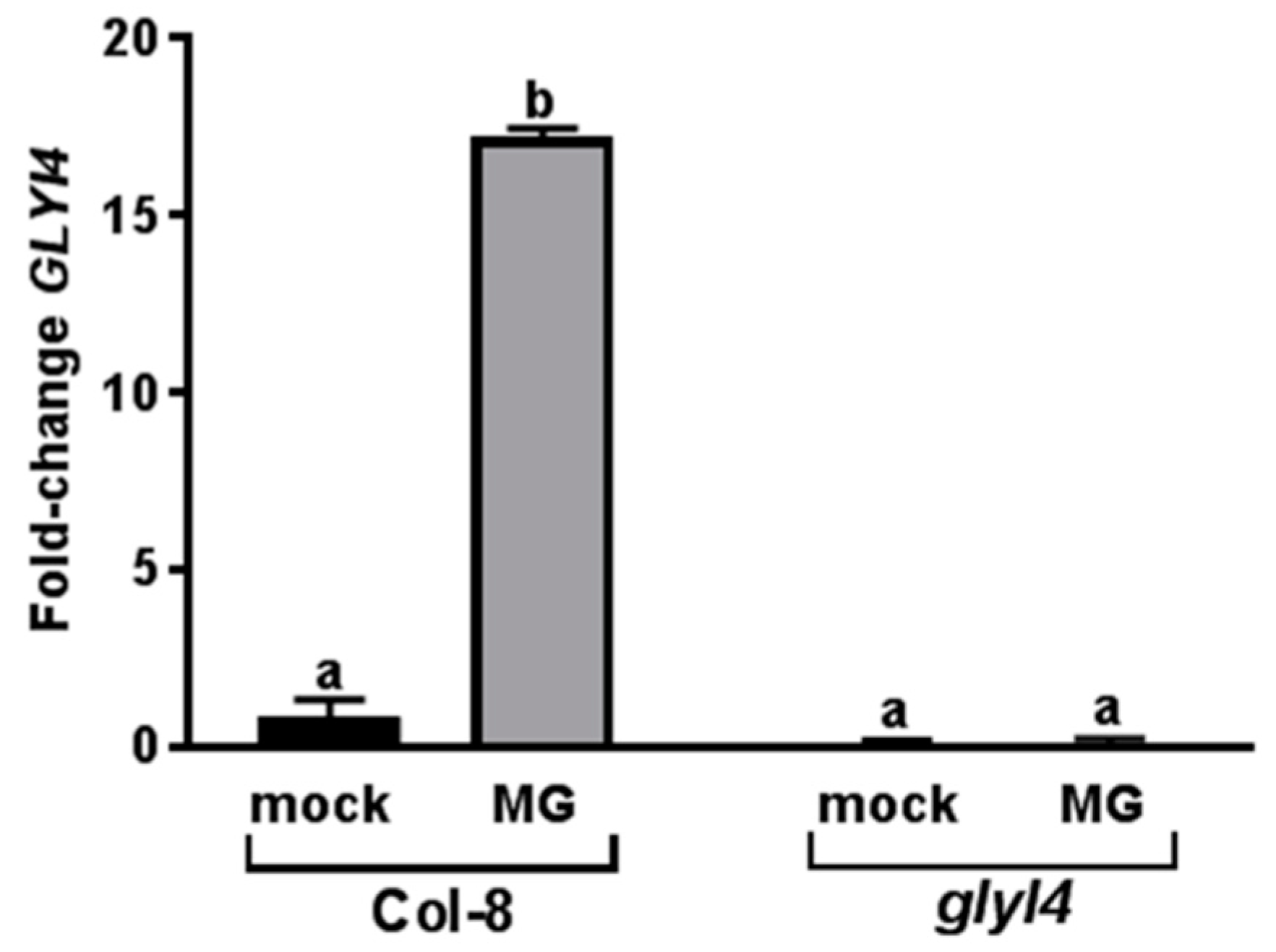
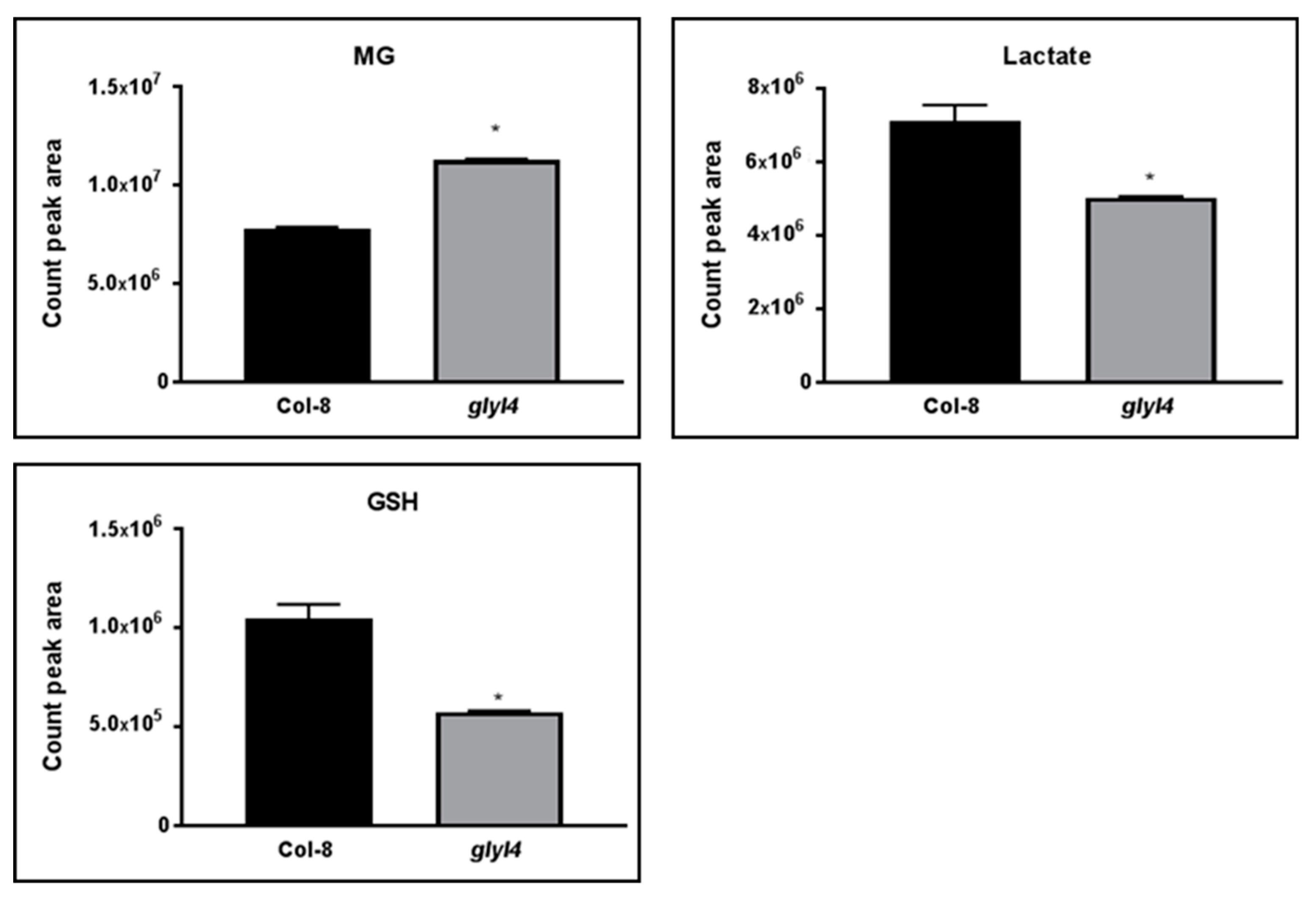
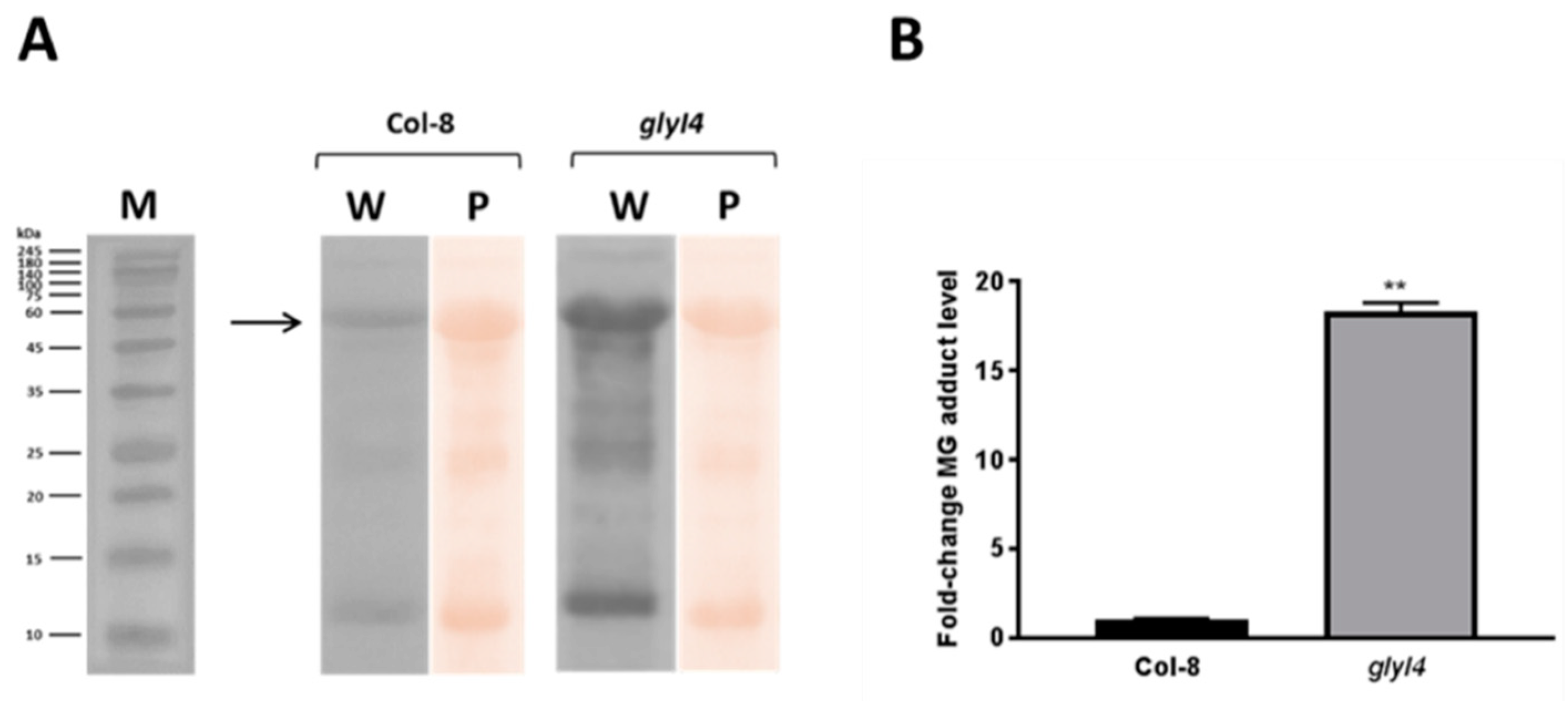
3.1. GLYI4 Acts in the MG Scavenging Pathway
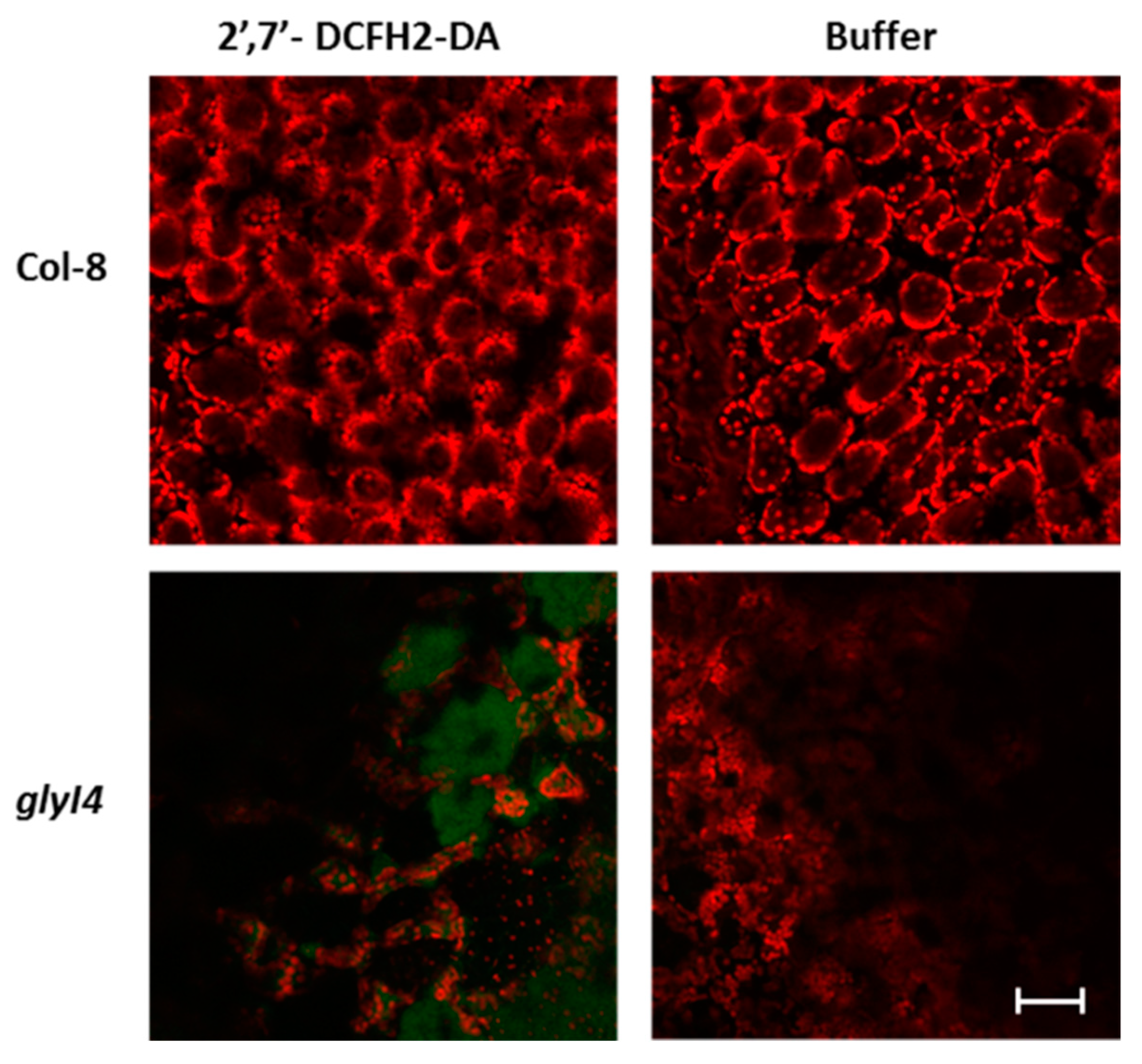
3.2. GLYI4 Impacts Fitness and Physiology of Arabidopsis Plants
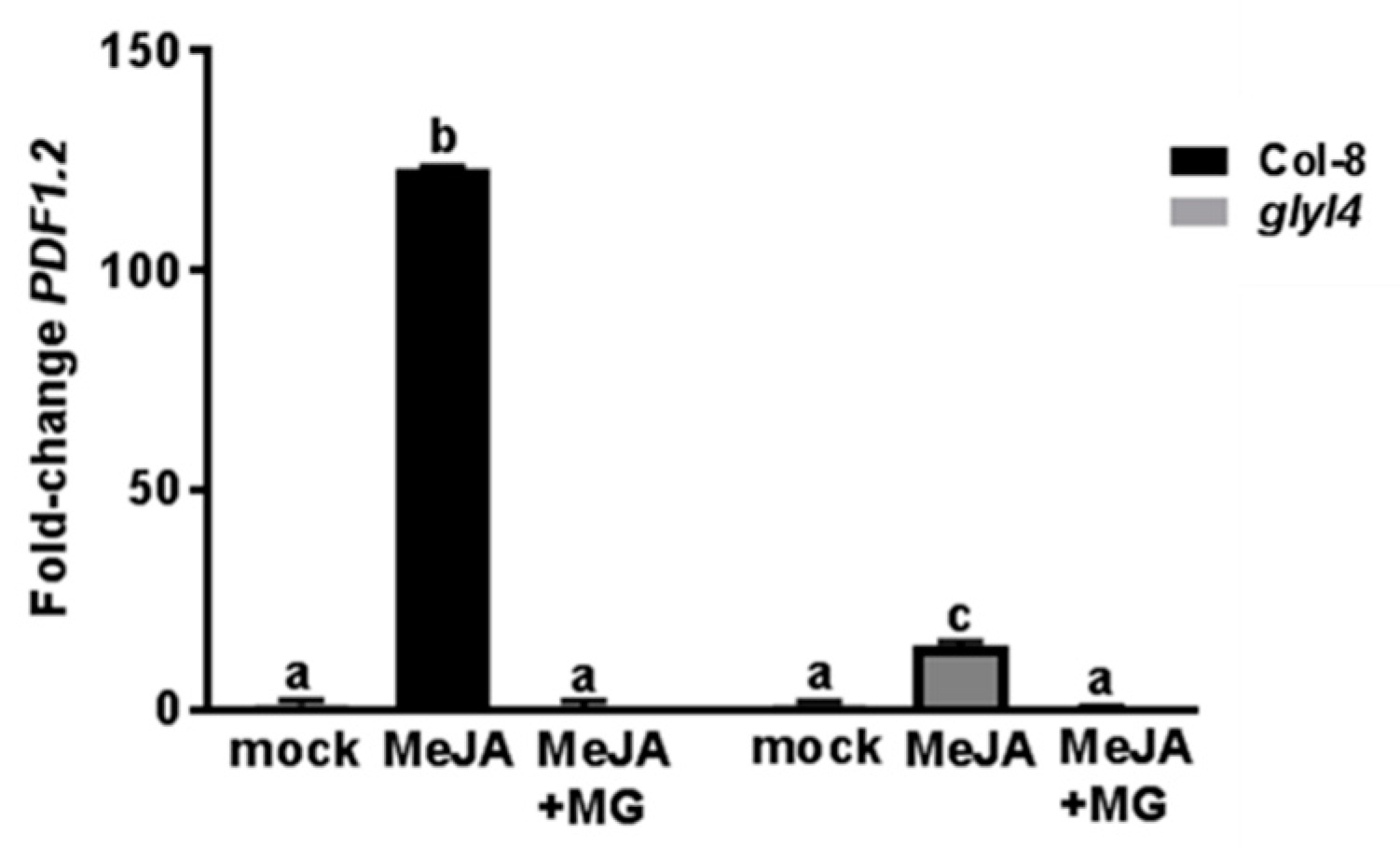
3.3. GLYI4 Affects JA-Dependent Defense Responses
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.A.; Benková, E. Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 2013, 4, 451. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone crosstalk in plant disease and defense: More than just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Lyons, R. Intervention of phytohormone pathways by pathogen effectors. Plant Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef]
- Hoque, T.S.; Okuma, E.; Uraji, M.; Furuichi, T.; Sasaki, T.; Hoque, M.A.; Nakamura, Y.; Murata, Y. Inhibitory effects of methylglyoxal on light-induced stomatal opening and inward KC channel activity in Arabidopsis. Biosci. Biotechnol. Biochem. 2012, 76, 617–619. [Google Scholar] [CrossRef]
- Hoque, T.S.; Uraji, M.; Tuya, A.; Nakamura, Y.; Murata, Y. Methylglyoxal inhibits seed germination and root elongation and up-regulates transcription of stress-responsive genes in ABA-dependent pathway in Arabidopsis. Plant Biol. 2012, 14, 854–858. [Google Scholar] [CrossRef]
- Hoque, T.S.; Uraji, M.; Ye, W.; Hossain, M.A.; Nakamura, Y.; Murata, Y. Methylglyoxal-induced stomatal closure accompanied by peroxidase-mediated ROS production in Arabidopsis. J. Plant Physiol. 2012, 169, 979–986. [Google Scholar] [CrossRef]
- Hoque, M.A.; Uraji, M.; Banu, M.N.A.; Mori, I.C.; Nakamura, Y.; Murata, Y. Methylglyoxal inhibition of cytosolic ascorbate peroxidase from Nicotiana tabacum. J. Biochem. Mol. Toxicol. 2012, 26, 315–321. [Google Scholar] [CrossRef]
- Mano, J. Reactive carbonyl species: Their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 2012, 59, 90–97. [Google Scholar] [CrossRef]
- Hoque, T.S.; Hossain, M.A.; Mostofa, M.G.; Burritt, D.J.; Fujita, M.; Tran, L.S. Methylglyoxal: An Emerging Signaling Molecule in Plant Abiotic Stress Responses and Tolerance. Front. Plant Sci. 2016, 7, 1341. [Google Scholar] [CrossRef]
- Jain, M.; Nagar, P.; Sharma, A.; Batth, R.; Aggarwal, S.; Kumari, S.; Mustafiz, A. GLYI and D-LDH play key role in methylglyoxal detoxification and abiotic stress tolerance. Sci. Rep. 2018, 8, 5451. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Sopory, S.K. An overview on the role of methylglyoxal and glyoxalases in plants. Drug Metabol. Drug Interact. 2008, 23, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Campbell, A.K.; Naseem, R.; Holland, I.B.; Matthews, S.B.; Wann, K.T. Methylglyoxal and other carbohydrate metabolites induce lanthanum-sensitive Ca2+ transients and inhibit growth in E coli. Arch. Biochem. Biophys. 2007, 468, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Takatsume, Y.; Izawa, S.; Inoue, Y. Methylglyoxal as a signal initiator for activation of the stress-activated protein kinase cascade in the fission yeast. Schizosaccharomyces pombe. J. Biol. Chem. 2006, 281, 9086–9092. [Google Scholar] [CrossRef]
- Nomura, W.; Maeta, K.; Kita, K.; Izawa, S.; Inoue, Y. Role of Gcn4 for adaptation to methylglyoxal in Saccharomyces cerevisiae: Methylglyoxal attenuates protein synthesis through phosphorylation of eIF2α. Biochem. Biophys. Res. Commun. 2008, 376, 738–742. [Google Scholar] [CrossRef]
- Akhand, A.A.; Hossain, K.; Mitsui, H.; Kato, M.; Miyata, T.; Inagi, R.; Du, J.; Takeda, K.; Kawamoto, Y.; Suzuki, H.; et al. Glyoxal and methylglyoxal trigger distinct signals for map family kinases and caspase activation in human endothelial cells. Free Radic. Biol. Med. 2001, 31, 20–30. [Google Scholar] [CrossRef]
- Fukunaga, M.; Miyata, S.; Higo, S.; Hamada, Y.; Ueyama, S.; Kasuga, M. Methylglyoxal induces apoptosis through oxidative stress-mediated activation of p38 mitogen-activated protein kinase in rat Schwann Cells. Ann. N. Y. Acad. Sci. 2005, 1043, 151–157. [Google Scholar] [CrossRef]
- Riboulet-Chavey, A.; Pierron, A.; Durand, I.; Murdaca, J.; Giudicelli, J.; Van Obberghen, E. Methylglyoxal impairs the insulin signaling pathways independently of the formation of intracellular reactive oxygen species. Diabetes 2006, 55, 1289–1299. [Google Scholar] [CrossRef]
- Jia, X.; Chang, T.; Wilson, T.W.; Wu, L. Methylglyoxal mediates adipocyte proliferation by increasing phosphorylation of Akt1. PLoS ONE 2012, 7, e36610. [Google Scholar] [CrossRef]
- Sharma, S.; Mustafiz, A.; Singla-Pareek, S.L.; Shankar Srivastava, P.; Sopory, S.K. Characterization of stress and methylglyoxal inducible triose phosphate isomerase (OscTPI) from rice. Plant Signal. Behav. 2012, 7, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. CRC. Crit. Rev. Plant Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Ray, A.; Ray, S.; Mukhopadhyay, S.; Ray, M. Methylglyoxal with glycine or succinate enhances differentiation and shoot morphogenesis in Nicotiana tabacum callus. Biol. Plant. 2013, 57, 219–223. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Jamshed, M.; Kumar, A.; Skori, L.; Scandola, S.; Wang, T.; Spiegel, D.; Samuel, M.A. Glyoxalase goes green: The expanding roles of glyoxalase in plants. Int. J. Mol. Sci. 2017, 18, 898. [Google Scholar] [CrossRef] [PubMed]
- Nokin, M.-J.; Durieux, F.; Peixoto, P.; Chiavarina, B.; Peulen, O.; Blomme, A.; Turtoi, A.; Costanza, B.; Smargiasso, N.; Baiwir, D.; et al. Methylglyoxal, a glycolysis side-product, induces Hsp90 glycation and YAP mediated tumor growth and metastasis. eLife 2016, 5, e19375. [Google Scholar] [CrossRef]
- Schmitz, J.; Rossoni, A.W.; Maurino, V.G. Dissecting the physiological function of plant glyoxalase I and glyoxalase I-like proteins. Front. Plant Sci. 2018, 9, 1618. [Google Scholar] [CrossRef]
- Hossain, M.A.; Fujita, M. Purification of glyoxalase I from onion bulbs and molecular cloning of its cDNA. Biosci. Biotechnol. Biochem. 2009, 73, 2007–2013. [Google Scholar] [CrossRef]
- Hossain, M.A.; Hossain, M.Z.; Fujita, M. Stress-induced changes of methylglyoxal level and glyoxalase I activity in pumpkin seedlings and cDNA cloning of glyoxalase I gene. Aust. J. Crop Sci. 2009, 3, 53–64. [Google Scholar]
- Ghosh, A.; Islam, T. Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 2016, 16, 87. [Google Scholar] [CrossRef]
- Singla-Pareek, S.; Reddy, M.; Sopory, S. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. USA 2003, 100, 14672–14677. [Google Scholar] [CrossRef]
- Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J. 2014, 80, 93–105. [Google Scholar] [CrossRef]
- Kaur, C.; Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Glyoxalases and stress tolerance in plants. Biochem. Soc. Trans. 2014, 42, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Mustafiz, A.; Ghosh, A.; Tripathi, A.K.; Kaur, C.; Ganguly, A.K.; Bhavesh, N.S.; Tripathi, J.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A unique Ni2+ -dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 2014, 78, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A. Genome-wide identification of glyoxalase genes in Medicago truncatula and their expression profiling in response to various developmental and environmental stimuli. Front. Plant Sci. 2017, 8, 836. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 2006, 140, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Alvarez Viveros, M.F.; Inostroza-Blancheteau, C.; Timmermann, T.; González, M.; Arce-Johnson, P. Overexpression of GlyI and GlyII genes in transgenic tomato (Solanum lycopersicum Mill.) plants confers salt tolerance by decreasing oxidative stress. Mol. Biol. Rep. 2013, 40, 3281–3290. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Moran, G.R. Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. J. Inorg. Biochem. 2011, 105, 1259–1272. [Google Scholar] [CrossRef]
- Mustafiz, A.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 2011, 11, 293–305. [Google Scholar] [CrossRef]
- Proietti, S.; Caarls, L.; Coolen, S.; Van Pelt, J.A.; Van Wees, S.C.M.; Pieterse, C.M.J. Genome-wide association study reveals novel players in defense hormone crosstalk in Arabidopsis. Plant Cell Environ. 2018, 41, 2342–2356. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water culture method for growing plants without soil. Calif. Agric. Exp. Stn. Bull. 1938, 347, 36–39. [Google Scholar]
- Oñate-Sánchez, L.; Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 2008, 1, 93. [Google Scholar]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from acrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Melamud, E.; Vastag, L.; Rabinowitz, J.D. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 2010, 82, 9818–9826. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Giangrande, C.; Amoresano, A.; Pucci, P.; Molinaro, A.; Bertini, L.; Caporale, C.; Caruso, C. Xanthomonas campestris lipooligosaccharides trigger innate immunity and oxidative burst in Arabidopsis. Plant Physiol. Biochem. 2014, 85, 51–62. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bertini, L.; Palazzi, L.; Proietti, S.; Pollastri, S.; Arrigoni, G.; Polverino de Laureto, P.; Caruso, C. Proteomic analysis of MeJa-induced defense responses in rice against wounding. Int. J. Mol. Sci. 2019, 20, 2525. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, N.; Hossain, M.S.; Mahmud, J.A.; Rahman, A.; Inafuku, M.; Oku, H.; Fujita, M. Coordinated actions of glyoxalase and antioxidant defense systems in conferring abiotic stress tolerance in plants. Int. J. Mol. Sci. 2017, 18, 200. [Google Scholar] [CrossRef]
- Sharma, C.; Kaur, A.; Thind, S.S.; Singh, B.; Raina, S. Advanced glycation End-products (AGEs): An emerging concern for processed food industries. J. Food Sci. Technol. 2015, 52, 7561–7576. [Google Scholar] [CrossRef]
- Møller, I.M.; Rogowska-Wrzesinska, A.; Rao, R.S. Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J. Proteom. 2011, 74, 2228–2242. [Google Scholar]
- Shumilina, J.; Kusnetsova, A.; Tsarev, A.; Janse van Rensburg, H.C.; Medvedev, S.; Demidchik, V.; Van den Ende, W.; Frolov, A. Glycation of plant proteins: Regulatory roles and interplay with sugar signalling? Int. J. Mol. Sci. 2019, 20, 2366. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, K.A.; Jensen, P.E.; Møller, I.M.; Schulz, A. Monitoring reactive oxygen species formation and localisation in living cells by use of the fluorescent probe CM-H(2)DCFDA and confocal laser microscopy. Physiol. Plant. 2009, 136, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Jiang, L.; Zhang, H.; Shimoda, L.A.; DeBerardinis, R.J.; Semenza, G.L. Analysis of hypoxia-induced metabolic reprogramming. Methods Enzymol. 2014, 542, 425–455. [Google Scholar]
- Ferguson, G.P.; McLaggan, D.; Booth, I.R. Potassium channel activation by glutathione-S-conjugates in Escherichia coli: Protection against methylglyoxal is mediated by cytoplasmic acidification. Mol Microbiol. 1995, 17, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Cook, L.J.; Davies, J.; Yates, A.P.; Elliott, A.C.; Lovell, J.; Joule, J.A.; Pemberton, P.; Thornalley, P.J.; Best, L. Effects of methylglyoxal on rat pancreatic beta-cells. Biochem Pharmacol. 1998, 55, 1361–1367. [Google Scholar] [CrossRef]
- Kniep, E.M.; Roehlecke, C.; Özkucur, N.; Steinberg, A.; Reber, F.; Knels, L.; Funk, R.H.W. Inhibition of apoptosis and reduction of intracellular pH decrease in retinal neural cell cultures by a blocker of carbonic anhydrase. Investig. Ophthalmol. Vis. Sci. 2006, 47, 1185–1192. [Google Scholar] [CrossRef]
- Chakraborty, S.; Gogoi, M.; Chakravortty, D. Lactoylglutathione lyase, a critical enzyme in methylglyoxal detoxification, contributes to survival of Salmonella in the nutrient rich environment. Virulence 2015, 6, 50–65. [Google Scholar] [CrossRef]
- Bak, G.; Lee, E.J.; Lee, Y.; Kato, M.; Segami, S.; Sze, H.; Maeshima, M.; Hwang, J.U.; Lee, Y. Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell. 2013, 25, 2202–2216. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Cai, G.; Jiang, S.; Sun, L.; Li, D. Response of tobacco to the Pseudomonas syringae pv. Tomato DC3000 is mainly dependent on salicylic acid signaling pathway. FEMS Microbiol. Lett. 2013, 344, 77–85. [Google Scholar] [CrossRef]
- Yan, C.; Xie, D. Jasmonate in plant defence: Sentinel or double agent? Plant Biotechnol. J. 2015, 13, 1233–1240. [Google Scholar] [CrossRef]
- Pétriacq, P.; Stassen, J.H.; Ton, J. Spore density determines infection strategy by the plant pathogenic fungus Plectosphaerella cucumerina. Plant Physiol. 2016, 170, 2325–2339. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.; Seo, S.; Han, J.Y.; Ki, S.H.; Shin, S.M. Resveratrol attenuates methylglyoxal-induced mitochondrial dysfunction and apoptosis by Sestrin2 induction. Toxicol. Appl. Pharmacol. 2014, 280, 2–314. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Thornalley, P.J. Dicarbonyl proteome and genome damage in metabolic and vascular disease. Biochem. Soc. Trans. 2014, 42, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Thelen, J.J. The plastid isoform of triose phosphate isomerase is required for the postgerminative transition from heterotrophic to autotrophic growth in Arabidopsis. Plant Cell 2010, 22, 77–90. [Google Scholar] [CrossRef]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 2005, 337, 61–67. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Niu, L.; Liao, W. Hydrogen Peroxide Signaling in Plant Development and Abiotic Responses: Crosstalk with Nitric Oxide and Calcium. Front. Plant Sci. 2016, 7, 230. [Google Scholar] [CrossRef]
- Khokon, A.R.; Okuma, E.; Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 2011, 34, 434–444. [Google Scholar] [CrossRef]
- Nokin, M.J.; Durieux, F.; Bellier, J.; Peulen, O.; Uchida, K.; Spiegel, D.A.; Cochrane, J.R.; Hutton, C.A.; Castronovo, V.; Bellahcène, A. Hormetic potential of methylglyoxal, a side-product of glycolysis, in switching tumours from growth to death. Sci. Rep. 2017, 7, 11722. [Google Scholar] [CrossRef]
- Chaplin, A.K.; Chernukhin, I.; Bechtold, U. Profiling of advanced glycation end products uncovers abiotic stress-specific target proteins in Arabidopsis. J. Exp. Bot. 2018, 70, 653–670. [Google Scholar] [CrossRef]
- Lee, E.J.; Park, J.H. Receptor for advanced glycation endproducts (RAGE), its ligands, and soluble RAGE: Potential biomarkers for diagnosis and therapeutic targets for human renal diseases. Genom. Inform. 2013, 11, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Land, W.G. Danger signals - damaged-self recognition across the tree of life. Front. Plant Sci. 2014, 5, 578. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Méndez, J.D.; Méndez-Valenzuela, V.; Aguilar-Hernández, M.M. Cellular signalling of the receptor for advanced glycation end products (RAGE). Cell Signal. 2013, 25, 2185–2197. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Díaz, M.; Lobos, T.; Cardemil, L.; Nunes-Nesi, A.; Retamales, J.; Jaakola, L.; Alberdi, M.; Ribera-Fonseca, A. Methyl Jasmonate: An alternative for improving the quality and health properties of fresh fruits. Molecules 2016, 21, 567. [Google Scholar]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Shin, R.; Alvarez, S.; Burch, A.Y.; Jez, J.M.; Schachtman, D.P. Phosphoproteomic identification of targets of the Arabidopsis sucrose non fermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc. Natl. Acad. Sci. USA 2007, 104, 6460–6465. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proietti, S.; Falconieri, G.S.; Bertini, L.; Baccelli, I.; Paccosi, E.; Belardo, A.; Timperio, A.M.; Caruso, C. GLYI4 Plays A Role in Methylglyoxal Detoxification and Jasmonate-Mediated Stress Responses in Arabidopsis thaliana. Biomolecules 2019, 9, 635. https://doi.org/10.3390/biom9100635
Proietti S, Falconieri GS, Bertini L, Baccelli I, Paccosi E, Belardo A, Timperio AM, Caruso C. GLYI4 Plays A Role in Methylglyoxal Detoxification and Jasmonate-Mediated Stress Responses in Arabidopsis thaliana. Biomolecules. 2019; 9(10):635. https://doi.org/10.3390/biom9100635
Chicago/Turabian StyleProietti, Silvia, Gaia Salvatore Falconieri, Laura Bertini, Ivan Baccelli, Elena Paccosi, Antonio Belardo, Anna Maria Timperio, and Carla Caruso. 2019. "GLYI4 Plays A Role in Methylglyoxal Detoxification and Jasmonate-Mediated Stress Responses in Arabidopsis thaliana" Biomolecules 9, no. 10: 635. https://doi.org/10.3390/biom9100635
APA StyleProietti, S., Falconieri, G. S., Bertini, L., Baccelli, I., Paccosi, E., Belardo, A., Timperio, A. M., & Caruso, C. (2019). GLYI4 Plays A Role in Methylglyoxal Detoxification and Jasmonate-Mediated Stress Responses in Arabidopsis thaliana. Biomolecules, 9(10), 635. https://doi.org/10.3390/biom9100635






