The Reverse Side of a Coin: “Factor-Free” Ribosomal Protein Synthesis In Vitro is a Consequence of the In Vivo Proofreading Mechanism
Abstract
1. Introduction
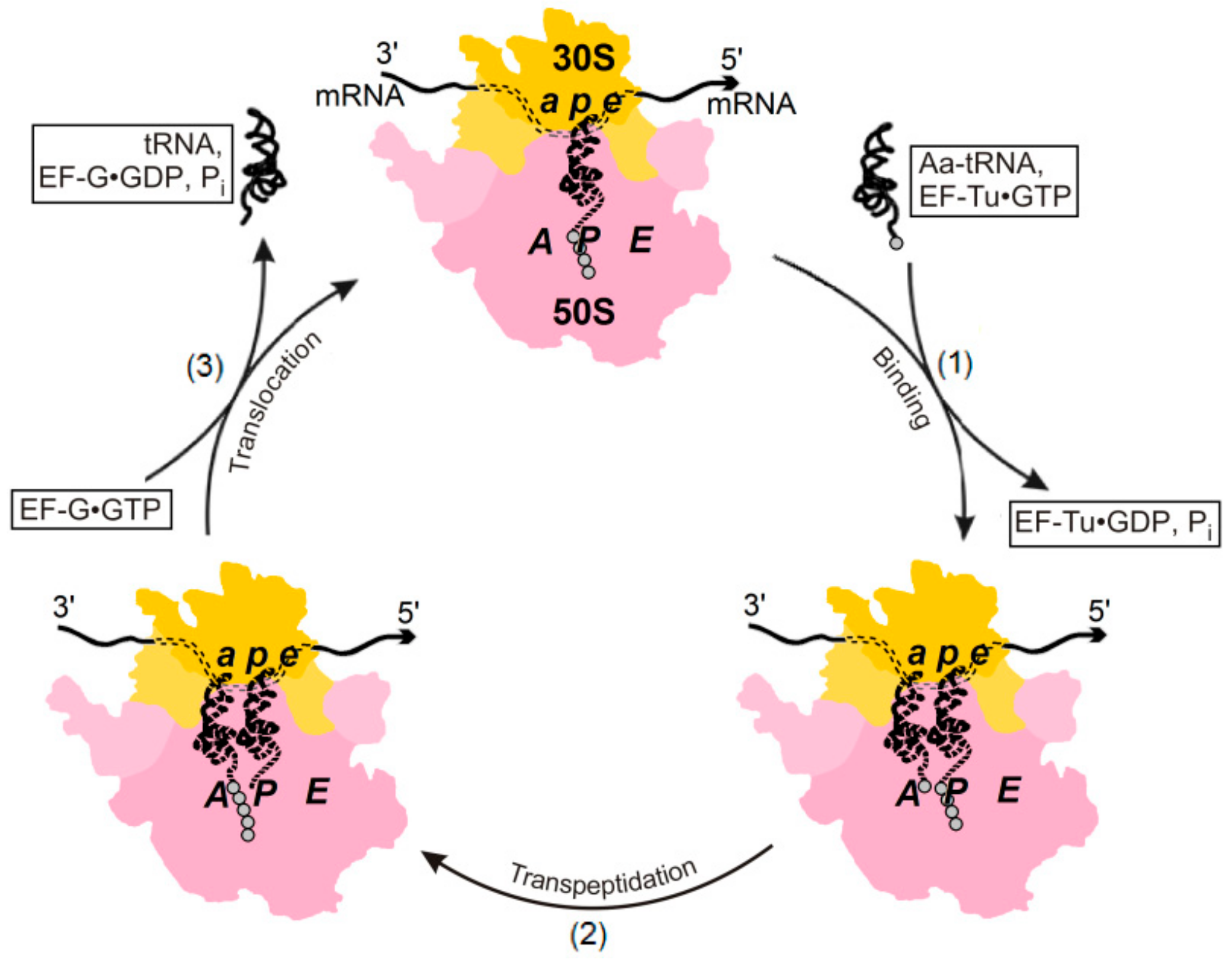
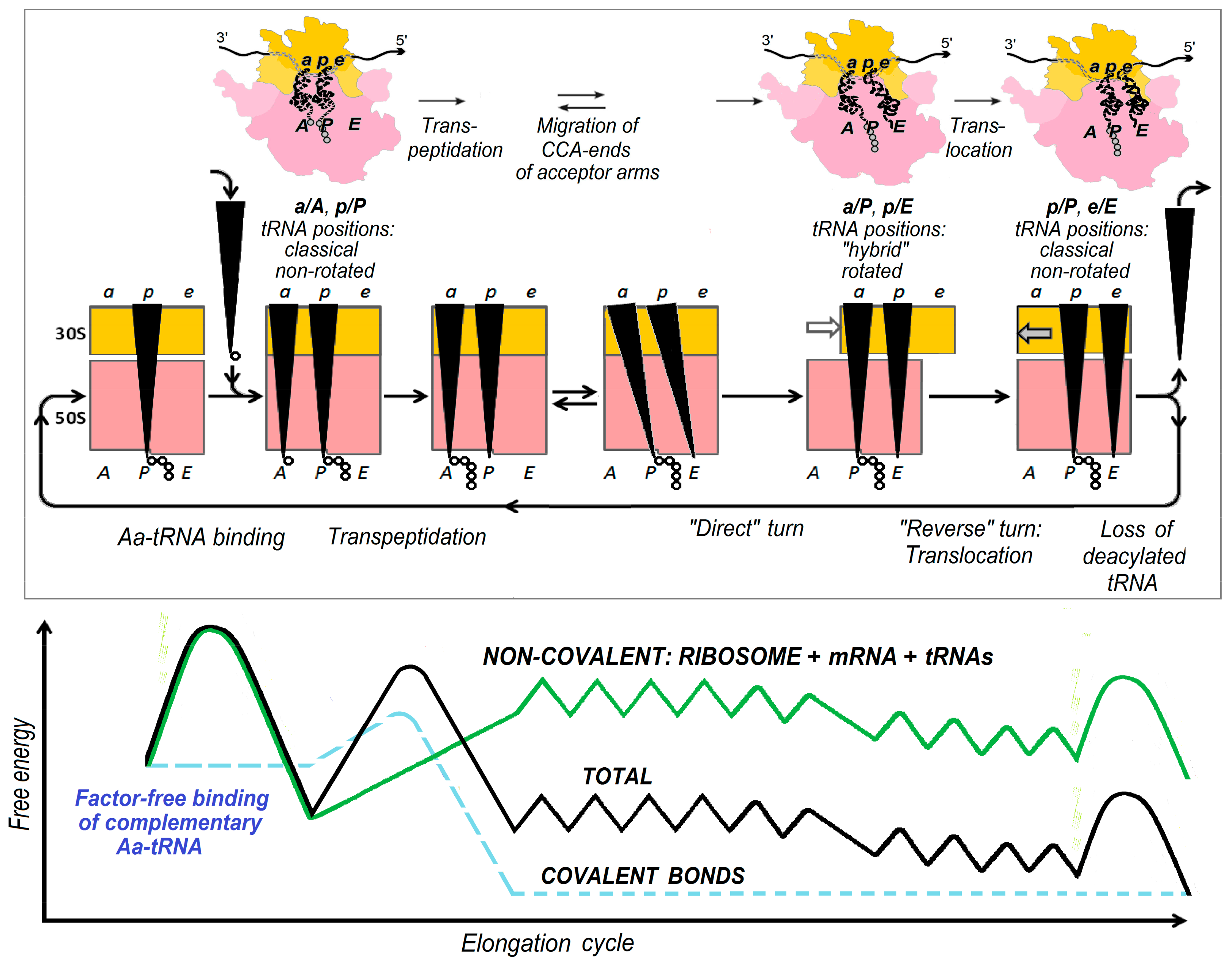
2. Energetics and Kinetics of Aa-tRNA Binding to the Ribosome
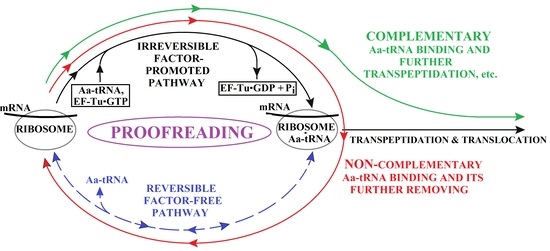
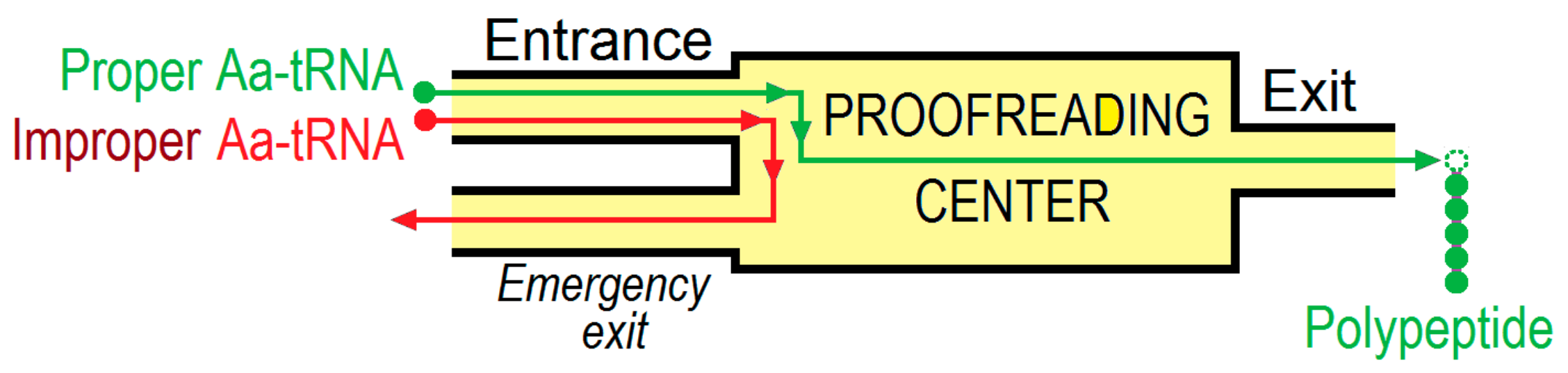
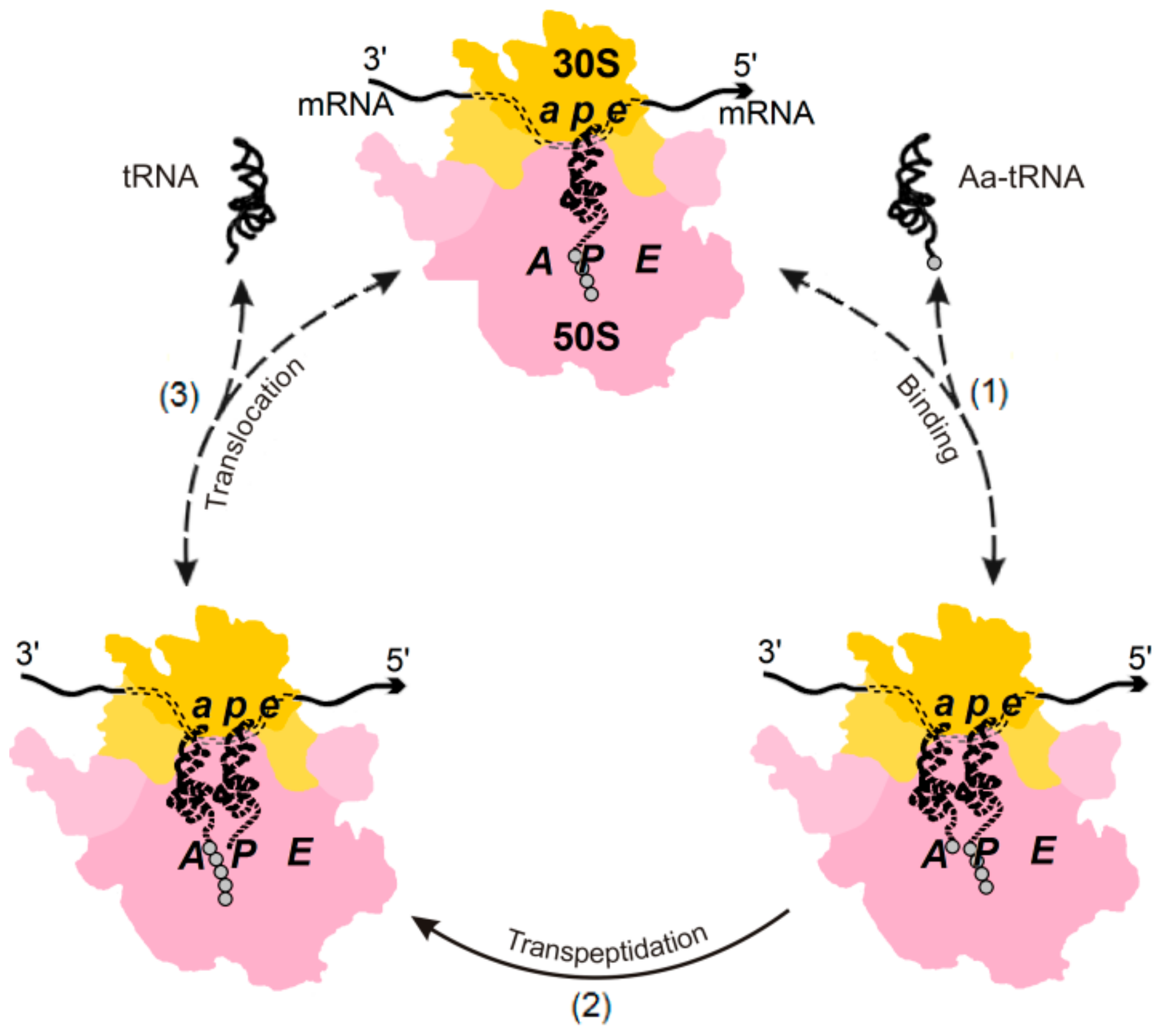
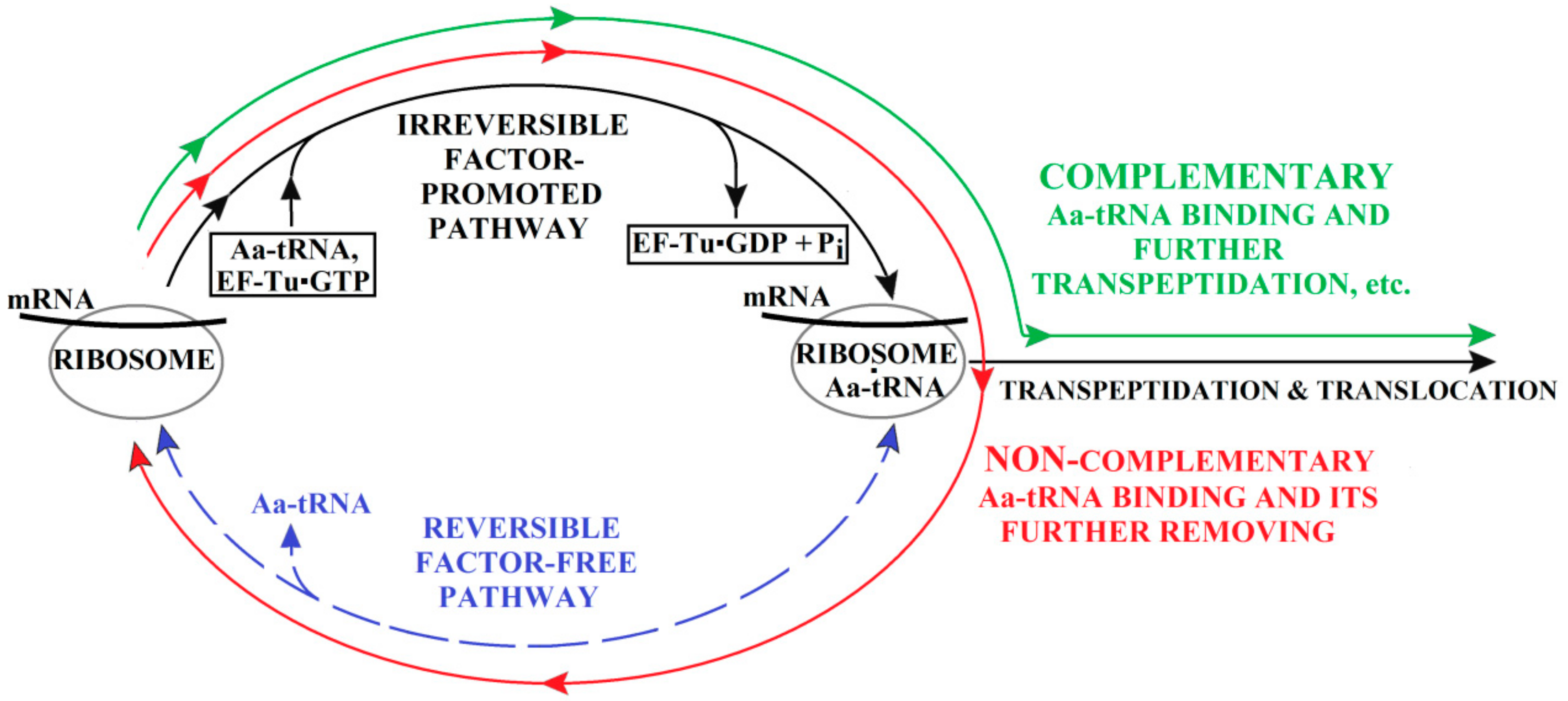
3. The (Ribosome•mRNA) Complex with Aa-tRNA Can be Initially Formed by the Factor-Promoted Pathway, but, If Unstable, It Then Decays Using A Reverse Move Along the Factor-Free Pathway
4. Discussion and Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding, 4th ed.; World Scientific: Hackensack, NJ, USA, 2017. [Google Scholar]
- Spirin, A.S. Ribosomes; Kluwer Acad. Publ./Plenum Press: New York, NY, USA, 1999. [Google Scholar]
- Moras, D. Proofreading in translation: Dynamics of the double-sieve model. Proc. Natl. Acad. Sci. USA 2010, 107, 21949–21950. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Kamarthapu, V.; Kruparani, S.P.; Deshmukh, M.V.; Sankaranarayanan, R. Mechanistic insights into cognate substrate discrimination during proofreading in translation. Proc. Natl. Acad. Sci. USA 2010, 107, 22117–22121. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, A.V.; Ptitsyn, O.B. Protein Physics. A Course of Lectures, 2nd ed.; Academic Press: New York, NY, USA, 2016; Lectures 8, 20, 25. [Google Scholar]
- Hopfield, J.J. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA 1974, 71, 4135–4139. [Google Scholar] [CrossRef] [PubMed]
- Leong, K.-W.; Uzun, Ü.; Selmer, M.; Ehrenberg, M. Two proofreading steps amplify the accuracy of genetic code translation. Proc. Natl. Acad. Sci. USA 2016, 113, 13744–13749. [Google Scholar]
- Gromadski, K.B.; Rodnina, M.V. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004, 12, 191–200. [Google Scholar] [CrossRef]
- Finkelstein, A.V.; Gavrilova, L.P. Protein biosynthesis proofreading is closely associated with existence of the factor-free ribosomal synthesis. Molec. Biol. (Mosk.) 2019, 53, 308–311. [Google Scholar] [CrossRef]
- Pestka, S. Studies on the formation of transfer ribonucleic acid-ribosome complexes III. The formation of peptide bonds by ribosomes in the absence of supernatant enzymes. J. Biol. Chem. 1968, 243, 2810–2820. [Google Scholar] [PubMed]
- Pestka, S. Studies on the formation of transfer ribonucleic acid-ribosome complexes VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J. Biol. Chem. 1969, 244, 1533–1539. [Google Scholar] [PubMed]
- Gavrilova, L.P.; Smolyaninov, V.V. Studies on the mechanism of translocation in ribosomes: I. Synthesis of polyphenylalanine in E. coli ribosomes in the absence of GTP and protein translation factors. Molec. Biol. (Mosk.) 1971, 5, 883–891. [Google Scholar]
- Gavrilova, L.P.; Spirin, A.S. Stimulation of “non-enzymic” translocation in ribosomes by ρ-chloromercuribenzoate. FEBS Lett. 1971, 17, 324–326. [Google Scholar] [CrossRef]
- Lucas-Lenard, J.; Lipmann, F. Protein biosynthesis. Annu. Rev. Biochem. 1971, 40, 409–448. [Google Scholar] [CrossRef] [PubMed]
- Gavrilova, L.P.; Spirin, A.S. A modification of the 30S ribosomal subparticle is responsible for stimulation of “non-enzymatic” translocation by p-chloromercuribenzoate. FEBS Lett. 1972, 22, 91–92. [Google Scholar] [CrossRef]
- Spirin, A.S.; Finkelstein, A.V. The ribosome as a Brownian ratchet machine. In Molecular Machines in Biology; Frank, J., Ed.; Cambridge Univ. Press: Cambridge, UK, 2011; pp. 158–190. [Google Scholar]
- Finkelstein, A.V.; Razin, S.V.; Spirin, A.S. Intersubunit Mobility of the Ribosome. Molec. Biol. (Mosk.) 2018, 52, 799–811. [Google Scholar] [CrossRef]
- Spirin, A.S. On the mechanism of ribosome function. The hypothesis of locking-unlocking of subparticles. Dokl. Akad. Nauk SSSR 1968, 179, 1467–1470. [Google Scholar] [PubMed]
- Spirin, A.S. A model of the functioning ribosome: Locking and unlocking of the ribosome subparticles. Cold Spring Harb. Symp. Quant. Biol. 1969, 34, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Gavrilova, L.P.; Rutkevitch, N.M. Ribosomal synthesis of polyleucine on polyuridylic acid as a template: Contribution of the elongation factors. FEBS Lett. 1980, 120, 135–140. [Google Scholar] [PubMed]
- Gavrilova, L.P.; Perminova, I.N.; Spirin, A.S. Elongation factor Tu can reduce translation errors in poly(U)-directed cell-free systems. J. Mol. Biol. 1981, 149, 69–78. [Google Scholar] [CrossRef]
- Noel, J.K.; Whitford, P.C. How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome. Nat. Commun. 2016, 7, 13314. [Google Scholar] [CrossRef] [PubMed]
- Gavrilova, L.P.; Kakhniashvili, D.G., Smailov. Stoichiometry of GTP hydrolysis in a poly(U)-dependent cell-free translation system. Determination of GTP/peptide bond ratios during codon-specific elongation and misreading. FEBS Lett. 1984, 178, 283–287. [Google Scholar] [CrossRef]
- Kakhniashvili, D.G.; Smailov, S.K., Gavrilova. The excess GTP hydrolyzed during mistranslation is expended at the stage of EF-Tu-promoted binding of non-cognate aminoacyl-tRNA. FEBS Lett 1986, 196, 103–107. [Google Scholar] [CrossRef]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Finkelstein, A.V. The Reverse Side of a Coin: “Factor-Free” Ribosomal Protein Synthesis In Vitro is a Consequence of the In Vivo Proofreading Mechanism. Biomolecules 2019, 9, 588. https://doi.org/10.3390/biom9100588
Finkelstein AV. The Reverse Side of a Coin: “Factor-Free” Ribosomal Protein Synthesis In Vitro is a Consequence of the In Vivo Proofreading Mechanism. Biomolecules. 2019; 9(10):588. https://doi.org/10.3390/biom9100588
Chicago/Turabian StyleFinkelstein, Alexei V. 2019. "The Reverse Side of a Coin: “Factor-Free” Ribosomal Protein Synthesis In Vitro is a Consequence of the In Vivo Proofreading Mechanism" Biomolecules 9, no. 10: 588. https://doi.org/10.3390/biom9100588
APA StyleFinkelstein, A. V. (2019). The Reverse Side of a Coin: “Factor-Free” Ribosomal Protein Synthesis In Vitro is a Consequence of the In Vivo Proofreading Mechanism. Biomolecules, 9(10), 588. https://doi.org/10.3390/biom9100588