Abstract
The plant hormone auxin controls almost all aspects of plant development through the gene regulatory properties of auxin response factors (ARFs) which bind so-called auxin responsive elements (AuxREs) in regulatory regions of their target genes. It has been proposed that ARFs interact and cooperate with other transcription factors (TFs) to bind to complex DNA-binding sites harboring cis-elements for several TFs. Complex DNA-binding sites have not been studied systematically for ARF target genes. ETTIN (ETT; ARF3) is a key regulator of gynoecium development. Cooperatively with its interacting partner INDEHISCENT (IND), ETT regulates PINOID (PID), a gene involved in the regulation gynoecium apical development (style development). Here, we mutated two ETT-bound AuxREs within the PID promoter and observed increased style length in gynoecia of plants carrying mutated promoter variants. Furthermore, mutating the AuxREs led to ectopic repression of PID in one developmental context while leading to ectopically upregulated PID expression in another stage. Our data also show that IND associates with the PID promoter in an auxin-sensitive manner. In summary, we demonstrate that targeted mutations of cis-regulatory elements can be used to dissect the importance of single cis-regulatory elements within complex regulatory regions supporting the importance of the ETT-IND interaction for PID regulation. At the same time, our work also highlights the challenges of such studies, as gene regulation is highly robust, and mutations within gene regulatory regions may only display subtle phenotypes.
Keywords:
auxin response; cis-regulation; plant development; ETTIN; PINOID; INDEHISCENT; gynoecium; gene regulation 1. Introduction
Over the last decades, great progress has been made in understanding how eukaryotic transcription factors (TFs) can specifically recognise the genes that they regulate [1,2,3]. Eukaryotic TFs recognise small specific DNA sequences present in regulatory regions of their target genes, called cis-regulatory elements. Loss or mutation of these cis-regulatory elements can dramatically impact an organism’s lifestyle and development [4,5]. Specific binding to cis-regulatory elements depends on protein sequence and structure of the TF and the range of biophysical interactions that the TF properties permit [6]. In many cases, members of TF families share affinity for the same DNA motif, yet they regulate different genes [6,7,8]. The mechanisms that provide target specificity in these TF families are largely unknown, and it is likely that specific DNA binding may require additional levels of control to ensure that regulation of target genes complies with developmental needs. One way to fine-tune the specificity of a DNA-TF interaction is the ability of the TF to interact and cooperate with other DNA binding proteins. In the case that a TF complex is heterotypic and, thus, consists of TFs from different families, the number as well as the sequence of cis-regulatory elements matters for binding specificity.
Auxin response factors (ARFs) are TFs that are responsible for regulating numerous developmental processes during a plant’s lifespan in response to the plant hormone auxin (indole-3-acetic acid or IAA). Impaired response to auxin can lead to severe flower defects and low fertility (e.g., arf6/8 and ettin (ett/arf3) mutants) or even to plant death as is the case in monoperteros (mp/arf5) mutants, which cannot form a root [9,10,11]. Through their B3 DNA-binding domain (DBD), ARF proteins bind to cis-regulatory elements composed of six base pairs that are called auxin responsive elements (AuxREs). The AuxRE motif has the sequence TGTCNN and was first discovered and characterised in the late 1990s in the promoter of the soybean GH3 auxin-responsive gene [12]. Since then, it has been studied in more detail with respect to its biophysical and regulatory properties [6,13,14,15,16,17,18]. Systematically mutating AuxREs in the auxin-responsive gene LEAFY confirmed the physiological importance of AuxREs for auxin response [5,6]. Despite these studies, complex DNA-binding sites and TF-DNA binding have not been characterised in depth for the ARF family.
The ETT protein has been implemented in several key developmental processes including gynoecium and fruit development [10,19,20]. Recent studies on protein–protein interactions identified a diverse range of proteins that directly interacts with ETT [21,22]. These proteins include several TFs from different families [21]. One of the identified interacting partners of ETT is the bHLH-type TF INDEHISCENT (IND). IND is required for the formation of the valve margins—a tissue that allows the Arabidopsis fruit to open and disperse the seeds upon maturation [23]. Additionally, IND was implemented in the control of polarity and symmetry transition at the apex of the gynoecium during development. In this process, IND cooperates with the bHLH TF SPATULA (SPT) [24,25]. In both processes, IND mediates its function at least in part by controlling auxin distribution [24,25,26]. IND coordinates directional auxin flux by direct repression of the PINOID (PID) gene, which encodes a serine–threonine kinase that is fundamental for proper symmetry establishment [25,26]. PID phosphorylates PIN-FORMED (PIN) auxin efflux carriers affecting their localisation and activity and thereby mediating polar auxin transport [27,28,29].
During gynoecium development, style formation is tightly controlled by the distribution of auxin in a spatial and temporal fashion. At stage 7, auxin accumulates in two lateral foci at the apex of developing gynoecia. At stage 8, two medial auxin foci emerge. These foci fuse to form an apical auxin ring at stage 9/10 that triggers a bilateral-to-radial symmetry switch, establishing the development of the radial style [25]. The generation of this apical auxin accumulation pattern relies on transcriptional repression of PID. After the bilateral-to-radial symmetry switch, PID becomes upregulated, and the apical auxin maximum disappears at stage 12 and is absent throughout style elongation [21,25].
Characterisation of the ETT–IND interaction has shown that these two proteins control polarity at the gynoecium apex by direct regulation of PID gene expression. Two AuxREs, in positions −429 and −447 with respect to the start codon, were identified as ETT-binding sites [21]. Firstly, this inverted AuxRE repeat lies within a region that is evolutionarily conserved among PID promoters in the Brassicaceae family [21]. Secondly, it is positioned in close proximity to two G-boxes and an E-box, which can be bound by SPT and IND, respectively [24,26]. Together, this makes the locus a complex regulatory patch within the PID promoter. Prior studies showed that the loss of either ETT or both ETT and IND function leads to misregulation of PID. In addition, in vitro studies using yeast one-hybrid (Y1H) experiments showed that mutating the identified AuxRE inverted repeat leads to loss of ETT binding [21]. Together, this suggests an important role for the two AuxREs in planta. Furthermore, the extensive knowledge on the regulation of the PID locus makes it a suitable model for in planta studies of cis-regulatory elements and their biological relevance in the context of complex regulatory regions and more broadly on gynoecium and fruit development.
Here, we evaluate the biological importance of these two specific AuxREs for the regulation of the PID gene in planta. We show that the two examined AuxRE sites play a role in determining style length. Furthermore, whilst providing in planta evidence that IND binds the previously identified E-box within the PID promoter, we also show that this binding is sensitive to increased levels of auxin.
2. Materials and Methods
2.1. Plant Materials
Plants were grown in soil at 22 °C in long-day conditions (16 h day/8 h dark). All transgenic plant lines, ind-2 and the pid-8 were in Columbia (Col-0) background.
2.2. Generation of Transgenic Lines
The transgenic rescue and reporter lines were generated using the Golden Gate method and transformed into Arabidopsis [30,31]. PID rescue constructs were transformed into pid-8 mutants, the pIND:IND:YFP construct was transformed into ind-2. Other while reporter constructs were transformed into Col-0. Primers and plasmids used for cloning can be found in Tables S1 and S2.
2.3. Phenotypic Analysis
For the silique and style length measurements, siliques were sampled from plants when they were mature. For wild-type, ten siliques from five plants (n = 50 siliques) were collected, while for transgenics, ten siliques from five independent lines per construct were collected and photographed. Silique and style length were then measured using ImageJ 1.48 [32]. The results were statistically analysed by one-way ANOVA followed by a Tukey’s multiple comparison test using GraphPad Prism Version 5.04 (La Jolla, CA, USA, www.graphpad.com).
2.4. Confocal Microscopy and Corrected Total Cell Fluorescence Quantification
Confocal images were taken using a Leica SP5 (HyD detector) confocal microscope (laser 20%, smart gain 30%, 200 hz, 8× line average, and a pinhole equivalent to 1.0× the airy disk diameter) using a 40× oil emersion lens. Excitation and detection of fluorophores were configured as follows: GFP was excited at 488 nm and detected at 498–530 nm; mCherry was excited at 561 nm and detected at 571–630 nm. For comparability the acquisition settings were set on the brightest sample and kept constant. Analysis was conducted in ImageJ 1.48 [32] using the corrected total cell fluorescence (CTCF) method [33] to quantify GFP and mCherry expression at the gynoecium apex. CTCFGFP-to-CTCFmCherry ratios were calculated and statistically analysed using one-way ANOVA followed by Tukey’s multiple comparison test using GraphPad Prism Version 5.04. Correlation between style length and CTCFGFP-to-CTCFmCherry ratios were analysed by linear regression analysis using GraphPad Prism Version 5.04.
2.5. Chromatin Immunoprecipitation Quantitative PCR
Transcription factor ChIP was performed in triplicate using the pIND:IND:YFP line, and data were analysed as described previously [21,34]. Immunoprecipitation (IP) was conducted using the anti-GFP antibody (Roche, Mannheim, Germany, 11814460001, Lot: 19958500), and Pierce Protein G magnetic beads (ThermoFisher, Waltham, MA, USA, 88847, Lot: SI253639). Enrichment for the E-box within the PID promoter was quantified using quantitative PCR (qPCR). In the ChIPqPCR, a region of the WUSCHEL (WUS) promoter that does not contain any E-box was used as a negative control (NC). ChIPqPCR data were analysed using one-way ANOVA followed by Tukey’s multiple comparison test using GraphPad Prism Version 5.04.
3. Results
3.1. Mutation of cis-Regulatory AuxREs within the PID Promoter Affects Style Length
To examine the relevance of the identified AuxRE sites for the regulation of PID, different constructs were generated and introduced into the pid-8 mutant. Each of these constructs carries 5 kb of the PID promoter region upstream of the PID genomic sequence fused to GFP. The 5-kb promoter region either carries a wild-type form or mutated versions of the two AuxREs. In the mutated versions, the G at the second positions of the TGTCNN AuxRE sequence was mutated into a T to prevent ARF binding, as previously described (Figure 1A) [6,21]. Using this approach, single mutants in the first (−434) and second −447) AuxRE as well as a double mutant for both sites were generated (named pPID1M, pPID2M, and pPID1+2M, respectively). In addition, the construct also carried a nuclear-localised mCherry gene under control of a constitutive ACTIN2 (ACT2) promoter.
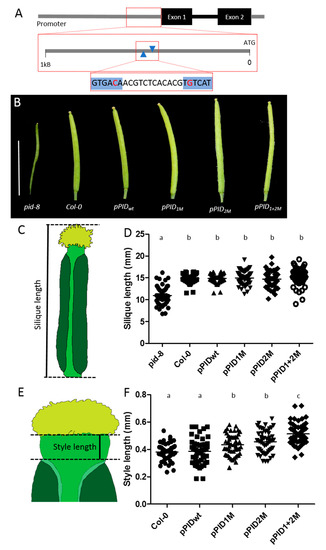
Figure 1.
All four promoter variants can rescue the pid-8 fruit phenotype. However, rescue lines differ significantly in style length. (A) Schematic representation of the regulatory region indicating the location and sequence of the two AuxREs (blue). The nucleotides that have been mutated are highlighted in red. (B) Representative siliques for each promoter variant construct. Scale bar = 10 mm. (C) Silique length describes the length of the fruit from the lower edge of the valves to the tip of the stigmatic tissue. (D) Silique length measurements show that all promoter variants can rescue the pid-8 fruit length phenotype, and none of the lines differs significantly from the wild type (N = 50). (E) Style length is measured as the distance from the most apical part of the valves to the underside of the stigmatic tissue. (F) Style length measurements show that styles are significantly longer in lines with mutated AuxRE elements in the PID promoter compared to wild-type and the positive pPIDwt line (N = 50). a,b indicate significant differences according to multiple comparison.
The pid-8 mutant phenotype shows a dramatic reduction in fruit size and a reduction of the valves [25]. Interestingly, all four constructs could rescue the pid-8 phenotype regarding the defects in valve development and silique length (Figure 1B–D), and this series of lines will, therefore, be referred to as ‘rescue lines’. PID has previously been shown to localise to the plasma membrane in an apolar fashion [35]. We found that this was also the case for cells at the tip of the developing gynoecium in all examined lines (Figure S1). Our macroscopic analysis of the complemented fruits suggested that the style was enlarged in lines carrying the mutated promoter variants. To test this in more detail, we measured the style lengths of the rescue lines under the microscope in comparison to wild type (Col-0). In five independent lines for each construct, we measured ten styles per line, and the results indeed revealed a significant difference in style length (Figure 1E,F). Specifically, whilst the constructs carrying the wild-type promoter allele (pPIDwt) were sufficient to fully rescue the pid-8 phenotype regarding both silique length and style length, mutating either the first (pPID1M) or the second (pPID2M) AuxRE moderately, yet significantly, increased the style length when compared to wild-type and pPIDwt lines. However, style lengths of the pPID1M and pPID2M lines did not differ significantly when compared to each other. In contrast, the double-mutant pPID1+2M lines had significantly longer styles than the Col-0, pPIDwt, pPID1M, and pPID2M lines. This indicates that both AuxREs in the examined region affected style length in an additive manner. The data also suggest that the two AuxRE had an equal contribution to style length. Finally, the results imply that the two AuxRE were indeed biologically important for the correct regulation of PID gene expression during style development.
3.2. Mutating ETT-Binding AuxRE Positively Affects PID Promoter Activity at Gynoecium Stage 12
To assess whether the mutation in the AuxRE sites affected the PID gene expression, we decided to quantify the PID promoter activity in the promoter mutant lines. We generated promoter-reporter lines, that contain the same 5-kb PID promoter variants as the rescue lines, but this time controlled the expression of a nuclear GFP. Additionally, these constructs carried a nuclear mCherry controlled by a constitutive ACTIN2 (ACT2) promoter for normalisation of the GFP signals and, thus, allowed quantitative studies of gene expression. When transformed into Col-0 plants, the ratio of GFP to mCherry corrected total cell fluorescence (CTCF) can be used as a measure of promoter activity and gene expression [33]. The promoter activity of the different variants was examined in stage 12 gynoecia (Figure 2A–D).
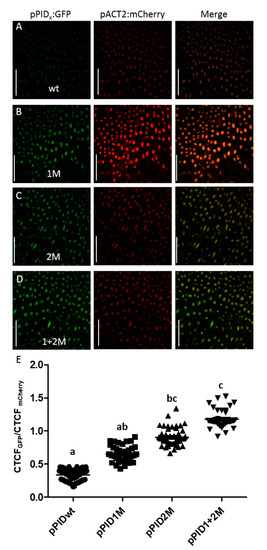
Figure 2.
Mutating AuxREs affect PID promoter activity in the style region. (A–D) Representative confocal images of the style region of stage 12 for the respective promoter variant reporter lines. Scale bars = 50 µM (E) Fluorescence quantification shows that mutating AuxREs lead to a gradual activation of the PID gene. The data indicate that the two AuxREs have an additive effect (N = 40). a,b,c indicate significant differences according to multiple comparison.
The results showed significant differences between lines of different promoter variants (Figure 2E), with weakest GFP fluorescence (CTCFGFP/CTCFmCherry) in lines carrying the wild-type promoter variant, while fluorescence in lines with the promoter mutated in the first AuxRE were moderately, but not significantly, stronger. GFP fluorescence in lines mutated in the second AuxRE was significantly increased compared to the wild-type but not compared to the single mutant in the first AuxRE. The highest relative fluorescence was measured in the doubly mutated promoter lines. For these lines, the expression was significantly higher compared to pPIDwt and pPID1M lines but not pPID2M.
Together with the style measurements, these data suggest that both the first and the second AuxRE affected PID promoter activity. Even though activity of pPID1M lines did not differ significantly from wild-type promoter or pPID2M promoter activity and appeared as intermediate between the two, combining mutations in the first and the second AuxRE had an additive effect on PID promoter activity. This suggests that both AuxREs are regulatory elements important for the repression of the PID gene. Comparing the PID promoter activity with the phenotypic style-length data, it becomes evident that high promoter activity at stage 12 correlated positively with longer styles (Figure S2, R2 = 0.978). Therefore, higher PID expression seems to promote style elongation at this stage.
3.3. Complex Transcription Factor Interactions Regulate PID Expression During Style Development
The presented data indicate that the two examined AuxREs affected PID promoter activity during style formation, as mutating any of the AuxREs increases the promoter activity significantly at stage 12. The region in which these AuxREs are found also contained other potential cis-elements. For instance, two bHLH binding motifs (G-boxes) were identified, which intriguingly overlapped with the AuxREs (Figure 3A). These G-boxes were previously shown to be candidates for binding sites for the bHLH TF, SPT [24]. In agreement with this, PID was ectopically expressed in spt mutants [25]. The IND bHLH TF has also been shown to directly bind the PID promoter [26], most likely via a so-called E-box positioned 57 bp downstream of the AuxRE-G-box patch (Figure 3A). The ETT–IND protein interaction is sensitive to auxin [21], however, whilst it has been demonstrated that ETT is bound to the PID promoter regardless of the auxin level [21], it is unknown whether this is also true for IND. To test this, ChIPqPCR was carried out using an IND reporter (pIND:IND:YFP), which rescued the defects of the strongly indehiscent ind-2 mutant. The ChIP experiment showed that interaction of the IND:YFP protein with the examined regulatory patch around 0.4 kb upstream of the PID coding region was significantly reduced after auxin treatment (Figure 3B). This implies that, in contrast to ETT, IND binds its target DNA sequence in an auxin-sensitive manner.
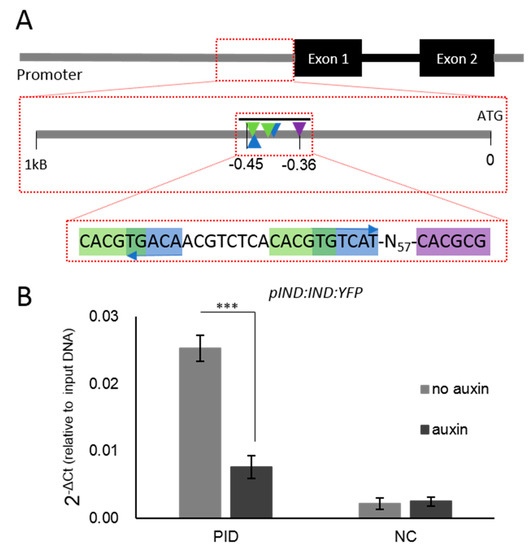
Figure 3.
The transcription factors ETT and IND can bind to the complex regulatory patch within the PID promoter. (A) Schematic representation of the regulatory region. The black bar above the zoomed region indicates the amplicon used in qPCR reactions. Green indicates G-boxes; blue indicates AuxREs; purple indicates the E-box. (B) IND can bind to the regulatory patch. Auxin reduces IND binding to the PID promoter (*** p-Values < 0.0001; shown are the averages of three biological replicates ± standard deviation). NC, negative control.
4. Discussion
Spatial and temporal control of gene expression is key for the regulation of most developmental processes. However, key regulatory genes are often subject to complex regulation that involves multiple transcription factors forming various protein complexes interacting with various cis-regulatory elements at a complex regulatory region. Often the composition of these complexes and their mode of action depends on the biological context and can differ within the same cell under different developmental stages or environmental conditions. This has complicated studies on the contribution of specific cis-regulatory elements to gene expression.
Here, we attempted to uncover the importance of two ETT-binding AuxRE sites within the promoter of the PID gene for expression during Arabidopsis gynoecium development. PID is a key regulator of polar auxin transport and the main driver of apical auxin accumulation in the gynoecium, which affects style establishment and potentially style elongation [25]. During style establishment, PID is repressed (developmental stages 7–10). This leads to auxin accumulation at the gynoecium apex, triggering a bilateral-to-radial symmetry switch to establish the radial style [25]. At later stages, during style elongation, auxin levels decrease, and PID is expressed [21].
The presented data show that both AuxREs identified in the PID promoter play a role in the regulation of PID gene expression during style elongation. Phenotypic analysis showed that the PID gene under control of any of the four examined promoter variants was able to rescue the pid-8 ovary defects (reduced valve development). This suggests that the examined AuxREs do not play a major role in ovary formation. Besides the two AuxREs studied here, the PID promoter contains several other cis-regulatory elements, including several AuxREs [21,24,26]. Although, the strongest ETT binding is associated with the two AuxREs studied here during gynoecium development, it is possible that other AuxREs are important in other developmental contexts. Further phenotypic examination, however, showed significant differences in style length between different promoter variants, suggesting that the main function of the two AuxREs is in the regulation of style development. Our data show that mutations in both AuxREs increase the promoter activity significantly at stage 12 in a dosage-dependent manner. Hence, mutating the AuxREs lead to de-repression of PID at developmental stage 12.
It is well established that, besides ETT, two bHLH transcription factors, SPT and IND, are essential regulators of PID expression during gynoecium development [25]. In spt mutants as well as in ett ind double mutants, the PID gene is ectopically expressed at the apex of developing gynoecia during early stages, indicating that these factors act as repressors of PID expression [21,25]. Sequence analysis has identified the presence of a putative IND-binding E-box and two SPT-binding G-boxes that partially overlap with the AuxREs studied here. ETT and SPT do not appear to interact with each other, but previous studies have identified that they both can form complexes with IND [24]. Moreover, while high auxin concentrations can disrupt ETT–IND interactions, this is not the case for SPT–IND interactions [21]. These observations open the possibility that SPT and ETT may compete to bind the PID promoter in this region, and that the effect of changing auxin levels may depend on whether ETT or SPT is bound.
We show that IND can bind the PID promoter in planta, presumably via the E-box element that IND has been shown to bind in yeast one-hybrid studies [24,26]. Intriguingly, auxin treatment leads to a disruption of this IND–DNA interaction (Figure 3B), whilst ETT remains bound to the PID promoter in the presence of auxin [21]. The mechanism by which auxin affects IND binding to its cis-element is not yet understood. Nevertheless, it is possible that IND has an active role in recruiting ETT proteins to their binding sites in the PID promoter to repress gene expression in low auxin levels. High auxin concentrations can disrupt ETT–IND interactions and IND-DNA interactions, releasing IND from the PID promoter and leading to an up-regulation of PID. In contrast, data for the mutated PID promoter lines presented here suggest that DNA-bound ETT maintains some residual repressor activity that fine-tunes PID expression during style elongation.
ETT and IND are expressed at the gynoecium apex and style from early stages throughout development [21,24,26]. In contrast, SPT is expressed until stage 11 but absent from stage 12 and onwards [24,35]. It will be interesting to address whether this differential expression of ETT and SPT contributes to the developmentally specific regulation of PID expression. Alternatively, it is possible that competitive protein–DNA interactions and/or complex protein–protein interactions are important. These possibilities are not mutually exclusive and will be addressed in future studies.
5. Conclusions
Here, we present an approach to test the contribution of two ETT-binding AuxREs in the promoter of an auxin-responsive gene (PID) in the developmental context of gynoecium development. This work shows that targeted mutations of potential cis-elements can be used to dissect the importance of cis-regulation within complex regulatory regions in planta. However, it also shows the challenges involved in assessing the contributions of these elements, as gene regulation is highly robust, and mutations within gene regulatory regions may display subtle phenotypes. To conclude, this study provides a starting point for a more in-depth study understanding cis-regulation of auxin-responsive genes, as the data still do not permit full elucidation of the mechanism by which ETT regulates PID expression. For example, it remains an open question how ETT is recruited to its cis-binding sites and whether ETT binds as a monomer, homo-, or heterodimer. In line with this, it is yet to be addressed whether IND acts as a pioneering factor and if ETT and SPT compete to bind their overlapping DNA binding sites. Nonetheless, this work indicates that cis-elements affect the complex formation at target loci, and their regulation reveals the advantage of studying cis-regulation in planta through focusing on a specific developmental process.
Supplementary Materials
The supplementary materials are available online.
Author Contributions
Conceptualization, A.K. and L.Ø.; methodology, A.K. and L.Ø.; analysis, A.K.; investigation, A.K., B.R. and B.T.-B.; resources, A.K.; data curation, A.K.; writing—original draft preparation, A.K.; writing—review and editing, A.K. and L.Ø.; visualization, A.K.; supervision, L.Ø.; project administration, L.Ø.; funding acquisition, L.Ø.
Funding
This research was funded by the UKRI Biotechnology and Biological Sciences Research Council, grant numbers BB/M011216/1 (Norwich Research Park Doctoral Training Partnership), BB/I017232/1 (Crop Improvement Research Club) and BB/P013511/1 (Institute Strategic Programme Grant).
Acknowledgments
We thank the following support services at the John Innes Centre: Bio-imaging and Horticulture for skillful assistance. We are grateful to Sara Simonini for producing the pIND:IND:YFP line and for sharing it prior to publication.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Furey, T.S. ChIP–seq and beyond: New and improved methodologies to detect and characterize protein–DNA interactions. Nat. Rev. Genet. 2012, 13, 840. [Google Scholar] [CrossRef] [PubMed]
- Godoy, M.; Franco-Zorrilla, J.M.; Pérez-Pérez, J.; Oliveros, J.C.; Lorenzo, Ó.; Solano, R. Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J. 2011, 66, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Jolma, A.; Kivioja, T.; Toivonen, J.; Cheng, L.; Wei, G.; Enge, M.; Taipale, M.; Vaquerizas, J.M.; Yan, J.; Sillanpaa, M.J.; et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010, 20, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Questa, J.I.; Song, J.; Geraldo, N.; An, H.L.; Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 2016, 353, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Wu, M.-F.; Winter, C.M.; Berns, M.C.; Nole-Wilson, S.; Yamaguchi, A.; Coupland, G.; Krizek, B.A.; Wagner, D. A Molecular Framework for Auxin-Mediated Initiation of Flower Primordia. Dev. Cell 2013, 24, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Boer, R.; Freire-Rios, A.; van den Berg, W.A.M.; Saaki, T.; Manfield, I.W.; Kepinski, S.; Lopez-Vidrieo, I.; Manuel Franco-Zorrilla, J.; de Vries, S.C.; Solano, R.; et al. Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors. Cell 2014, 156, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 2014, 111, 2367–2372. [Google Scholar] [CrossRef]
- Merika, M.; Orkin, S.H. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 1993, 13, 3999–4010. [Google Scholar] [CrossRef]
- Berleth, T.; Jurgens, G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 1993, 118, 575–587. [Google Scholar]
- Sessions, R.A. Arabidopsis (Brassicaceae) flower development and gynoecium patterning in wild type and Ettin mutants. Am. J. Bot. 1997, 84, 1179–1191. [Google Scholar] [CrossRef]
- Tabata, R.; Ikezaki, M.; Fujibe, T.; Aida, M.; Tian, C.; Ueno, Y.; Yamamoto, K.T.; Machida, Y.; Nakamura, K.; Ishiguro, S. Arabidopsis AUXIN RESPONSE FACTOR6 and 8 Regulate Jasmonic Acid Biosynthesis and Floral Organ Development via Repression of Class 1 KNOX Genes. Plant Cell Physiol. 2009, 51, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a Transcription Factor That Binds to Auxin Response Elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Dimerization and DNA binding of auxin response factors. Plant J. 1999, 19, 309–319. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, R.C.; Huang, S.S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Stigliani, A.; Martin-Arevalillo, R.; Lucas, J.; Bessy, A.; Vinos-Poyo, T.; Mironova, V.; Vernoux, T.; Dumas, R.; Parcy, F. Capturing Auxin Response Factors Syntax Using DNA Binding Models. Mol. Plant 2019, 12, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Omelyanchuk, N.A.; Wiebe, D.S.; Novikova, D.D.; Levitsky, V.G.; Klimova, N.; Gorelova, V.; Weinholdt, C.; Vasiliev, G.V.; Zemlyanskaya, E.V.; Kolchanov, N.A.; et al. Auxin regulates functional gene groups in a fold-change-specific manner in Arabidopsis thaliana roots. Sci. Rep. 2017, 7, 2489. [Google Scholar] [CrossRef] [PubMed]
- Cherenkov, P.; Novikova, D.; Omelyanchuk, N.; Levitsky, V.; Grosse, I.; Weijers, D.; Mironova, V. Diversity of cis-regulatory elements associated with auxin response in Arabidopsis thaliana. J. Exp. Bot. 2018, 69, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Sessions, A.; Nemhauser, J.L.; McColl, A.; Roe, J.L.; Feldmann, K.A.; Zambryski, P.C. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 1997, 124, 4481–4491. [Google Scholar] [PubMed]
- Sessions, A.Z. Arabidopsis gynoecium structure in the wild type and ettin mutants. Development 1995, 121, 1519–1532. [Google Scholar] [PubMed]
- Simonini, S.; Deb, J.; Moubayidin, L.; Stephenson, P.; Valluru, M.; Freire-Rios, A.; Sorefan, K.; Weijers, D.; Friml, J.; Ostergaard, L. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 2016, 30, 2286–2296. [Google Scholar] [CrossRef] [PubMed]
- Simonini, S.; Bencivenga, S.; Trick, M.; Ostergaard, L. Auxin-Induced Modulation of ETTIN Activity Orchestrates Gene Expression in Arabidopsis. Plant Cell 2017. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J.; Roeder, A.H.K.; Kempin, S.A.; Gremski, K.; Ostergaard, L.; Guimil, S.; Reyes, D.K.; Yanofsky, M.F. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 2004, 116, 843–853. [Google Scholar] [CrossRef]
- Girin, T.; Paicu, T.; Stephenson, P.; Fuentes, S.; Koerner, E.; O’Brien, M.; Sorefan, K.; Wood, T.A.; Balanza, V.; Ferrandiz, C.; et al. INDEHISCENT and SPATULA Interact to Specify Carpel and Valve Margin Tissue and Thus Promote Seed Dispersal in Arabidopsis. Plant Cell 2011, 23, 3641–3653. [Google Scholar] [CrossRef] [PubMed]
- Moubayidin, L.; Ostergaard, L. Dynamic Control of Auxin Distribution Imposes a Bilateral-to-Radial Symmetry Switch during Gynoecium Development. Curr. Biol. 2014, 24, 2743–2748. [Google Scholar] [CrossRef] [PubMed]
- Sorefan, K.; Girin, T.; Liljegren, S.J.; Ljung, K.; Robles, P.; Galvan-Ampudia, C.S.; Offringa, R.; Friml, J.; Yanofsky, M.F.; Ostergaard, L. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 2009, 459, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Benjamins, R.; Quint, A.; Weijers, D.; Hooykaas, P.; Offringa, R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 2001, 128, 4057–4067. [Google Scholar] [PubMed]
- Friml, J.; Yang, X.; Michniewicz, M.; Weijers, D.; Quint, A.; Tietz, O.; Benjamins, R.; Ouwerkerk, P.B.F.; Ljung, K.; Sandberg, G.; et al. A PINOID-Dependent Binary Switch in Apical-Basal PIN Polar Targeting Directs Auxin Efflux. Science 2004, 306, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Cho, H.-T. PINOID Positively Regulates Auxin Efflux in Arabidopsis Root Hair Cells and Tobacco Cells. Plant Cell 2006, 18, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Youles, M.; Gruetzner, R.; Ehnert, T.M.; Werner, S.; Jones, J.D.G.; Patron, N.J.; Marillonnet, S. A Golden Gate Modular Cloning Toolbox for Plants. ACS Synt. Biol. 2014, 3, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- McCloy, R.A.; Rogers, S.; Caldon, C.E.; Lorca, T.; Castro, A.; Burgess, A. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 2014, 13, 1400–1412. [Google Scholar] [CrossRef] [PubMed]
- Schiessl, K.; Muiño, J.M.; Sablowski, R. Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc. Natl. Acad. Sci. USA 2014, 111, 2830–2835. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.; Gaillochet, C.; Lohmann, J.U. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 2015, 142, 3343–3350. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).