Searching for Chymase Inhibitors among Chamomile Compounds Using a Computational-Based Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Structure-Based Pharmacophore Models and Ligand Screening
2.2. Molecular Docking Simulations
2.3. Molecular Dynamics Simulations
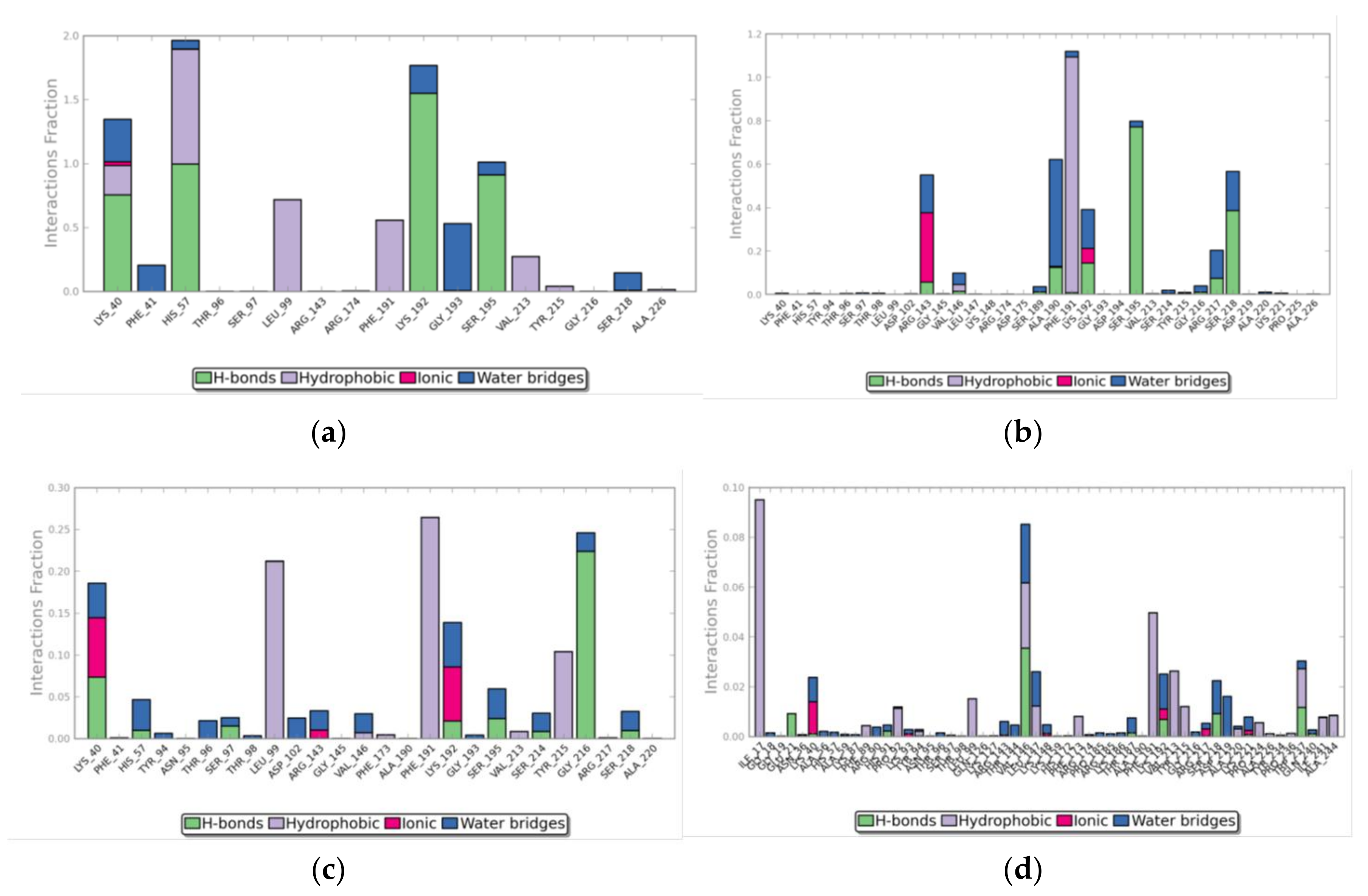
3. Results
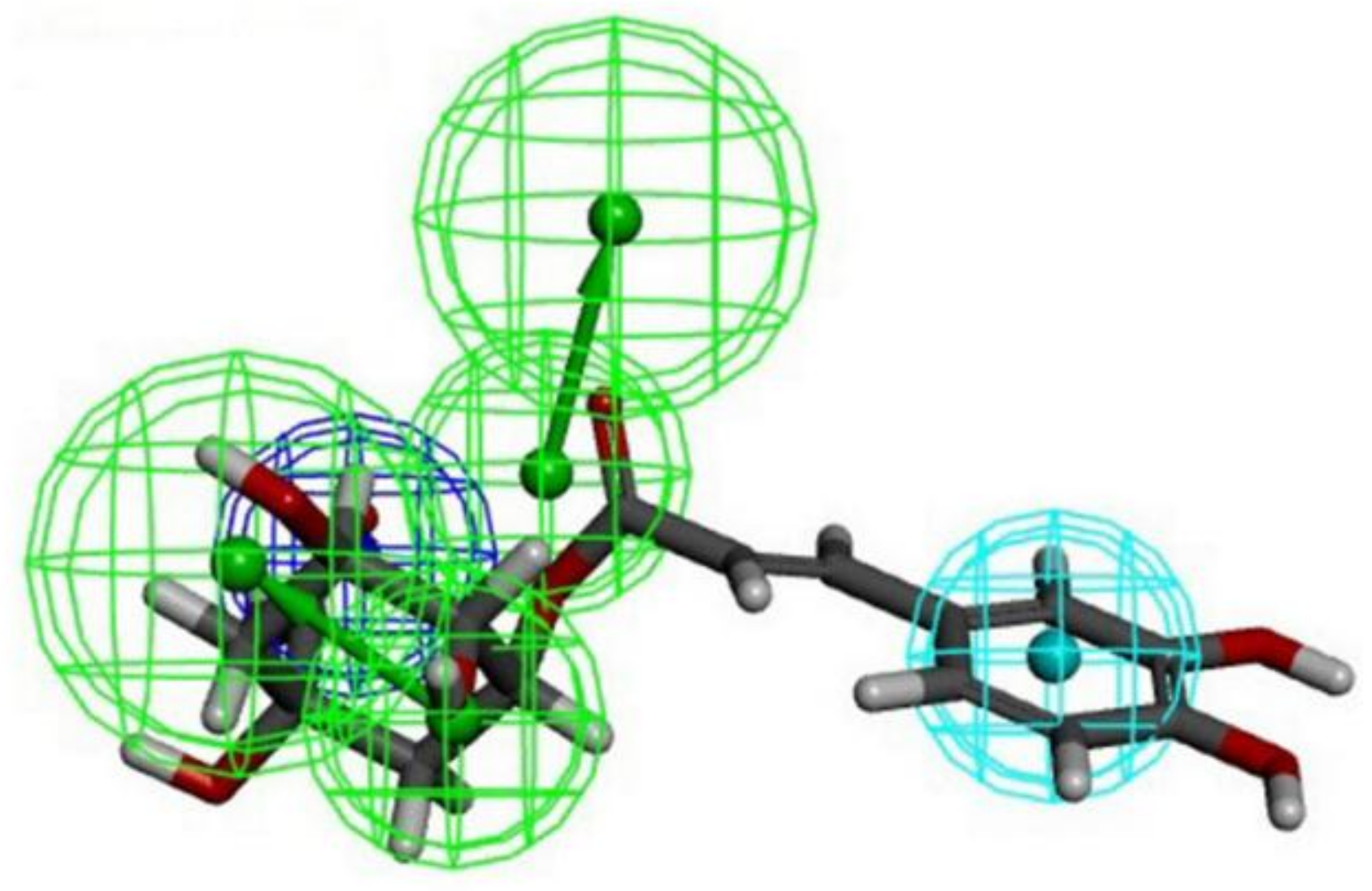
3.1. Structure-Based Pharmacophore Modeling and Ligand Screening
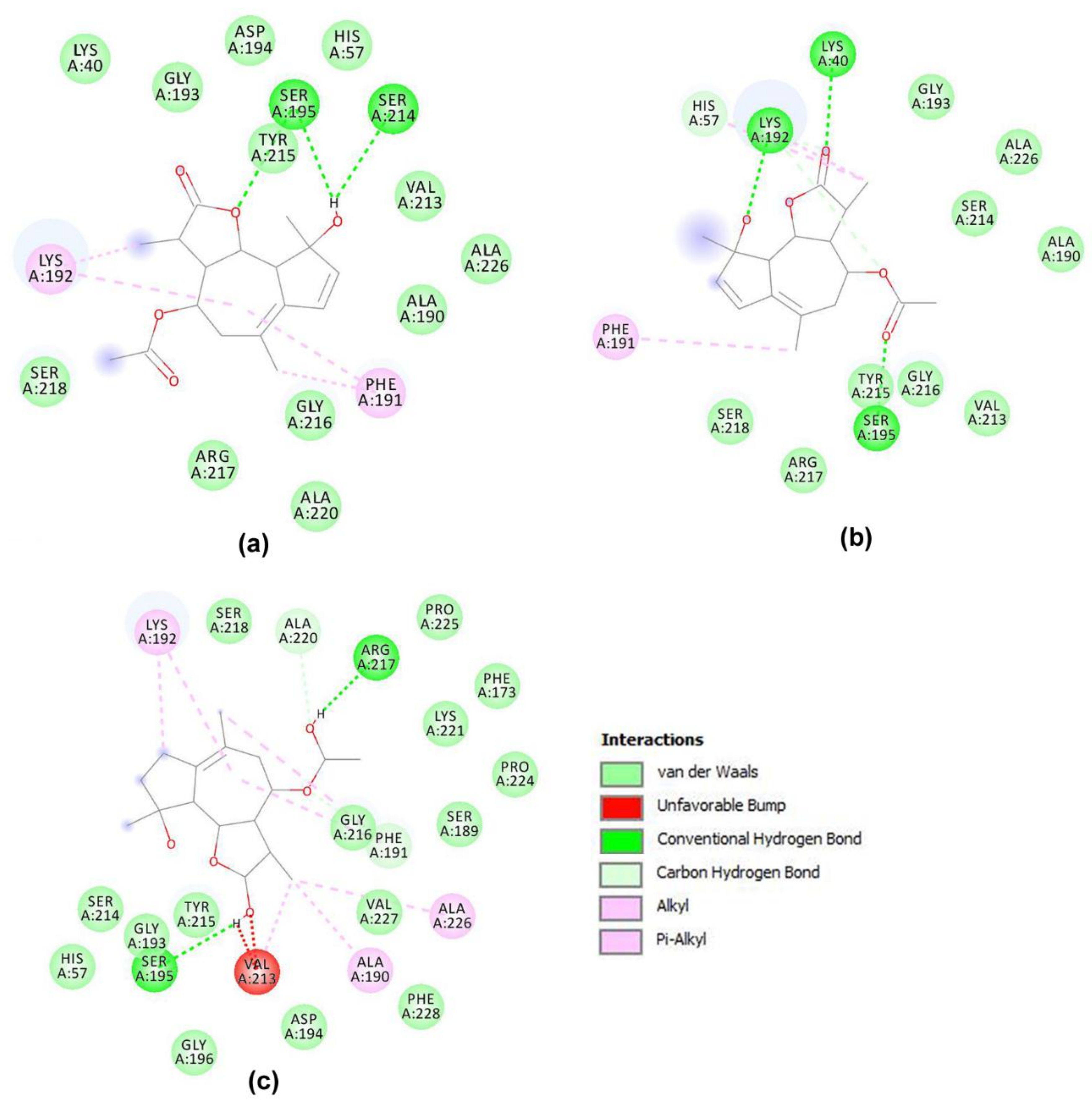
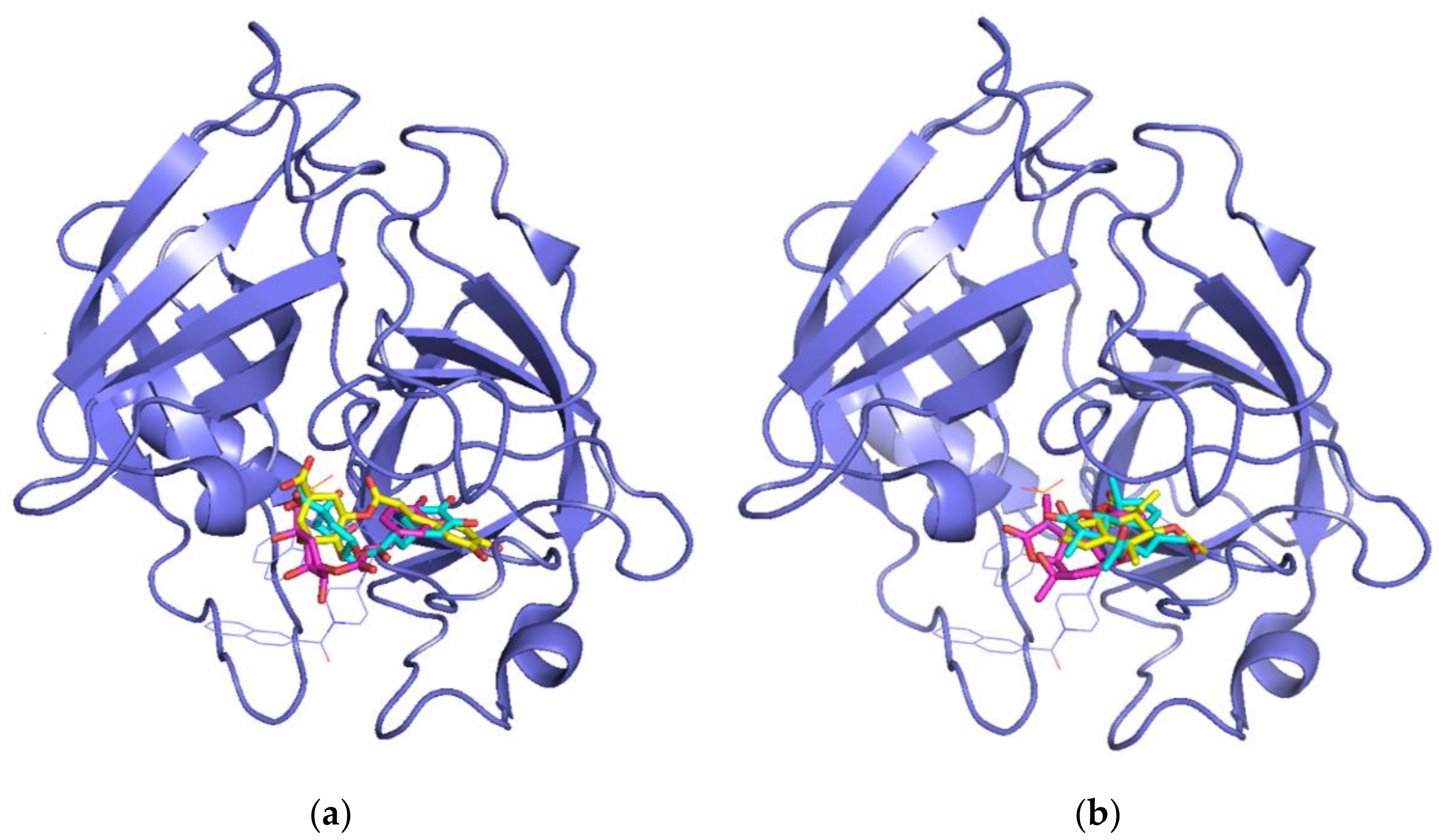
3.2. Molecular Docking Simulations
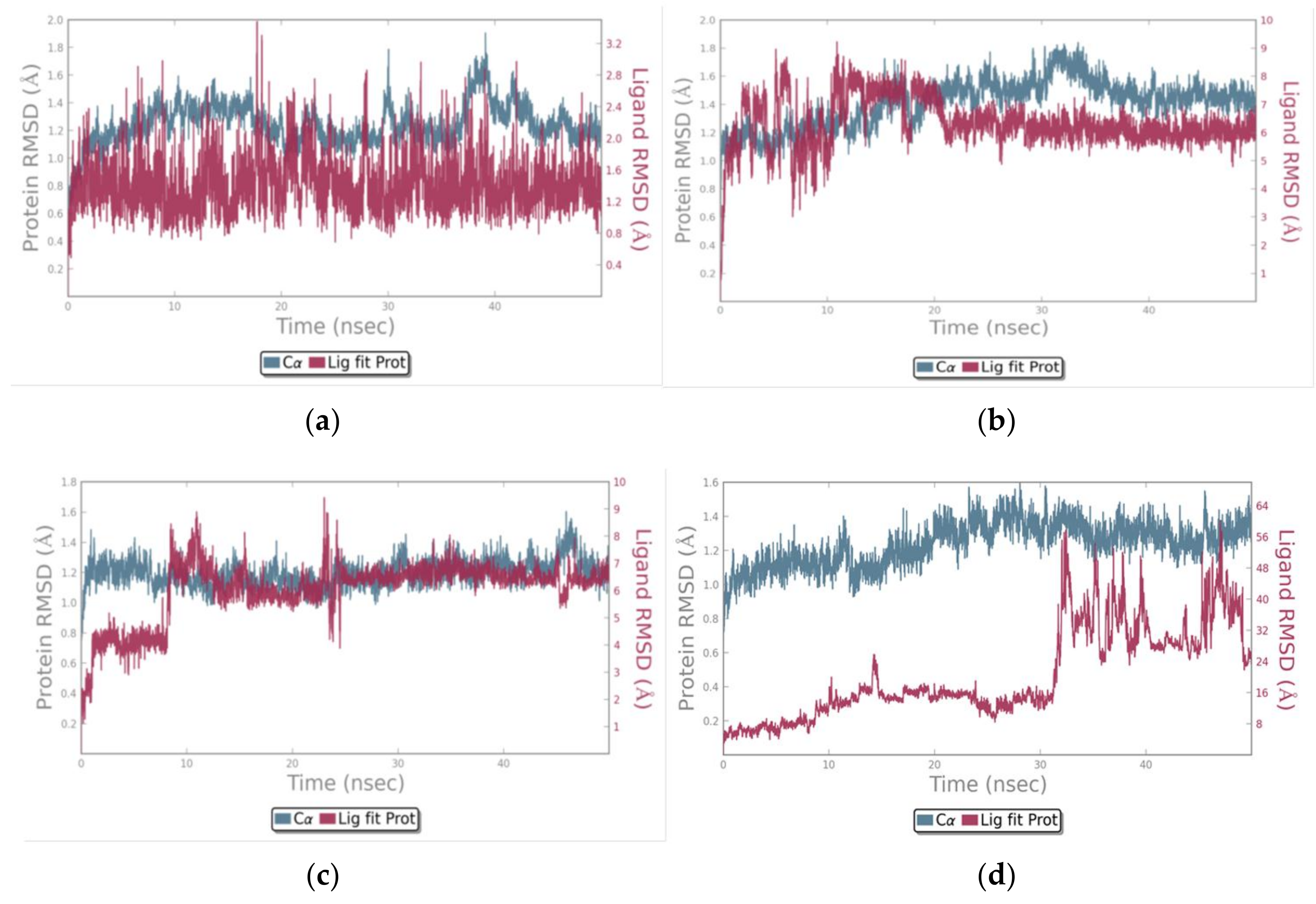
3.3. Molecular Dynamics Simulations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Douhaier, J.; Succar, J.; Lancerotto, L.; Gurish, M.F.; Orgill, D.P.; Hamilton, M.J.; Krilis, S.A.; Stevens, R.L. Development of mast cells and importance of their tryptase and chymase serine proteases in inflammation and wound healing. Adv. Immunol. 2014, 122, 211–252. [Google Scholar] [CrossRef]
- Ahmad, S.; Simmons, T.; Varagic, J.; Moniwa, N.; Chappell, M.C.; Ferrario, C.M. Chymase-dependent generation of Angiotensin II from Angiotensin-(1-12) in human atrial tissue. PLoS ONE 2011, 6, e28501. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, E.J.; Virani, S.S.; Callaway, C.W.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Chiuve, S.E.; Cushman, M.; Delling, F.N.; Deo, R.; et al. Heart disease and stroke statistics-2018 update: A report from the American Heart Association. Circulation 2018, 137, e62–e492. [Google Scholar] [CrossRef] [PubMed]
- Takai, S.; Jin, D. Improvement of cardiovascular remodelling by chymase inhibitor. Clin. Exp. Pharmacol. Physiol. 2016, 43, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Takai, S.; Jin, D.; Chen, H.; Li, W.; Yamamoto, H.; Yamanishi, K.; Miyazaki, M.; Higashino, H.; Yamanishi, H.; Okamura, H. Chymase inhibition improves vascular dysfunction and survival in stroke-prone spontaneously hypertensive rats. J. Hypertens. 2014, 32, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Bastos, M.; Maeji, N.J.; Abeles, R.H. Inhibitors of human heart chymase based on a peptide library. Proc. Natl. Acad. Sci. USA 1995, 92, 6738–6742. [Google Scholar] [CrossRef] [PubMed]
- De Garavilla, L.; Greco, M.N.; Sukumar, N.; Chen, Z.W.; Pineda, A.O.; Mathews, F.S.; Di Cera, E.; Giardino, E.C.; Wells, G.I.; Haertlein, B.J.; et al. A novel, potent dual inhibitor of the leukocyte proteases cathepsin G and chymase molecular mechanisms and anti-inflammatory activity in vivo. J. Biol. Chem. 2005, 280, 18001–18007. [Google Scholar] [CrossRef]
- Tausch, L.; Henkel, A.; Siemoneit, U.; Poeckel, D.; Kather, N.; Franke, L.; Hofmann, B.; Schneider, G.; Angioni, C.; Geisslinger, G.; et al. Identification of human cathepsin G as a functional target of boswellic acids from the anti-inflammatory remedy frankincen. J. Immunol. 2009, 183, 3433–3442. [Google Scholar] [CrossRef]
- Futamura-Takahashi, J.; Tanaka, T.; Sugawara, H.; Iwashita, S.; Imajo, S.; Oyama, Y.; Muto, T. Structure-based design, synthesis, and binding mode analysis of novel and potent chymase inhibitors. Bioorg. Med. Chem. Lett. 2018, 28, 188–192. [Google Scholar] [CrossRef]
- Ahmad, S.; Ferrario, C.M. Chymase inhibitors for the treatment of cardiac diseases: A patent review (2010–2018). Expert Opin. Ther. Pat. 2018, 28, 755–764. [Google Scholar] [CrossRef]
- Aoyama, Y.; Konoike, T.; Kanda, A.; Naya, N.; Nakajima, M. Total synthesis of human chymase inhibitor methyllinderone and structure-activity relationships of its derivatives. Bioorg. Med. Chem. Lett. 2001, 11, 1695–1697. [Google Scholar] [CrossRef]
- Arooj, M.; Kim, S.; Sakkiah, S.; Cao, G.P.; Lee, Y.; Lee, K.W. Molecular modeling study for inhibition mechanism of human chymase and its application in inhibitor design. PLoS ONE 2013, 8, 62740. [Google Scholar] [CrossRef] [PubMed]
- Arooj, M.; Sakkiah, S.; Kim, S.; Arulalapperumal, V.; Lee, K.W. A combination of receptor-based pharmacophore modeling & QM techniques for identification of human chymase inhibitors. PLoS ONE 2013, 8, 63030. [Google Scholar] [CrossRef]
- Li, L.; Zhou, X.; Li, N.; Sun, M.; Lv, J.; Xu, Z. Herbal drugs against cardiovascular disease: Traditional medicine and modern development. Drug Discov. Today 2015, 20, 1074–1086. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Chen, H. Modern bioinformatics meets traditional Chinese medicine. Brief. Bioinform. 2014, 15, 984–1003. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.; Marabotti, A.; Ramteke, P.W.; Facchiano, A. In silico approach to find chymase inhibitors among biogenic compounds. Future Med. Chem. 2016, 8, 841–851. [Google Scholar] [CrossRef] [PubMed]
- De Souza Silva, J.E.; Santos Souza, C.A.; da Silva, T.B.; Gomes, I.A.; Brito Gde, C.; de Souza Araújo, A.A.; de Lyra-Júnior, D.P.; da Silva, W.B.; da Silva, F.A. Use of herbal medicines by elderly patients: A systematic review. Arch. Gerontol. Geriatr. 2014, 59, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.; Marabotti, A.; Ramteke, P.W.; Facchiano, A. Interaction of human chymase with ginkgolides, terpene trilactones of Ginkgo biloba investigated by molecular docking simulations. Biochem. Biophys. Res. Commun. 2016, 473, 449–454. [Google Scholar] [CrossRef]
- Miraj, S.; Alesaeidi, S. A systematic review study of therapeutic effects of Matricaria recutita chamomile (Chamomile). Electron. Phys. 2016, 8, 3024–3031. [Google Scholar] [CrossRef]
- Orav, A.; Raal, A.; Arak, E. Content and composition of the essential oil of Chamomilla recutita (L.) Rauschert from some European countries. Nat. Prod. Res. 2010, 24, 48–55. [Google Scholar] [CrossRef]
- Nováková, L.; Vildová, A.; Mateus, J.P.; Gonçalves, T.; Solich, P. Development and application of UHPLC-MS/MS method for the determination of phenolic compounds in Chamomile flowers and Chamomile tea extracts. Talanta 2010, 82, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Haghi, G.; Hatami, A.; Safaei, A.; Mehran, M. Analysis of phenolic compounds in Matricaria chamomilla and its extracts by UPLC-UV. Res. Pharm. Sci. 2014, 9, 31–37. [Google Scholar] [PubMed]
- Gould, L.; Reddy, C.V.; Gomprecht, R.F. Cardiac effects of chamomile tea. J. Clin. Pharmacol. 1973, 13, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Hertog, M.G.; Feskens, E.J.; Hollman, P.C.; Katan, M.B.; Kromhout, D. Dietary antioxidant flavonoids and risk of coronary heart disease: The Zutphen Elderly Study. Lancet 1993, 342, 1007–1011. [Google Scholar] [CrossRef]
- Hertog, M.G.; Kromhout, D.; Aravanis, C.; Blackburn, H.; Buzina, R.; Fidanza, F.; Giampaoli, S.; Jansen, A.; Menotti, A.; Nedeljkovic, S.; et al. Flavonoid intake and long-term risk of coronary heart disease and cancer in the Seven Countries Study. Arch. Intern. Med. 1995, 155, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Knekt, P.; Jarvinen, R.; Reunanen, A.; Maatela, J. Flavonoid intake and coronary mortality in Finland: A cohort study. Br. Med. J. 1996, 312, 478–481. [Google Scholar] [CrossRef]
- Rose, P.W.; Prlic, A.; Bi, C.; Bluhm, W.F.; Christie, C.H.; Dutta, S.; Green, R.K.; Goodsell, D.S.; Westbrook, J.D.; Woo, J.; et al. The RCSB Protein Data Bank: Views of structural biology for basic and applied research and education. Nucl. Acids Res. 2015, 43, D345–D356. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucl. Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Repasky, M.P.; Shelley, M.; Friesner, R.A. Flexible ligand docking with Glide. Curr. Protoc. Bioinform. 2007, 8. [Google Scholar] [CrossRef]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef] [PubMed]
- Bernett, M.J.; Blaber, S.I.; Scarisbrick, I.A.; Dhanarajan, P.; Thompson, S.M.; Blaber, M. Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system. J. Biol. Chem. 2002, 277, 24562–24570. [Google Scholar] [CrossRef] [PubMed]
- McGrath, M.E.; Sprengeler, P.A.; Hirschbein, B.; Somoza, J.R.; Lehoux, I.; Janc, J.W.; Gjerstad, E.; Graupe, M.; Estiarte, A.; Venkataramani, C.; et al. Structure-guided design of peptide-based tryptase inhibitor. Biochemistry 2006, 45, 5964–5973. [Google Scholar] [CrossRef] [PubMed]
- Von Nussbaum, F.; Li, V.M. Neutrophil elastase inhibitors for the treatment of (cardio)pulmonary diseases: Into clinical testing with pre-adaptive pharmacophores. Bioorg. Med. Chem. Lett. 2015, 25, 4370–4381. [Google Scholar] [CrossRef]
- Shelley, J.C.; Cholleti, A.; Frye, L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comp. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Olsson, M.H.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef]
- Martyna, G.J.; Klein, M.L.; Tuckerman, M. Nosé–Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992, 97, 2635–2643. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Kräutler, V.; van Gunsteren, W.F.; Hünenberger, P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Humphreys, D.D.; Friesner, R.A.; Berne, B.J. A multiple-time-step molecular dynamics algorithm for macromolecules. J. Chem. Phys. 1994, 98, 6885–6892. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, J.; Ballevre, O.; Luo, H.; Zhang, W. Antihypertensive effects and mechanisms of chlorogenic acids. Hypertens. Res. 2012, 35, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Onakpoya, I.J.; Spencer, E.A.; Thompson, M.J.; Heneghan, C.J. The effect of chlorogenic acid on blood pressure: A systematic review and meta-analysis of randomized clinical trials. J. Hum. Hypertens. 2015, 29, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, M.; Goeters, S.; Watzer, B.; Krause, E.; Lohmann, K.; Bauer, R.; Hempel, B.; Imming, P. Chamazulene carboxylic acid and matricin: A natural profen and its natural prodrug, identified through similarity to synthetic drug substances. J. Nat. Prod. 2006, 69, 1041–1045. [Google Scholar] [CrossRef] [PubMed]

| AutoDock | Glide | Molegro Virtual Docker | ||||
|---|---|---|---|---|---|---|
| Chamomile Compounds (PubChem ID) | Best-Predicted Binding Energy (kcal/mol) | No. of Poses in the Cluster with Best-Predicted Energy | Glide Score | Glide Emodel | Moldock Score | Rerank Score |
| Alpha-Bisabolol (10586) | −8.28 | 85 | −4.915 | −32.667 | −109.629 | −92.408 |
| Alpha-Farnesene (5281516) | −7.57 | 72 | −1.071 | −26.181 | −120.466 | −97.4505 |
| Alpha-Pinene (6654) | −5.35 | 100 | −4.194 | −18.417 | −49.1045 | −45.0675 |
| Bisabolol (1549992) | −8.27 | 85 | −4.800 | −33.463 | −110.496 | −93.6185 |
| Caffeic Acid (689043) | −6.62 | 76 | −6.192 | −60.275 | −115.909 | −98.1313 |
| Chamazulene (10719) | −8.43 | 100 | −5.294 | −32.017 | −124.609 | −75.8608 |
| Chlorogenic Acid (1794427) | −8.57 | 28 | −7.037 | −79.332 | −138.276 | −10.5578 |
| Herniarin (10748) | −6.9 | 100 | −5.809 | −37.800 | −96.5088 | −77.6553 |
| Matricin (92265) | −9.12 | 85 | −5.040 | −37.582 | −139.206 | −48.1241 |
| Nobilin (11953937) | −7.9 | 95 | −4.835 | −41.410 | −129.981 | −37.3249 |
| Patuletin (5281678) | −7.44 | 36 | −5.969 | −53.612 | −120.525 | −53.6831 |
| Salicylic Acid (338) | −5.26 | 76 | −5.832 | −44.952 | −82.5318 | −66.5609 |
| Umbelliferone (5281426) | −6.72 | 82 | −5.868 | −37.074 | −90.6394 | −70.215 |
| OHH (self-docking with crystallographic inhibitor) | −14.84 | 90 | −7.101 | −78.669 | −209.86 | −105.051 |
| Methyllinderone (21953547) | −6.62 | 43 | 0.469 | −22.132 | −114.237 | −93.903 |
| Protease (with PDB ID) and Best Chamomile Compounds (with PubChem ID) | AutoDock | Glide | Molegro Virtual Docker | |||
|---|---|---|---|---|---|---|
| Best-Predicted Binding Energy (kcal/mol) | No. of Poses in the Cluster with Best-Predicted Energy | Glide Score | Glide Emodel | Moldock Score | Rerank Score | |
| Kallikrein (1LO6) | ||||||
| Chlorogenic Acid (1794427) | −7.94 | 32 | −7.111 | −69.452 | −116.975 | −108.579 |
| Matricin (92265) | −8.51 | 75 | −6.618 | −46.227 | −126.84 | −21.8832 |
| Chymase Crystallographic Inhibitor OHH | −10.03 | 21 | −7.439 | −68.327 | −174.073 | −3.06207 |
| Tryptase (2FPZ) | ||||||
| Chlorogenic Acid (1794427) | −6.24 | 41 | −6.985 | −59.835 | −105.543 | −84.9801 |
| Matricin (92265) | −6.74 | 76 | −5.863 | −37.119 | −117.111 | −109.58 |
| Chymase Crystallographic Inhibitor OHH | −9.77 | 29 | −7.441 | −76.763 | −157.822 | 6.93001 |
| Elastase (5ABW) | ||||||
| Chlorogenic Acid (1794427) | −6.25 | 20 | −6.301 | −54.401 | −108.314 | −95.7808 |
| Matricin (92265) | −6.34 | 7 | −5.476 | −37.090 | −110.134 | 90.7393 |
| Chymase Crystallographic Inhibitor OHH | −12.2 | 12 | −6.237 | −52.337 | −153.169 | −85.1654 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubey, A.; Dotolo, S.; Ramteke, P.W.; Facchiano, A.; Marabotti, A. Searching for Chymase Inhibitors among Chamomile Compounds Using a Computational-Based Approach. Biomolecules 2019, 9, 5. https://doi.org/10.3390/biom9010005
Dubey A, Dotolo S, Ramteke PW, Facchiano A, Marabotti A. Searching for Chymase Inhibitors among Chamomile Compounds Using a Computational-Based Approach. Biomolecules. 2019; 9(1):5. https://doi.org/10.3390/biom9010005
Chicago/Turabian StyleDubey, Amit, Serena Dotolo, Pramod W. Ramteke, Angelo Facchiano, and Anna Marabotti. 2019. "Searching for Chymase Inhibitors among Chamomile Compounds Using a Computational-Based Approach" Biomolecules 9, no. 1: 5. https://doi.org/10.3390/biom9010005
APA StyleDubey, A., Dotolo, S., Ramteke, P. W., Facchiano, A., & Marabotti, A. (2019). Searching for Chymase Inhibitors among Chamomile Compounds Using a Computational-Based Approach. Biomolecules, 9(1), 5. https://doi.org/10.3390/biom9010005






