Complementary LC-MS/MS-Based N-Glycan, N-Glycopeptide, and Intact N-Glycoprotein Profiling Reveals Unconventional Asn71-Glycosylation of Human Neutrophil Cathepsin G
Abstract
:1. Introduction
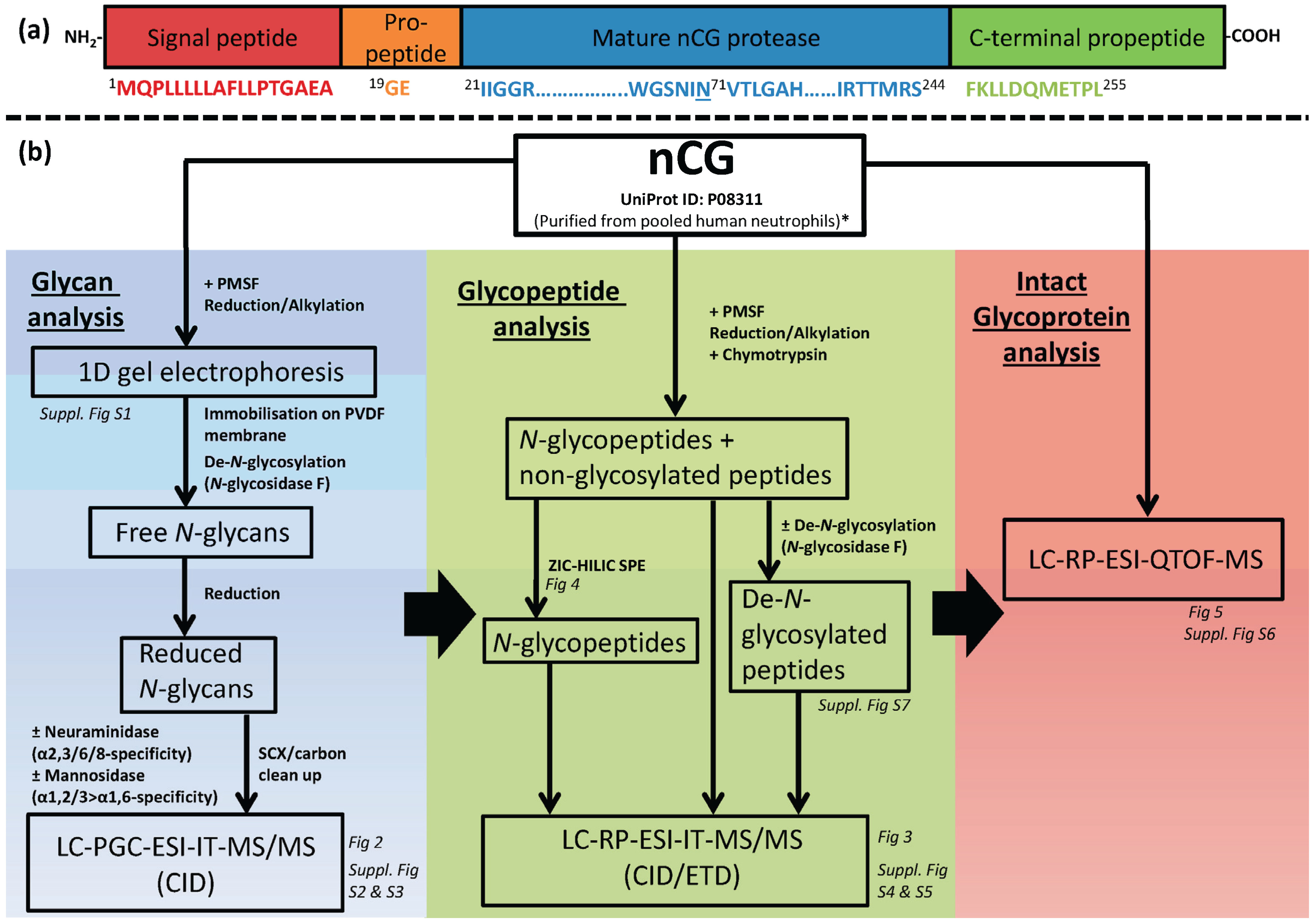
2. Results and Discussion
2.1. Design of Study—Three Analysis Levels were Used to Complete the nCG Glycoprofiling
2.2. N-Glycome Profiling Indicates Unconventional nCG N-Glycosylation
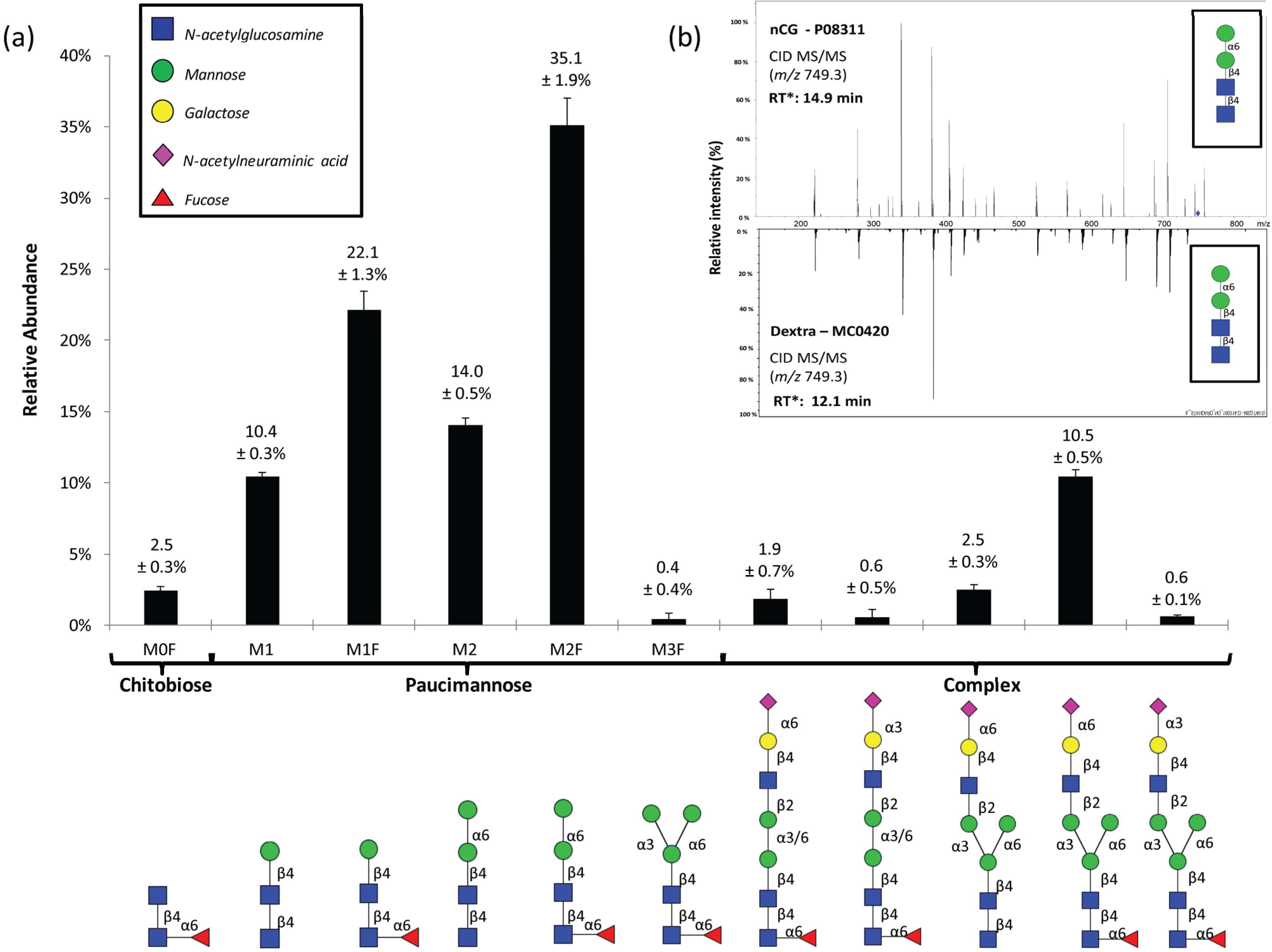
2.3. Site-Specific Asn71-Glycopeptide Profiling Uncovers Single GlcNAcβ and Fucα1,6GlcNAcβ on nCG and Reveals Significant Presence of Other Interfering N-Glycoproteins
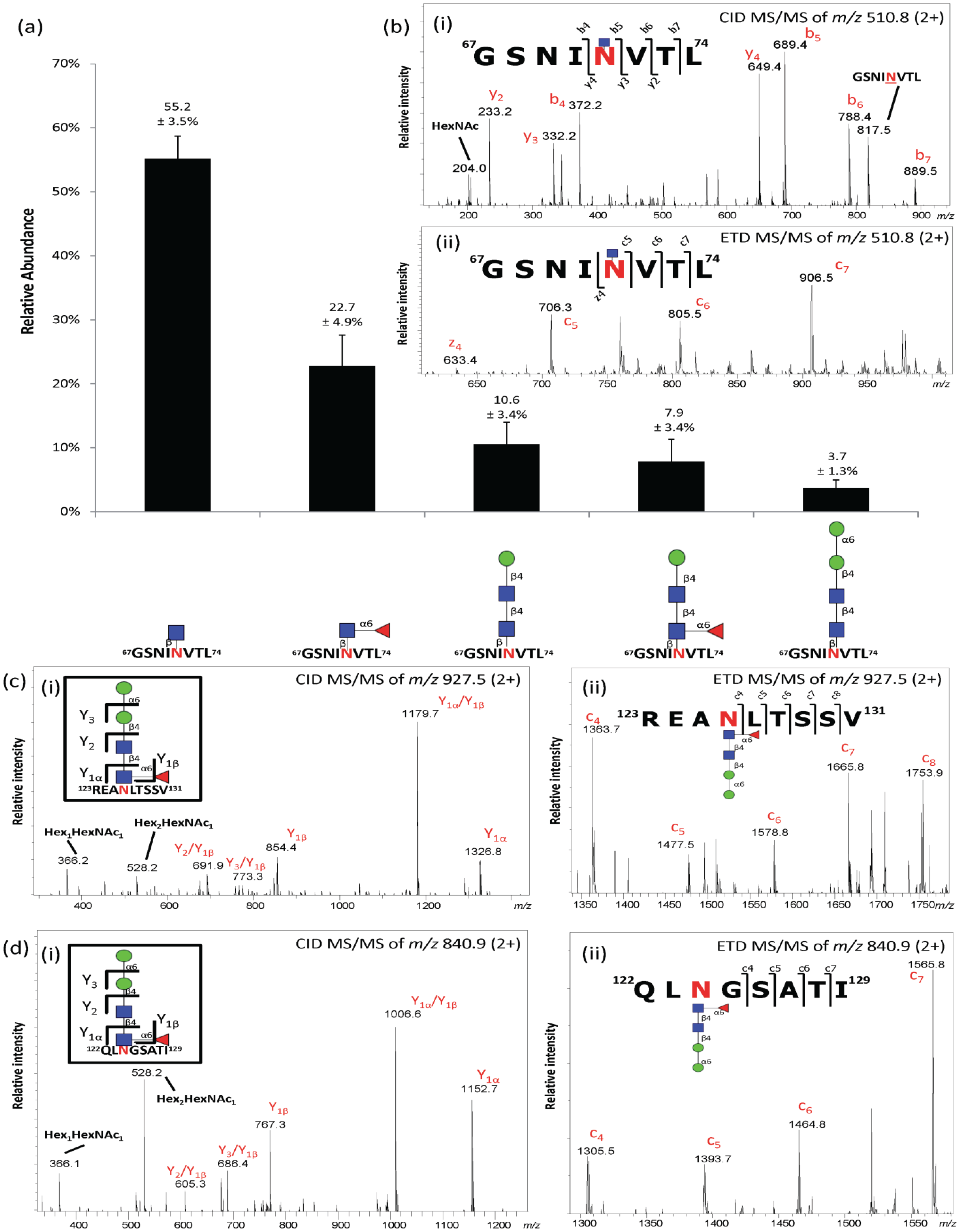
| N-glycan Structure | nCG | Azurocidin | NE | ||
|---|---|---|---|---|---|
| Asn71 | Asn126 | Asn171 | Asn124 | Asn173 | |
| GlcNAcβ | xxxx | ||||
| Fucα1,6GlcNAcβ | xxxx | ||||
| Manβ1,4GlcNAcβ1,4GlcNAcβ (M1) | xxx | ||||
| Manβ1,4GlcNAcβ1,4(Fucα1,6) GlcNAcβ (M1F) | xxx | xx | |||
| Manα1,6Manβ1,4GlcNAcβ1,4 GlcNAcβ (M2) | xxx | ||||
| Manα1,6Manβ1,4GlcNAcβ1,4 (Fucα1,6)GlcNAcβ (M2F) | x | xxxx | xxxx | xxxx | xxxx |
| Trimannosyl-chitobiose core monoantennary core fucosylated α2,6-monosialylated | x | ||||
| Trimannosyl-chitobiose core monoantennary core fucosylated α2,3-monosialylated | x | ||||
| Trimannosyl-chitobiose core monoantennary α2,6-monosialylated | x | ||||
| Bimannosyl-chitobiose core monoantennary core fucosylated α2,6-monosialylated | x | ||||
| Bimannosyl-chitobiose core monoantennary core fucosylated α2,3-monosialylated | x | ||||
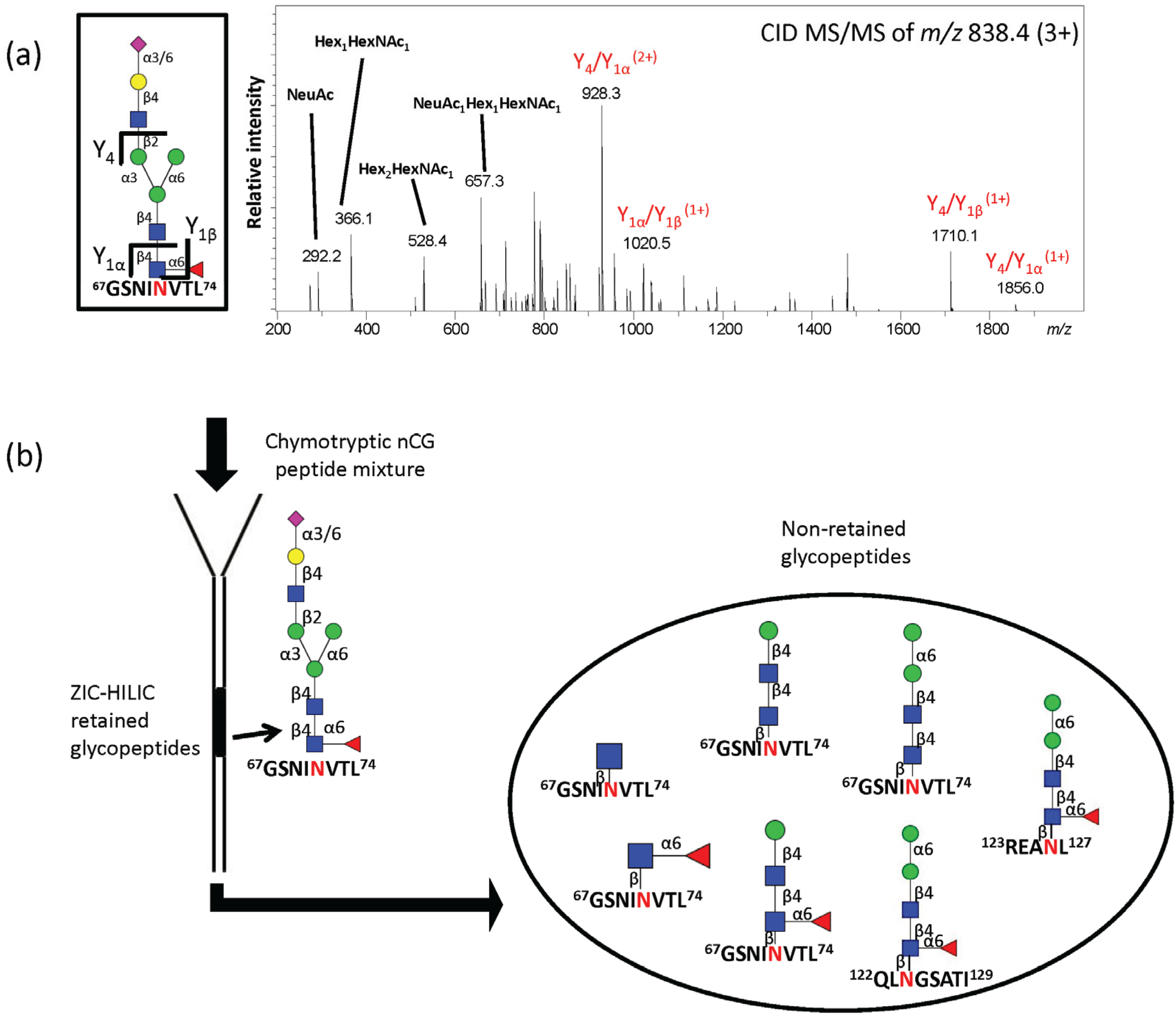
2.4. ZIC-HILIC SPE Enrichment of nCG N-Glycopeptides Favors Complex Glycoforms
2.5. Intact nCG Profiling Maps the Asn71-Glycosylation and Other PTMs
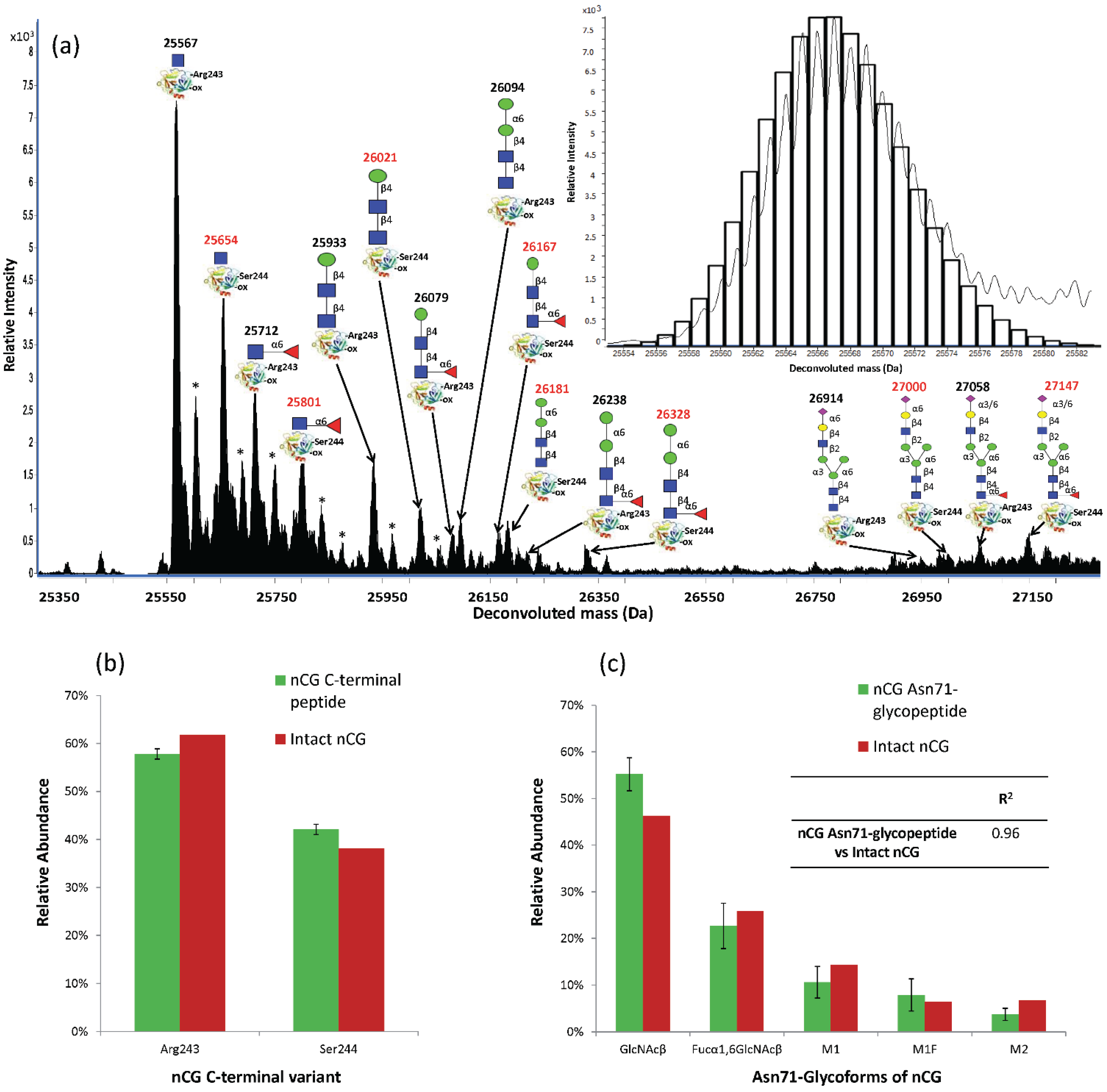
2.6. Establishing Asn71 Occupancy Level and N-Glycosidase F-Resistant Glycoforms of nCG
2.7. The Proximal, but Short, Asn71-Glycans are not Obstructing the Active Site of nCG
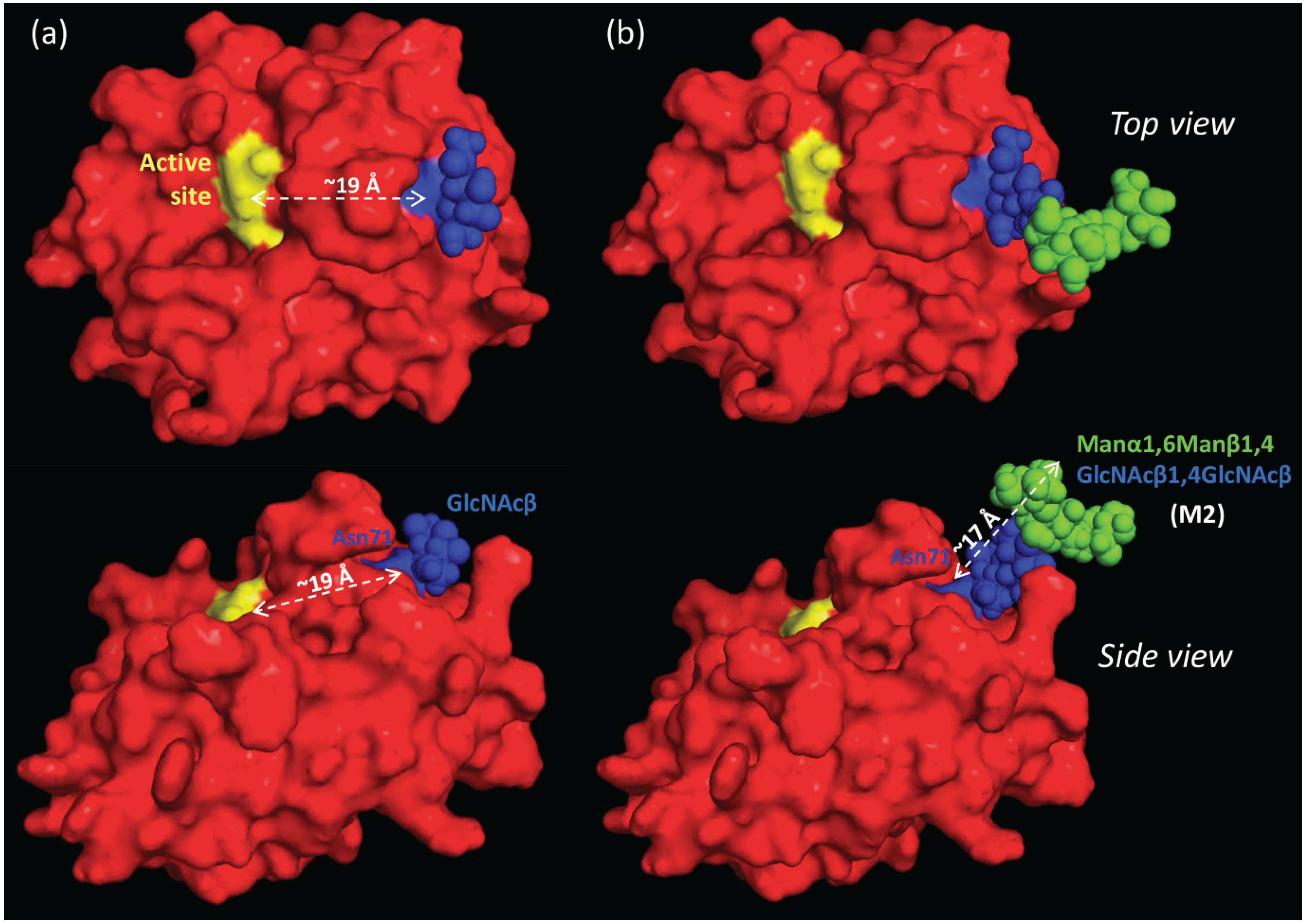
2.8. Spatial Considerations of nCG Structure Advance our Understanding of the Unconventional Asn71-Glycosylation
2.9. Subcellular-Specific N-Glycosylation of nCG in Human Neutrophils
3. Experimental Section
3.1. Origin and Initial Handling of nCG
3.2. N-Glycan Release and Handling
3.3. Exoglycosidase Treatment of Released N-Glycans
3.4. PGC-LC-ESI-MS/MS-Based N-Glycome Profiling
3.5. In-Solution Glycopeptide Generation, Enrichment, and Deglycosylation
3.6. LC-MS/MS-Based N-Glycopeptide Analysis
3.7. Intact nCG Profiling
3.8. Profiling nCG N-Glycans, N-Glycopeptides, and Intact Glycoprotein
3.9. Glycoprotein Modeling
3.10. Statistics
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Garwicz, D.; Lennartsson, A.; Jacobsen, S.E.; Gullberg, U.; Lindmark, A. Biosynthetic profiles of neutrophil serine proteases in a human bone marrow-derived cellular myeloid differentiation model. Haematologica 2005, 90, 38–44. [Google Scholar] [PubMed]
- Korkmaz, B.; Moreau, T.; Gauthier, F. Neutrophil elastase, proteinase 3 and cathepsin G: Physicochemical properties, activity and physiopathological functions. Biochimie 2008, 90, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.T. Neutrophil serine proteases: Specific regulators of inflammation. Nat. Rev. Immunol. 2006, 6, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Sambrano, G.R.; Huang, W.; Faruqi, T.; Mahrus, S.; Craik, C.; Coughlin, S.R. Cathepsin G activates protease-activated receptor-4 in human platelets. J. Biol. Chem. 2000, 275, 6819–6823. [Google Scholar] [CrossRef] [PubMed]
- Guyot, N.; Wartelle, J.; Malleret, L.; Todorov, A.A.; Devouassoux, G.; Pacheco, Y.; Jenne, D.E.; Belaaouaj, A. Unopposed cathepsin G, neutrophil elastase, and proteinase 3 cause severe lung damage and emphysema. Am. J. Pathol. 2014, 184, 2197–2210. [Google Scholar] [CrossRef] [PubMed]
- Garwicz, D.; Lindmark, A.; Persson, A.M.; Gullberg, U. On the role of the proform-conformation for processing and intracellular sorting of human cathepsin G. Blood 1998, 92, 1415–1422. [Google Scholar] [PubMed]
- Salvesen, G.; Enghild, J.J. An unusual specificity in the activation of neutrophil serine proteinase zymogens. Biochemistry 1990, 29, 5304–5308. [Google Scholar] [CrossRef] [PubMed]
- Hof, P.; Mayr, I.; Huber, R.; Korzus, E.; Potempa, J.; Travis, J.; Powers, J.C.; Bode, W. The 1.8 A crystal structure of human cathepsin G in complex with Suc-Val-Pro-PheP-(OPh)2: A Janus-faced proteinase with two opposite specificities. EMBO J. 1996, 15, 5481–5491. [Google Scholar] [PubMed]
- Korkmaz, B.; Horwitz, M.S.; Jenne, D.E.; Gauthier, F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol. Rev. 2010, 62, 726–759. [Google Scholar] [CrossRef] [PubMed]
- Burster, T.; Macmillan, H.; Hou, T.; Boehm, B.O.; Mellins, E.D. Cathepsin G: Roles in antigen presentation and beyond. Mol. Immunol. 2010, 47, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Salvesen, G.; Farley, D.; Shuman, J.; Przybyla, A.; Reilly, C.; Travis, J. Molecular cloning of human cathepsin G: Structural similarity to mast cell and cytotoxic T lymphocyte proteinases. Biochemistry 1987, 26, 2289–2293. [Google Scholar] [CrossRef] [PubMed]
- Watorek, W.; Halbeek, H.; Travis, J. The isoforms of human neutrophil elastase and cathepsin G differ in their carbohydrate side chain structures. Biol. Chem. Hoppe-Seyler 1993, 374, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Garwicz, D.; Lindmark, A.; Gullberg, U. Human cathepsin G lacking functional glycosylation site is proteolytically processed and targeted for storage in granules after transfection to the rat basophilic/mast cell line RBL or the murine myeloid cell line 32D. J. Biol. Chem. 1995, 270, 28413–28418. [Google Scholar] [PubMed]
- Venkatakrishnan, V.; Thaysen-Andersen, M.; Chen, S.C.; Nevalainen, H.; Packer, N.H. Cystic fibrosis and bacterial colonization define the sputum N-glycosylation phenotype. Glycobiology 2015, 25, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Thaysen-Andersen, M.; Venkatakrishnan, V.; Loke, I.; Laurini, C.; Diestel, S.; Parker, B.L.; Packer, N.H. Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J. Biol. Chem. 2015, 290, 8789–8802. [Google Scholar] [CrossRef] [PubMed]
- Rorvig, S.; Ostergaard, O.; Heegaard, N.H.; Borregaard, N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: Correlation with transcriptome profiling of neutrophil precursors. J. Leukocyte Biol. 2013, 94, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Ruhaak, L.R.; Stroble, C.; Underwood, M.A.; Lebrilla, C.B. Detection of milk oligosaccharides in plasma of infants. Anal. Bioanal. Chem. 2014, 406, 5775–5784. [Google Scholar] [CrossRef] [PubMed]
- Thomsson, K.A.; Karlsson, N.G.; Hansson, G.C. Liquid chromatography-electrospray mass spectrometry as a tool for the analysis of sulfated oligosaccharides from mucin glycoproteins. J. Chromatogr. A 1999, 854, 131–139. [Google Scholar] [CrossRef]
- Stavenhagen, K.; Kolarich, D.; Wuhrer, M. Clinical glycomics employing graphitized carbon liquid chromatography-mass spectrometry. Chromatographia 2015, 78, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.H.; Karlsson, N.G.; Kolarich, D.; Packer, N.H. Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 2012, 7, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Pabst, M.; Bondili, J.S.; Stadlmann, J.; Mach, L.; Altmann, F. Mass + retention time = structure: A strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal. Chem. 2007, 79, 5051–5057. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.Y.; Thaysen-Andersen, M.; Baker, M.S.; Packer, N.H.; Hancock, W.S.; Fanayan, S. Comprehensive N-glycome profiling of cultured human epithelial breast cells identifies unique secretome N-glycosylation signatures enabling tumorigenic subtype classification. J. Proteome Res. 2014, 13, 4783–4795. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, G.; Larsen, M.R.; Packer, N.H.; Thaysen-Andersen, M. Structural analysis of glycoprotein sialylation—Part II: LC-MS based detection. RSC Adv. 2013, 3. [Google Scholar] [CrossRef]
- Olczak, M.; Watorek, W. Structural analysis of N-glycans from human neutrophil azurocidin. Biochem. Biophys. Res. Commun. 2002, 293, 213–219. [Google Scholar] [CrossRef]
- Sumer-Bayraktar, Z.; Nguyen-Khuong, T.; Jayo, R.; Chen, D.D.; Ali, S.; Packer, N.H.; Thaysen-Andersen, M. Micro- and macroheterogeneity of N-glycosylation yields size and charge isoforms of human sex hormone binding globulin circulating in serum. Proteomics 2012, 12, 3315–3327. [Google Scholar] [CrossRef] [PubMed]
- Kolarich, D.; Jensen, P.H.; Altmann, F.; Packer, N.H. Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 2012, 7, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Hogan, J.M.; Pitteri, S.J.; Chrisman, P.A.; McLuckey, S.A. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J. Proteome Res. 2005, 4, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Wuhrer, M.; Catalina, M.I.; Deelder, A.M.; Hokke, C.H. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 849, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Peterman, S.M.; Mulholland, J.J. A novel approach for identification and characterization of glycoproteins using a hybrid linear ion trap/FT-ICR mass spectrometer. J. Am. Soc. Mass Spectrom. 2006, 17, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Stavenhagen, K.; Hinneburg, H.; Thaysen-Andersen, M.; Hartmann, L.; Silva, D.V.; Fuchser, J.; Kaspar, S.; Rapp, E.; Seeberger, P.H.; Kolarich, D. Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: An evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom. 2013, 48, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Hägglund, P.; Bunkenborg, J.; Elortza, F.; Jensen, O.N.; Roepstorff, P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 2004, 3, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Stimson, E.; Hope, J.; Chong, A.; Burlingame, A.L. Site-specific characterization of the N-linked glycans of murine prion protein by high-performance liquid chromatography/electrospray mass spectrometry and exoglycosidase digestions. Biochemistry 1999, 38, 4885–4895. [Google Scholar] [CrossRef] [PubMed]
- Leymarie, N.; Griffin, P.J.; Jonscher, K.; Kolarich, D.; Orlando, R.; McComb, M.; Zaia, J.; Aguilan, J.; Alley, W.R.; Altmann, F.; et al. Interlaboratory study on differential analysis of protein glycosylation by mass spectrometry: The ABRF glycoprotein research multi-institutional study 2012. Mol. Cell. Proteom. 2013, 12, 2935–2951. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Costello, C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjate J. 1988, 5, 397–409. [Google Scholar] [CrossRef]
- Everest-Dass, A.V.; Abrahams, J.L.; Kolarich, D.; Packer, N.H.; Campbell, M.P. Structural feature ions for distinguishing N- and O-linked glycan isomers by LC-ESI-IT MS/MS. J. Am. Soc. Mass Spectrom. 2013, 24, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Harvey, D.J. Fragmentation of negative ions from carbohydrates: Part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 2005, 16, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Winchester, B. Lysosomal metabolism of glycoproteins. Glycobiology 2005, 15, 1R–15R. [Google Scholar] [CrossRef] [PubMed]
- Balog, C.I.; Stavenhagen, K.; Fung, W.L.; Koeleman, C.A.; McDonnell, L.A.; Verhoeven, A.; Mesker, W.E.; Tollenaar, R.A.; Deelder, A.M.; Wuhrer, M. N-glycosylation of colorectal cancer tissues: A liquid chromatography and mass spectrometry-based investigation. Mol. Cell. Proteomics 2012, 11, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Dahmen, A.C.; Fergen, M.T.; Laurini, C.; Schmitz, B.; Loke, I.; Thaysen-Andersen, M.; Diestel, S. Paucimannosidic glycoepitopes are functionally involved in proliferation of neural progenitor cells in the subventricular zone. Glycobiology 2015, 25, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Hashii, N.; Kawasaki, N.; Itoh, S.; Nakajima, Y.; Kawanishi, T.; Yamaguchi, T. Alteration of N-glycosylation in the kidney in a mouse model of systemic lupus erythematosus: Relative quantification of N-glycans using an isotope-tagging method. Immunology 2009, 126, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Ravnsborg, T.; Houen, G.; Hojrup, P. The glycosylation of myeloperoxidase. Biochim. Biophys. Acta 2010, 1804, 2046–2053. [Google Scholar] [CrossRef] [PubMed]
- Zoega, M.; Ravnsborg, T.; Hojrup, P.; Houen, G.; Schou, C. Proteinase 3 carries small unusual carbohydrates and associates with alpha-defensins. J. Proteomics 2012, 75, 1472–1485. [Google Scholar] [CrossRef] [PubMed]
- Babu, P.; North, S.J.; Jang-Lee, J.; Chalabi, S.; Mackerness, K.; Stowell, S.R.; Cummings, R.D.; Rankin, S.; Dell, A.; Haslam, S.M. Structural characterisation of neutrophil glycans by ultra sensitive mass spectrometric glycomics methodology. Glycoconjugate J. 2009, 26, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.K. Requirements of cleavage of high mannose oligosaccharides in glycoproteins by peptide N-glycosidase F. J. Biol. Chem. 1986, 261, 172–177. [Google Scholar] [PubMed]
- Medzihradszky, K.F.; Kaasik, K.; Chalkley, R.J. Tissue-specific glycosylation at the glycopeptide level. Mol. Cell. Proteomics 2015, 14, 2103–2110. [Google Scholar] [CrossRef] [PubMed]
- Trinidad, J.C.; Schoepfer, R.; Burlingame, A.L.; Medzihradszky, K.F. N- and O-glycosylation in the murine synaptosome. Mol. Cell. Proteomics 2013, 12, 3474–3488. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Udeshi, N.D.; Slawson, C.; Compton, P.D.; Sakabe, K.; Cheung, W.D.; Shabanowitz, J.; Hunt, D.F.; Hart, G.W. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Jahren, N.; Stone, M.D.; Udeshi, N.D.; Markowski, T.W.; Witthuhn, B.A.; Shabanowitz, J.; Hunt, D.F.; Olszewski, N.E. Identification and origin of N-linked β-d-N-acetylglucosamine monosaccharide modifications on arabidopsis proteins. Plant Physiol. 2013, 161, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Klement, E.; Lipinszki, Z.; Kupihar, Z.; Udvardy, A.; Medzihradszky, K.F. Enrichment of O-GlcNAc modified proteins by the periodate oxidation-hydrazide resin capture approach. J. Proteome Res. 2010, 9, 2200–2206. [Google Scholar] [CrossRef] [PubMed]
- Campanelli, D.; Detmers, P.A.; Nathan, C.F.; Gabay, J.E. Azurocidin and a homologous serine protease from neutrophils. Differential antimicrobial and proteolytic properties. J. Clin. Invest. 1990, 85, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Lominadze, G.; Powell, D.W.; Luerman, G.C.; Link, A.J.; Ward, R.A.; McLeish, K.R. Proteomic analysis of human neutrophil granules. Mol. Cell. Proteomics 2005, 4, 1503–1521. [Google Scholar] [CrossRef] [PubMed]
- Thaysen-Andersen, M.; Mysling, S.; Hojrup, P. Site-specific glycoprofiling of N-linked glycopeptides using MALDI-TOF MS: Strong correlation between signal strength and glycoform quantities. Anal. Chem. 2009, 81, 3933–3943. [Google Scholar] [CrossRef] [PubMed]
- Mysling, S.; Palmisano, G.; Hojrup, P.; Thaysen-Andersen, M. Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 2010, 82, 5598–5609. [Google Scholar] [CrossRef] [PubMed]
- Lindmark, A.; Gullberg, U.; Lindgren, G.; Persson, A.M.; Nilsson, E.; Olsson, I. Carboxyl-terminal prodomain-deleted human leukocyte elastase and cathepsin G are efficiently targeted to granules and enzymatically activated in the rat basophilic/mast cell line RBL. J. Biol. Chem. 1995, 270, 12912–12918. [Google Scholar] [CrossRef]
- Shao, B.; Belaaouaj, A.; Verlinde, C.L.; Fu, X.; Heinecke, J.W. Methionine sulfoxide and proteolytic cleavage contribute to the inactivation of cathepsin G by hypochlorous acid: An oxidative mechanism for regulation of serine proteinases by myeloperoxidase. J. Biol. Chem. 2005, 280, 29311–29321. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cournoyer, J.J.; Lin, C.; O’Connor, P.B. Use of 18 O labels to monitor deamidation during protein and peptide sample processing. J. Am. Soc. Mass Spectrom. 2008, 19, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Thaysen-Andersen, M.; Packer, N.H. Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology 2012, 22, 1440–1452. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.Y.; Lin, C.H.; Fanayan, S.; Packer, N.H.; Thaysen-Andersen, M. Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front. Immunol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Borregaard, N.; Cowland, J.B. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood 1997, 89, 3503–3521. [Google Scholar] [PubMed]
- Borregaard, N.; Sorensen, O.E.; Theilgaard-Monch, K. Neutrophil granules: A library of innate immunity proteins. Trends Immunol. 2007, 28, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Ceroni, A.; Maass, K.; Geyer, H.; Geyer, R.; Dell, A.; Haslam, S.M. Glycoworkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 2008, 7, 1650–1659. [Google Scholar] [CrossRef] [PubMed]
- Peri, S.; Steen, H.; Pandey, A. GPMAW—A software tool for analyzing proteins and peptides. Trends Biochem. Sci. 2001, 26, 687–689. [Google Scholar] [CrossRef]
- Leymarie, N.; Zaia, J. Effective use of mass spectrometry for glycan and glycopeptide structural analysis. Anal. Chem. 2012, 84, 3040–3048. [Google Scholar] [CrossRef] [PubMed]
- Bohne-Lang, A.; von der Lieth, C.W. GlyProt: In silico glycosylation of proteins. Nucleic Acids Res. 2005, 33, W214–W219. [Google Scholar] [CrossRef] [PubMed]
- Hubber, S.J.; Thornton, J.M. NACCESS Computer Program, Department of Biochemistry and Molecular Biology; University College London: London, UK, 1993. [Google Scholar]
- Lee, B.; Richards, F.M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971, 55, 379–400. [Google Scholar] [CrossRef]
- Thaysen-Andersen, M.; Packer, N.H. Advances in LC-MS/MS-based glycoproteomics: Getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim. Biophys. Acta 2014, 1844, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Kolli, V.; Schumacher, K.N.; Dodds, E.D. Engaging challenges in glycoproteomics: Recent advances in MS-based glycopeptide analysis. Bioanalysis 2015, 7, 113–131. [Google Scholar] [CrossRef] [PubMed]
- Rosati, S.; Yang, Y.; Barendregt, A.; Heck, A.J. Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat. Protoc. 2014, 9, 967–976. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loke, I.; Packer, N.H.; Thaysen-Andersen, M. Complementary LC-MS/MS-Based N-Glycan, N-Glycopeptide, and Intact N-Glycoprotein Profiling Reveals Unconventional Asn71-Glycosylation of Human Neutrophil Cathepsin G. Biomolecules 2015, 5, 1832-1854. https://doi.org/10.3390/biom5031832
Loke I, Packer NH, Thaysen-Andersen M. Complementary LC-MS/MS-Based N-Glycan, N-Glycopeptide, and Intact N-Glycoprotein Profiling Reveals Unconventional Asn71-Glycosylation of Human Neutrophil Cathepsin G. Biomolecules. 2015; 5(3):1832-1854. https://doi.org/10.3390/biom5031832
Chicago/Turabian StyleLoke, Ian, Nicolle H. Packer, and Morten Thaysen-Andersen. 2015. "Complementary LC-MS/MS-Based N-Glycan, N-Glycopeptide, and Intact N-Glycoprotein Profiling Reveals Unconventional Asn71-Glycosylation of Human Neutrophil Cathepsin G" Biomolecules 5, no. 3: 1832-1854. https://doi.org/10.3390/biom5031832
APA StyleLoke, I., Packer, N. H., & Thaysen-Andersen, M. (2015). Complementary LC-MS/MS-Based N-Glycan, N-Glycopeptide, and Intact N-Glycoprotein Profiling Reveals Unconventional Asn71-Glycosylation of Human Neutrophil Cathepsin G. Biomolecules, 5(3), 1832-1854. https://doi.org/10.3390/biom5031832






