Structural Biology of Bacterial RNA Polymerase
Abstract
:1. Early Research on the Structure of Bacterial RNA Polymerase
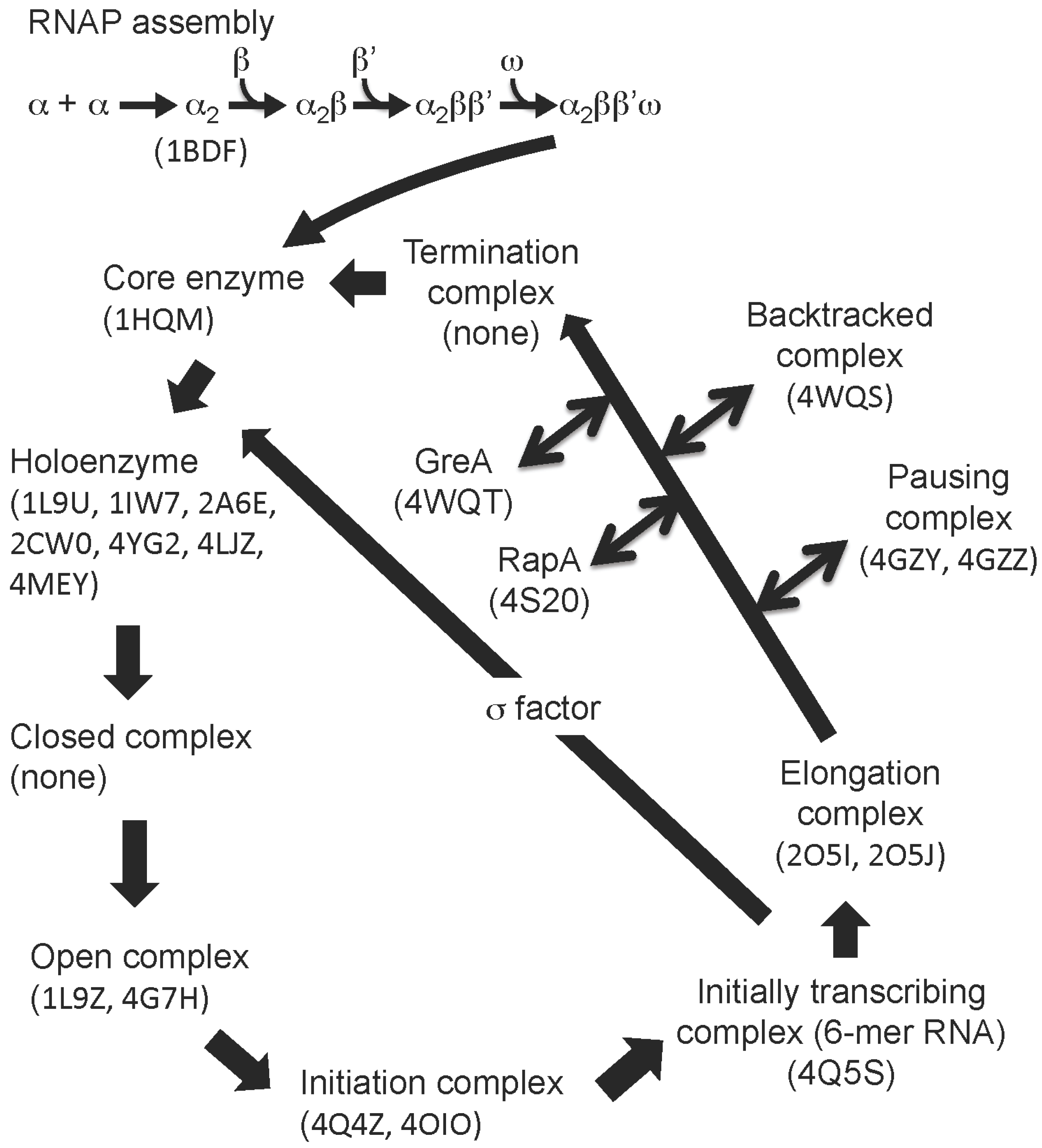
| Structure | PDB code | Reference | Source |
|---|---|---|---|
| Subunit and domain | |||
| α subunit NTD | 1BDFX | [7] | A |
| α subunit NTD | 4NOIX | none | F |
| α subunit CTD | 1COON, 3K4GX | [5,8] | A |
| α subunit CTD | 1DOQN | [9] | B |
| α subunit CTD | 2MAXN | [10] | E |
| β subunit 2/i4 domains | 3LTIX | [11] | A |
| β subunit flap domain | 2LY7N | none | C |
| β subunit 1/2 domains | 4KBJX | [12] | I |
| β' subunit i2 domain | 2AUJX | [13] | B |
| β' subunit i6 domain | 2AUKX, 4IQZX | [13] | A |
| σ region 1.1 | 2K6XN | [14] | G, a |
| σ70 domain2 | 1SIGX | [15] | A, a |
| σA domains2 and 3 | 1KU2X | [16] | B, a |
| σA domain4 | 1KU3X | [16] | B, a |
| σA domain4–DNA (−35 element) | 1KU7X | [16] | B, a |
| σA domain4 | 1TTYN | [17] | G, a |
| σA domain2–DNA (−10 element) | 3UGOX, 3UGPX | [18] | B, a |
| σA domain4–αCTD–DNA | 3N97X | none | B, a, A |
| σNRpoN–DNA (−24 element) | 2O8KN, 2O9LN | [19] | D, d |
| σN core binding domain | 2K9MN | [20] | D, d |
| σNRpoN domain | 2AHQN | [21] | D, d |
| σEdomain4–DNA (−35 element) | 2H27X | [22] | A, c |
| σC domain2 | 2O7GX | [23] | I, c |
| σC domain4 | 2O8XX | [23] | I, c |
| σD domain4 | 3VFZX | [24] | I, c |
| δ subunit NTD | 2KRCN, 4NC7X, 4NC8X, 2M4KN, 2KRCN | [25,26,27] | C |
| ε subunit | 4NJCX | [28] | C |
| RNAP | |||
| Core enzyme | 1HQMX | [29,30] | B |
| Core enzyme (Δω subunit) | 2GHOX | [31] | B |
| Holoenzyme | 1L9UX, 1IW7X, 2A6EX, 2CW0X | [32,33,34] | B |
| Holoenzyme | 4YG2X, 4LJZX, 4MEYX | [35,36,37] | A |
| Holoenzyme–DNA (−41 ~ −7) | 1L9ZX | [38] | B |
| Holoenzyme–DNA (−12 ~ +12) | 4G7HX, 4G7OX | [39] | B |
| de novo initiation complex | 4Q4ZX, 4OIOX | [40,41] | B |
| Initially transcribing complex | 4Q5SX | [40] | B |
| Elongation complex | 2O5IX, 2O5JX | [42,43] | B |
| Paused elongation complex | 4GZYX, 4GZZX | [44] | B |
| Backtracked elongation complex | 4WQSX | [45] | B |
2. An Explosion of Structural Information on Bacterial RNA Polymerase
3. Structural Basis of Transcription Elongation
4. Promoter-Dependent Transcription: How RNA Polymerase Recognizes Promoter DNA Sequences and Initiates Transcription
5. Structures of Alternative σ Factors
| Structure | PDB | References | Source |
|---|---|---|---|
| σ/anti-σ complex | |||
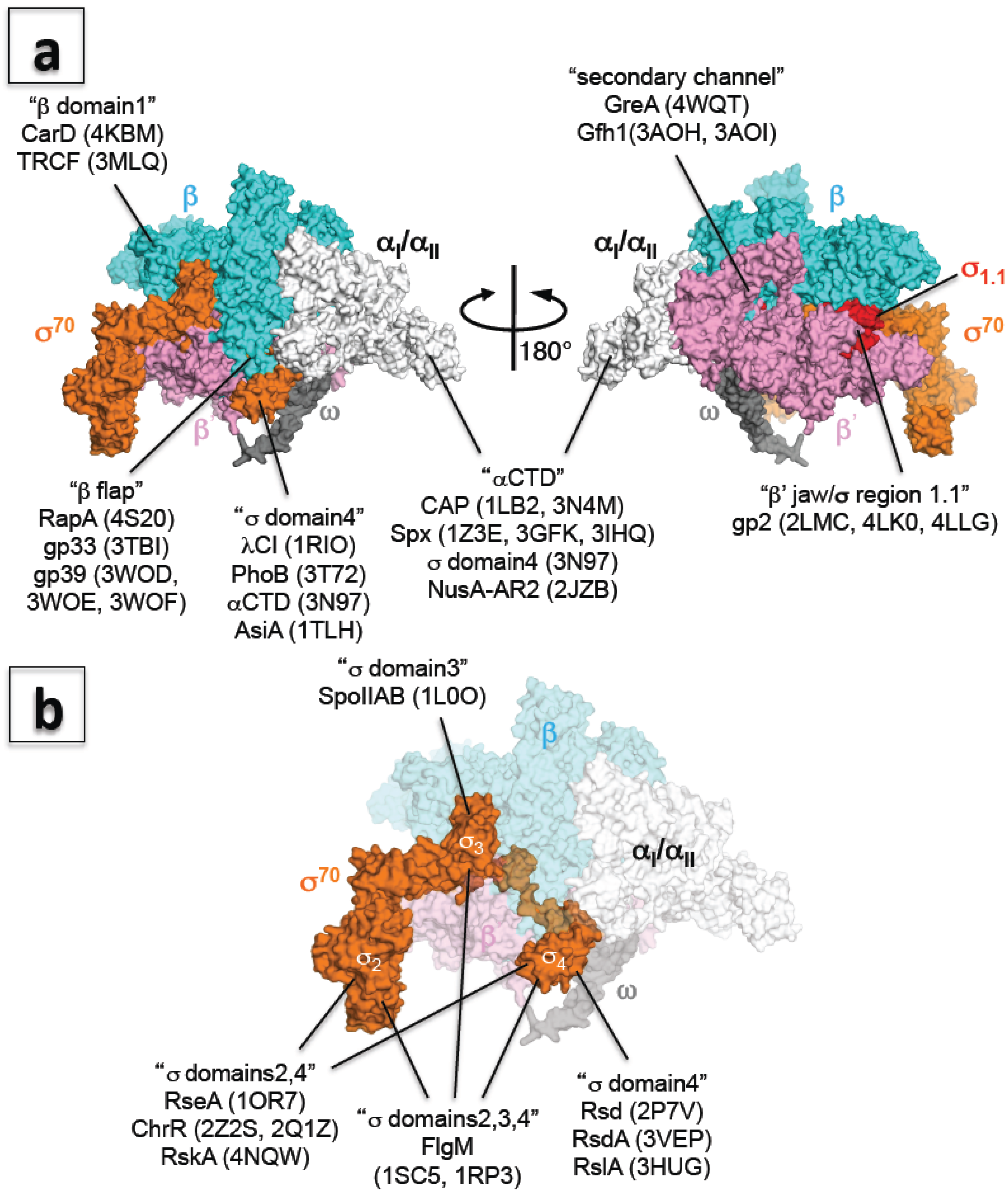
| σF domains2, 3, 4/FlgM | 1SC5X, 1RP3X | [54] | D, b |
| σE domains2, 4/ChrR | 2Z2SX, 2Q1ZX | [55] | J, c |
| σE domains2, 4/RseA | 1OR7X | [56] | A, c |
| σK domains2, 4/RskA | 4NQWX | [57] | I, c |
| σF domain3/SpoIIAB | 1L0OX | [58] | C, b |
| σ70 domain4/Rsd | 2P7VX | [59] | A, a |
| σD domain4/RsdA | 3VEPX | [24] | I, c |
| σL domain4/RslA | 3HUGX | [60] | I, c |
| Transcription factor complex | |||
| αCTD–CAP–DNA | 1LB2X, 3N4MX | [61] | A |
| αCTD–Spx | 1Z3EX, 3GFKX, 3IHQX | [62,63,64] | C |
| αCTD–NusA (AR2) | 2JZBN | none | H, A |
| β subunit 1/2 domains–CarD | 4KBMX | [12] | I |
| β subunit 1 domain–TRCF | 3MLQX | [65] | B |
| σA domain4–λcI–DNA | 1RIOX | [66] | B |
| σ70 domain4–PhoB–β flap–DNA | 3T72X | [67] | A |
| Holoenzyme–CAP–DNA | 3IYDC | [68] | A |
| Core enzyme–GreA/Gfh1 | 4WQTX | [45] | B |
| Elongation complex–Gfh1 | 3AOHX, 3AOIX | [69] | B |
| Elongation complex –RapA | 4S20X | [53] | A |
| Phage factor complex | |||
| Holoenzyme–gp2 | 4LK0X, 4LLGX | [36] | A |
| β' subunit jaw domain–gp2 | 2LMCN | [70] | A |
| β subunit flap domain–gp33 | 3TBIX | [71] | A |
| Holoenzyme–gp39 | 3WODX | [72] | B |
| β subunit flap domain –gp39 | 3WOFX, 3WOEX | [72] | B |
| σ70 domain4–AsiA | 1TLHN | [17] | A, a |
| σA domain4–gp67 | 4G8XX | [73] | K, a |
6. A New Era of Structural Study of Bacterial Transcription Using E. coli RNA Polymerase
7. Transcription Regulation: How RNA Polymerase Communicates with Transcription Factors
8. How Small Molecules Inhibit RNA Transcription
| Structure | PDB | Reference | Source |
|---|---|---|---|
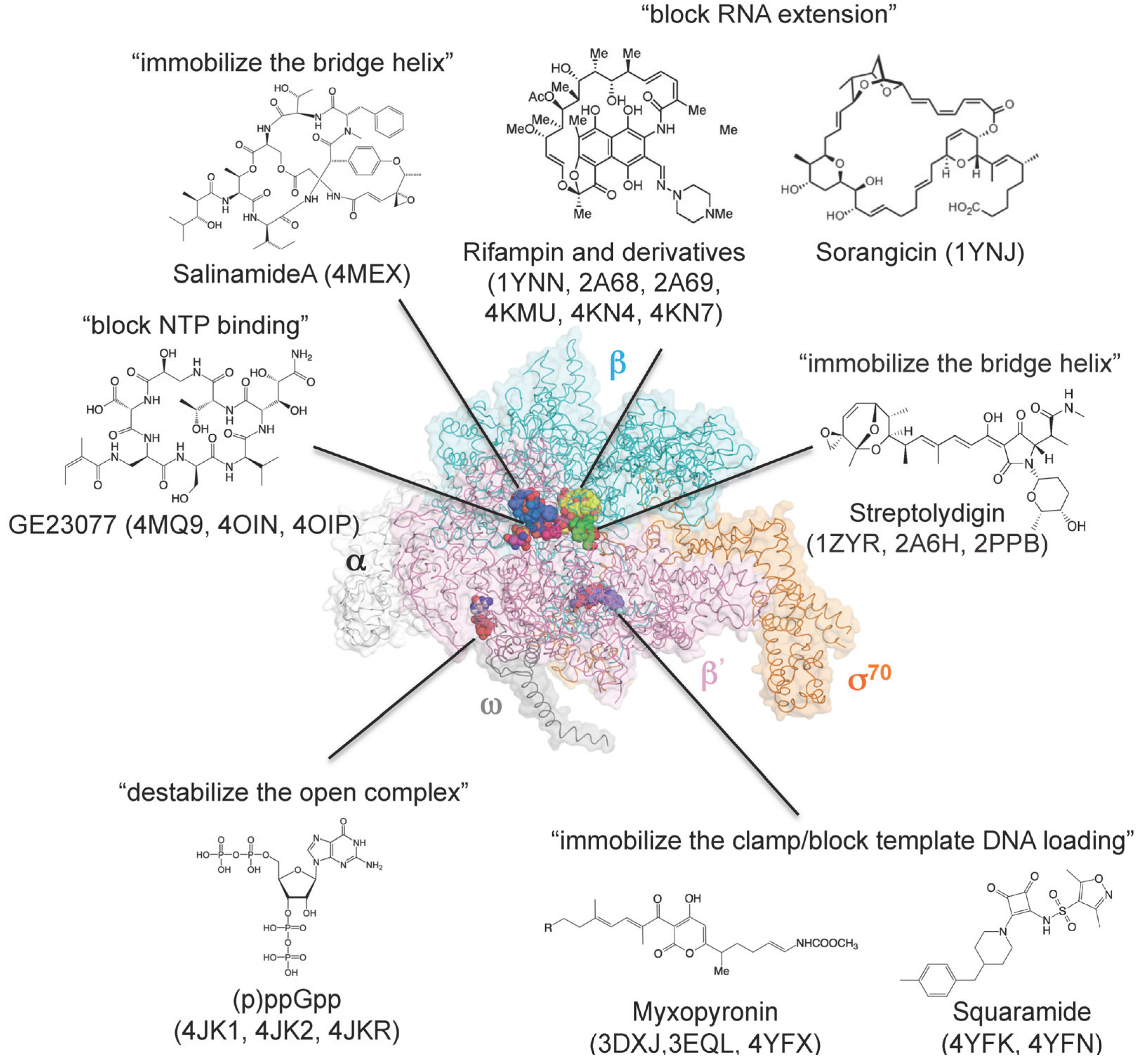
| Core enzyme–rifampin | 1YNN | [80] | B |
| Holoenzyme–rifampin | 4KMU | [81] | A |
| Holoenzyme–rifampin derivatives | 2A68, 2A69 | [83] | B |
| Holoenzyme–rifampin derivatives | 4KN4, 4KN7 | [81] | A |
| Holoenzyme–ppGpp | 1SMY | [76] | B |
| Holoenzyme–ppGpp | 4JK1, 4JKR | [74,75] | A |
| Holoenzyme–pppGpp | 4JK2 | [74] | A |
| Core enzyme–sorangicin | 1YNJ | [82] | B |
| Holoenzyme–streptolydigin | 1ZYR, 2A6H | [34,84] | B |
| Elongation complex–streptolydigin | 2PPB | [42] | B |
| Holoenzyme–myxopyronin | 3DXJ, 3EQL | [85,86] | B |
| Holoenzyme–myxopyronin | 4YFX | [87] | A |
| Holoenzyme–GE23077 | 4MQ9 | [41] | B |
| Holoenzyme–DNA–GE23077 | 4OIN, 4OIP | [41] | B |
| Holoenzyme–DNA–GE23077/rifamycin SV | 4OIR | [41] | B |
| Holoenzyme–salinamide A | 4MEX | [37] | A |
| Holoenzyme–squaramide | 4YFK, 4YFN | [87] | A |
Acknowledgements
Conflicts of Interest
References
- Murakami, K.S.; Darst, S.A. Bacterial RNA polymerases: The wholo story. Curr. Opin. Struct. Biol. 2003, 13, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.H.; Warner, B.A.; Murakami, K.S. RNA polymerase reaction in bacteria. In Encyclopedia of Biological Chemistry; Lane, W.J.L.D., Ed.; Academic Press: Waltham, MA, USA, 2013; pp. 167–172. [Google Scholar]
- Fuchs, E.; Zillig, W.; Hofschneider, P.H.; Preuss, A. Preparation and properties of RNA-polymerase particles. J. Mol. Biol. 1964, 10, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, S.; Suzuki, K.; Imahori, K. Crystallisation of DNA-dependent RNA polymerase from Thermus thermophilus HB 8. Nature 1976, 261, 725–726. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.H.; Negishi, T.; Shirakawa, M.; Yamazaki, T.; Fujita, N.; Ishihama, A.; Kyogoku, Y. Solution structure of the activator contact domain of the RNA polymerase alpha subunit. Science 1995, 270, 1495–1497. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, A. Role of the RNA polymerase alpha subunit in transcription activation. Mol. Microbiol. 1992, 6, 3283–3288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Darst, S.A. Structure of the Escherichia coli RNA polymerase alpha subunit amino-terminal domain. Science 1998, 281, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Lara-Gonzalez, S.; Birktoft, J.J.; Lawson, C.L. Structure of the Escherichia coli RNA polymerase alpha subunit C-terminal domain. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 806–812. [Google Scholar] [CrossRef]
- Wada, T.; Yamazaki, T.; Kyogoku, Y. The structure and the characteristic DNA binding property of the C-terminal domain of the RNA polymerase alpha subunit from Thermus thermophilus. J. Biol. Chem. 2000, 275, 16057–16063. [Google Scholar] [CrossRef] [PubMed]
- Borin, B.N.; Tang, W.; Krezel, A.M. Helicobacter pylori RNA polymerase alpha-subunit C-terminal domain shows features unique to varepsilon-proteobacteria and binds NikR/DNA complexes. Protein Sci. 2014, 23, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Opalka, N.; Brown, J.; Lane, W.J.; Twist, K.A.; Landick, R.; Asturias, F.J.; Darst, S.A. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLOS Biol. 2010, 8, e1000483. [Google Scholar] [CrossRef] [PubMed]
- Gulten, G.; Sacchettini, J.C. Structure of the MTB CarD/RNAP beta-lobes complex reveals the molecular basis of interaction and presents a distinct DNA-binding domain for MTB CarD. Structure 2013, 21, 1859–1869. [Google Scholar] [CrossRef] [PubMed]
- Chlenov, M.; Masuda, S.; Murakami, K.S.; Nikiforov, V.; Darst, S.A.; Mustaev, A. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase beta' subunit. J. Mol. Biol. 2005, 353, 138–154. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.C.; Shekhtman, A.; Dutta, K.; Pratt, M.R.; Cowburn, D.; Darst, S.; Muir, T.W. A full-length group 1 bacterial sigma factor adopts a compact structure incompatible with DNA binding. Chem. Biol. 2008, 15, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, A.; Severinova, E.; Darst, S.A. Crystal structure of a sigma70 subunit fragment from E. coli RNA polymerase. Cell 1996, 87, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Muzzin, O.; Chlenov, M.; Sun, J.L.; Olson, C.A.; Weinman, O.; Trester-Zedlitz, M.L.; Darst, S.A. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 2002, 9, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Lambert, L.J.; Wei, Y.; Schirf, V.; Demeler, B.; Werner, M.H. T4 AsiA blocks DNA recognition by remodeling sigma70 region 4. EMBO J. 2004, 23, 2952–2962. [Google Scholar] [CrossRef]
- Feklistov, A.; Darst, S.A. Structural basis for promoter −10 element recognition by the bacterial RNA polymerase sigma subunit. Cell 2011, 147, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Doucleff, M.; Pelton, J.G.; Lee, P.S.; Nixon, B.T.; Wemmer, D.E. Structural basis of DNA recognition by the alternative sigma-factor, sigma54. J. Mol. Biol. 2007, 369, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Doucleff, M.; Wemmer, D.E. Structure of the RNA polymerase core-binding domain of sigma54 reveals a likely conformational fracture point. J. Mol. Biol. 2009, 390, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Doucleff, M.; Malak, L.T.; Pelton, J.G.; Wemmer, D.E. The C-terminal RpoN domain of sigma54 forms an unpredicted helix-turn-helix motif similar to domains of sigma70. J. Biol. Chem. 2005, 280, 41530–41536. [Google Scholar] [CrossRef] [PubMed]
- Lane, W.J.; Darst, S.A. The structural basis for promoter −35 element recognition by the group IV sigma factors. PLoS Biol. 2006, 4, e269. [Google Scholar] [CrossRef] [PubMed]
- Thakur, K.G.; Joshi, A.M.; Gopal, B. Structural and biophysical studies on two promoter recognition domains of the extra-cytoplasmic function sigma factor SigmaC from Mycobacterium tuberculosis. J. Biol. Chem. 2007, 282, 4711–4718. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, R.K.; Prabha, T.S.; Manjeera, G.; Gopal, B. Mycobacterium tuberculosis RsdA provides a conformational rationale for selective regulation of sigma-factor activity by proteolysis. Nucleic Acids Res. 2013, 41, 3414–3423. [Google Scholar] [CrossRef] [PubMed]
- Motackova, V.; Sanderova, H.; Zidek, L.; Novacek, J.; Padrta, P.; Svenkova, A.; Korelusova, J.; Jonak, J.; Krasny, L.; Sklenar, V. Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs. Proteins 2010, 78, 1807–1810. [Google Scholar] [PubMed]
- Demo, G.; Papouskova, V.; Komarek, J.; Kaderavek, P.; Otrusinova, O.; Srb, P.; Rabatinova, A.; Krasny, L.; Zidek, L.; Sklenar, V.; et al. X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase. J. Struct. Biol. 2014, 187, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Papouskova, V.; Kaderavek, P.; Otrusinova, O.; Rabatinova, A.; H, S.S.; Novacek, J.; Krasny, L.; Sklenar, V.; Zidek, L. Structural study of the partially disordered full-length delta subunit of RNA polymerase from Bacillus subtilis. Chembiochem 2013, 14, 1772–1779. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.N.; Yang, X.; Wiedermannova, J.; Delumeau, O.; Krasny, L.; Lewis, P.J. Epsilon, a new subunit of RNA polymerase found in gram-positive bacteria. J. Bacteriol. 2014, 196, 3622–3632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Campbell, E.A.; Minakhin, L.; Richter, C.; Severinov, K.; Darst, S.A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 1999, 98, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Minakhin, L.; Bhagat, S.; Brunning, A.; Campbell, E.A.; Darst, S.A.; Ebright, R.H.; Severinov, K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit Rpb6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA 2001, 98, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Kuznedelov, K.; Lamour, V.; Patikoglou, G.; Chlenov, M.; Darst, S.A.; Severinov, K. Recombinant Thermus aquaticus RNA polymerase for structural studies. J. Mol. Biol. 2006, 359, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.S.; Masuda, S.; Darst, S.A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 2002, 296, 1280–1284. [Google Scholar] [CrossRef] [PubMed]
- Vassylyev, D.G.; Sekine, S.; Laptenko, O.; Lee, J.; Vassylyeva, M.N.; Borukhov, S.; Yokoyama, S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 2002, 417, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Tuske, S.; Sarafianos, S.G.; Wang, X.; Hudson, B.; Sineva, E.; Mukhopadhyay, J.; Birktoft, J.J.; Leroy, O.; Ismail, S.; Clark, A.D., Jr.; et al. Inhibition of bacterial RNA polymerase by streptolydigin: Stabilization of a straight-bridge-helix active-center conformation. Cell 2005, 122, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.S. X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme. J. Biol. Chem. 2013, 288, 9126–9134. [Google Scholar] [CrossRef] [PubMed]
- Bae, B.; Davis, E.; Brown, D.; Campbell, E.A.; Wigneshweraraj, S.; Darst, S.A. Phage T7 gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma70 domain 1.1. Proc. Natl. Acad. Sci. USA 2013, 110, 19772–19777. [Google Scholar] [CrossRef] [PubMed]
- Degen, D.; Feng, Y.; Zhang, Y.; Ebright, K.Y.; Ebright, Y.W.; Gigliotti, M.; Vahedian-Movahed, H.; Mandal, S.; Talaue, M.; Connell, N.; et al. Transcription inhibition by the depsipeptide antibiotic Salinamide A. eLife 2014, 3, e02451. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.S.; Masuda, S.; Campbell, E.A.; Muzzin, O.; Darst, S.A. Structural basis of transcription initiation: An RNA polymerase holoenzyme-DNA complex. Science 2002, 296, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, Y.; Chatterjee, S.; Tuske, S.; Ho, M.X.; Arnold, E.; Ebright, R.H. Structural basis of transcription initiation. Science 2012, 338, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.S.; Warner, B.A.; Molodtsov, V.; Pupov, D.; Esyunina, D.; Fernandez-Tornero, C.; Kulbachinskiy, A.; Murakami, K.S. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 2014, 289, 24549–24559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Degen, D.; Ho, M.X.; Sineva, E.; Ebright, K.Y.; Ebright, Y.W.; Mekler, V.; Vahedian-Movahed, H.; Feng, Y.; Yin, R.; et al. GE23077 binds to the RNA polymerase “I” and “I+1” sites and prevents the binding of initiating nucleotides. eLife 2014, 3, e02450. [Google Scholar] [PubMed]
- Vassylyev, D.G.; Vassylyeva, M.N.; Zhang, J.; Palangat, M.; Artsimovitch, I.; Landick, R. Structural basis for substrate loading in bacterial RNA polymerase. Nature 2007, 448, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Vassylyev, D.G.; Vassylyeva, M.N.; Perederina, A.; Tahirov, T.H.; Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 2007, 448, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Weixlbaumer, A.; Leon, K.; Landick, R.; Darst, S.A. Structural basis of transcriptional pausing in bacteria. Cell 2013, 152, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Murayama, Y.; Svetlov, V.; Nudler, E.; Yokoyama, S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol. Cell 2015, 57, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, G.M.; Steitz, T.A. Structure of a transcribing T7 RNA polymerase initiation complex. Science 1999, 286, 2305–2309. [Google Scholar] [CrossRef] [PubMed]
- Steitz, T.A.; Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6498–6502. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Tornero, C.; Moreno-Morcillo, M.; Rashid, U.J.; Taylor, N.M.; Ruiz, F.M.; Gruene, T.; Legrand, P.; Steuerwald, U.; Muller, C.W. Crystal structure of the 14-subunit RNA polymerase I. Nature 2013, 502, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Engel, C.; Sainsbury, S.; Cheung, A.C.; Kostrewa, D.; Cramer, P. RNA polymerase I structure and transcription regulation. Nature 2013, 502, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Cramer, P.; Bushnell, D.A.; Kornberg, R.D. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science 2001, 292, 1863–1876. [Google Scholar] [CrossRef] [PubMed]
- Hirata, A.; Klein, B.J.; Murakami, K.S. The X-ray crystal structure of RNA polymerase from archaea. Nature 2008, 451, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Imashimizu, M.; Shimamoto, N.; Oshima, T.; Kashlev, M. Transcription elongation: Heterogeneous tracking of RNA polymerase and its biological implications. Transcription 2014, 5, e28285. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zuo, Y.; Steitz, T.A. Structural basis for transcription reactivation by RapA. Proc. Natl. Acad. Sci. USA 2015, 112, 2006–2010. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, M.K.; Ray, S.S.; Darst, S.A. Crystal structure of the flagellar sigma/anti-sigma complex sigma28/flgm reveals an intact sigma factor in an inactive conformation. Mol. Cell 2004, 14, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Greenwell, R.; Anthony, J.R.; Wang, S.; Lim, L.; Das, K.; Sofia, H.J.; Donohue, T.J.; Darst, S.A. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell 2007, 27, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Tupy, J.L.; Gruber, T.M.; Wang, S.; Sharp, M.M.; Gross, C.A.; Darst, S.A. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell 2003, 11, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Shukla, J.; Gupta, R.; Thakur, K.G.; Gokhale, R.; Gopal, B. Structural basis for the redox sensitivity of the Mycobacterium tuberculosis SigK-RskA sigma-anti-sigma complex. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1026–1036. [Google Scholar] [CrossRef]
- Campbell, E.A.; Masuda, S.; Sun, J.L.; Muzzin, O.; Olson, C.A.; Wang, S.; Darst, S.A. Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor sigmaF. Cell 2002, 108, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Patikoglou, G.A.; Westblade, L.F.; Campbell, E.A.; Lamour, V.; Lane, W.J.; Darst, S.A. Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J. Mol. Biol. 2007, 372, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Thakur, K.G.; Praveena, T.; Gopal, B. Structural and biochemical bases for the redox sensitivity of Mycobacterium tuberculosis RslA. J. Mol. Biol. 2010, 397, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Benoff, B.; Yang, H.; Lawson, C.L.; Parkinson, G.; Liu, J.; Blatter, E.; Ebright, Y.W.; Berman, H.M.; Ebright, R.H. Structural basis of transcription activation: The CAP-alpha CTD-DNA complex. Science 2002, 297, 1562–1566. [Google Scholar] [CrossRef] [PubMed]
- Newberry, K.J.; Nakano, S.; Zuber, P.; Brennan, R.G. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. USA 2005, 102, 15839–15844. [Google Scholar] [CrossRef] [PubMed]
- Lamour, V.; Westblade, L.F.; Campbell, E.A.; Darst, S.A. Crystal structure of the in vivo-assembled Bacillus subtilis spx/RNA polymerase alpha subunit C-terminal domain complex. J. Struct. Biol. 2009, 168, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.M.; Lin, A.; Zuber, C.S.; Newberry, K.J.; Brennan, R.G.; Zuber, P. Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit. PLOS ONE 2010, 5, e8664. [Google Scholar] [CrossRef] [PubMed]
- Westblade, L.F.; Campbell, E.A.; Pukhrambam, C.; Padovan, J.C.; Nickels, B.E.; Lamour, V.; Darst, S.A. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010, 38, 8357–8369. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Nickels, B.E.; Sun, L.; Hochschild, A.; Darst, S.A. Structure of a ternary transcription activation complex. Mol. Cell 2004, 13, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.G.; Canals, A.; Bernues, J.; Sola, M.; Coll, M. The structure of a transcription activation subcomplex reveals how sigma70 is recruited to PhoB promoters. EMBO J. 2011, 30, 3776–3785. [Google Scholar] [CrossRef] [PubMed]
- Hudson, B.P.; Quispe, J.; Lara-Gonzalez, S.; Kim, Y.; Berman, H.M.; Arnold, E.; Ebright, R.H.; Lawson, C.L. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc. Natl. Acad. Sci. USA 2009, 106, 19830–19835. [Google Scholar] [CrossRef] [PubMed]
- Tagami, S.; Sekine, S.; Kumarevel, T.; Hino, N.; Murayama, Y.; Kamegamori, S.; Yamamoto, M.; Sakamoto, K.; Yokoyama, S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 2010, 468, 978–982. [Google Scholar] [CrossRef] [PubMed]
- James, E.; Liu, M.; Sheppard, C.; Mekler, V.; Camara, B.; Liu, B.; Simpson, P.; Cota, E.; Severinov, K.; Matthews, S.; et al. Structural and mechanistic basis for the inhibition of Escherichia coli RNA polymerase by T7 gp2. Mol. Cell 2012, 47, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Twist, K.A.; Campbell, E.A.; Deighan, P.; Nechaev, S.; Jain, V.; Geiduschek, E.P.; Hochschild, A.; Darst, S.A. Crystal structure of the bacteriophage T4 late-transcription coactivator gp33 with the beta-subunit flap domain of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA 2011, 108, 19961–19966. [Google Scholar] [CrossRef] [PubMed]
- Tagami, S.; Sekine, S.; Minakhin, L.; Esyunina, D.; Akasaka, R.; Shirouzu, M.; Kulbachinskiy, A.; Severinov, K.; Yokoyama, S. Structural basis for promoter specificity switching of RNA polymerase by a phage factor. Genes Dev. 2014, 28, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Osmundson, J.; Montero-Diez, C.; Westblade, L.F.; Hochschild, A.; Darst, S.A. Promoter-specific transcription inhibition in Staphylococcus aureus by a phage protein. Cell 2012, 151, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Mechold, U.; Potrykus, K.; Murphy, H.; Murakami, K.S.; Cashel, M. Differential regulation by ppgpp versus ppGpp in Escherichia coli. Nucleic Acids Res. 2013, 41, 6175–6189. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Wang, Y.; Steitz, T.A. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell 2013, 50, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Patlan, V.; Sekine, S.; Vassylyeva, M.N.; Hosaka, T.; Ochi, K.; Yokoyama, S.; Vassylyev, D.G. Structural basis for transcription regulation by alarmone ppGpp. Cell 2004, 117, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Vrentas, C.E.; Gaal, T.; Berkmen, M.B.; Rutherford, S.T.; Haugen, S.P.; Vassylyev, D.G.; Ross, W.; Gourse, R.L. Still looking for the magic spot: The crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 2008, 377, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Ishihama, A. Bipartite functional map of the E. coli RNA polymerase alpha subunit: Involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell 1991, 65, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Molodtsov, V.; Nawarathne, I.N.; Scharf, N.T.; Kirchhoff, P.D.; Showalter, H.D.; Garcia, G.A.; Murakami, K.S. X-ray crystal structures of the Escherichia coli RNA polymerase in complex with benzoxazinorifamycins. J. Med. Chem. 2013, 56, 4758–4763. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Pavlova, O.; Zenkin, N.; Leon, F.; Irschik, H.; Jansen, R.; Severinov, K.; Darst, S.A. Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase. EMBO J. 2005, 24, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Vassylyeva, M.N.; Svetlov, D.; Svetlov, V.; Perederina, A.; Igarashi, N.; Matsugaki, N.; Wakatsuki, S.; Tahirov, T.H.; Vassylyev, D.G. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell 2005, 122, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Temiakov, D.; Zenkin, N.; Vassylyeva, M.N.; Perederina, A.; Tahirov, T.H.; Kashkina, E.; Savkina, M.; Zorov, S.; Nikiforov, V.; Igarashi, N.; et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell 2005, 19, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, J.; Das, K.; Ismail, S.; Koppstein, D.; Jang, M.; Hudson, B.; Sarafianos, S.; Tuske, S.; Patel, J.; Jansen, R.; et al. The RNA polymerase “switch region” is a target for inhibitors. Cell 2008, 135, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Belogurov, G.A.; Vassylyeva, M.N.; Sevostyanova, A.; Appleman, J.R.; Xiang, A.X.; Lira, R.; Webber, S.E.; Klyuyev, S.; Nudler, E.; Artsimovitch, I.; et al. Transcription inactivation through local refolding of the RNA polymerase structure. Nature 2009, 457, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Molodtsov, V.; Fleming, P.R.; Eyermann, C.J.; Ferguson, A.D.; Foulk, M.A.; McKinney, D.C.; Masse, C.E.; Buurman, E.T.; Murakami, K.S. X-ray crystal structures of Escherichia coli RNA polymerase with switch region binding inhibitors enable rational design of squaramides with an improved fraction unbound to human plasma protein. J. Med. Chem. 2015, 58, 3156–3171. [Google Scholar] [CrossRef] [PubMed]
- Buurman, E.T.; Foulk, M.A.; Gao, N.; Laganas, V.A.; McKinney, D.C.; Moustakas, D.T.; Rose, J.A.; Shapiro, A.B.; Fleming, P.R. Novel rapidly diversifiable antimicrobial RNA polymerase switch region inhibitors with confirmed mode of action in Haemophilus influenzae. J. Bacteriol. 2012, 194, 5504–5512. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murakami, K.S. Structural Biology of Bacterial RNA Polymerase. Biomolecules 2015, 5, 848-864. https://doi.org/10.3390/biom5020848
Murakami KS. Structural Biology of Bacterial RNA Polymerase. Biomolecules. 2015; 5(2):848-864. https://doi.org/10.3390/biom5020848
Chicago/Turabian StyleMurakami, Katsuhiko S. 2015. "Structural Biology of Bacterial RNA Polymerase" Biomolecules 5, no. 2: 848-864. https://doi.org/10.3390/biom5020848
APA StyleMurakami, K. S. (2015). Structural Biology of Bacterial RNA Polymerase. Biomolecules, 5(2), 848-864. https://doi.org/10.3390/biom5020848





