PARP1 and PARG Are the Draft Horses for Polycomb-Trithorax Chromatin Regulator Machinery
Abstract
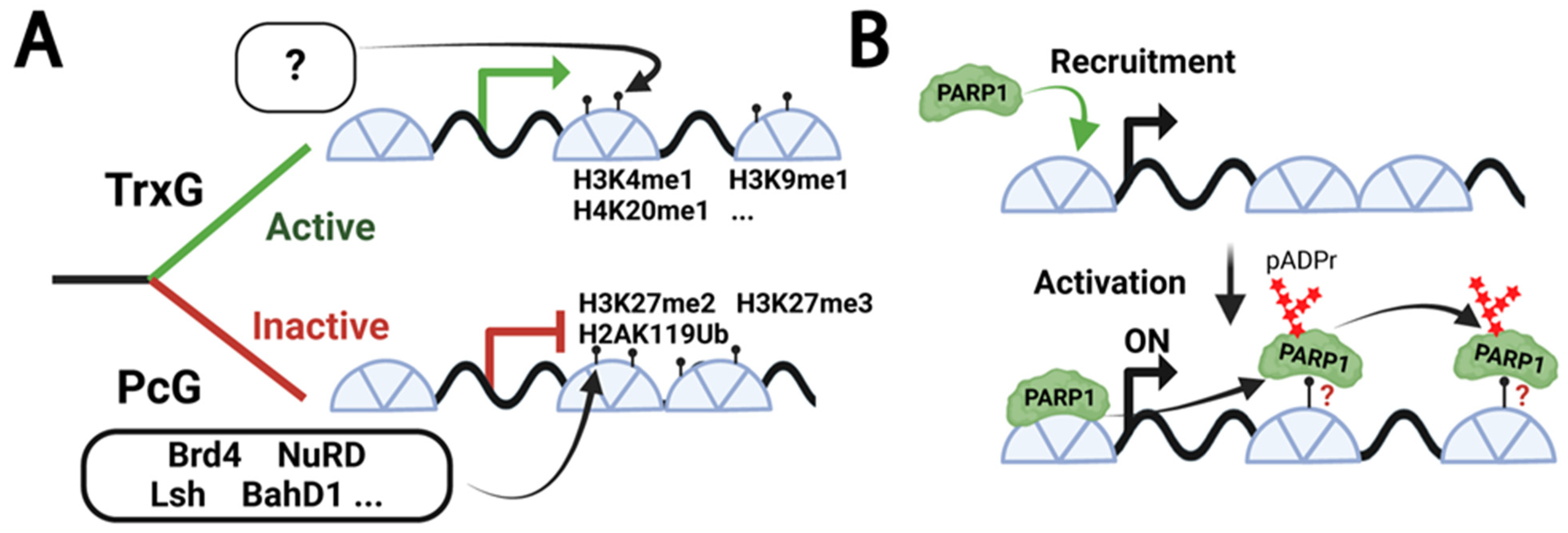
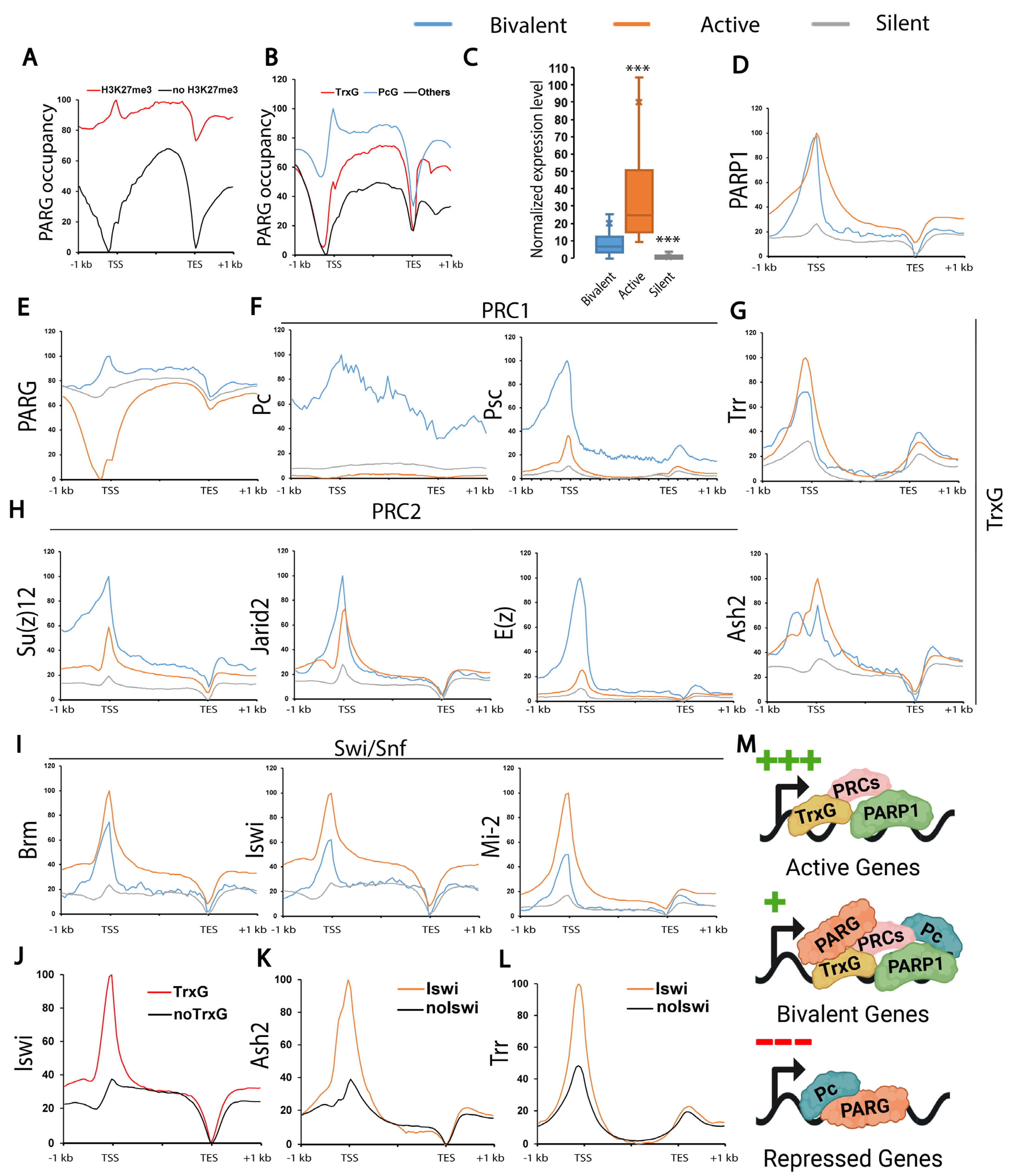
1. Introduction
2. Materials and Methods
2.1. Histone Peptide Array
2.2. ChIP-Seq Analysis
2.3. ATAC-Seq Analysis
2.4. Genome-Wide Datasets
2.5. Statistical Analysis
3. Results
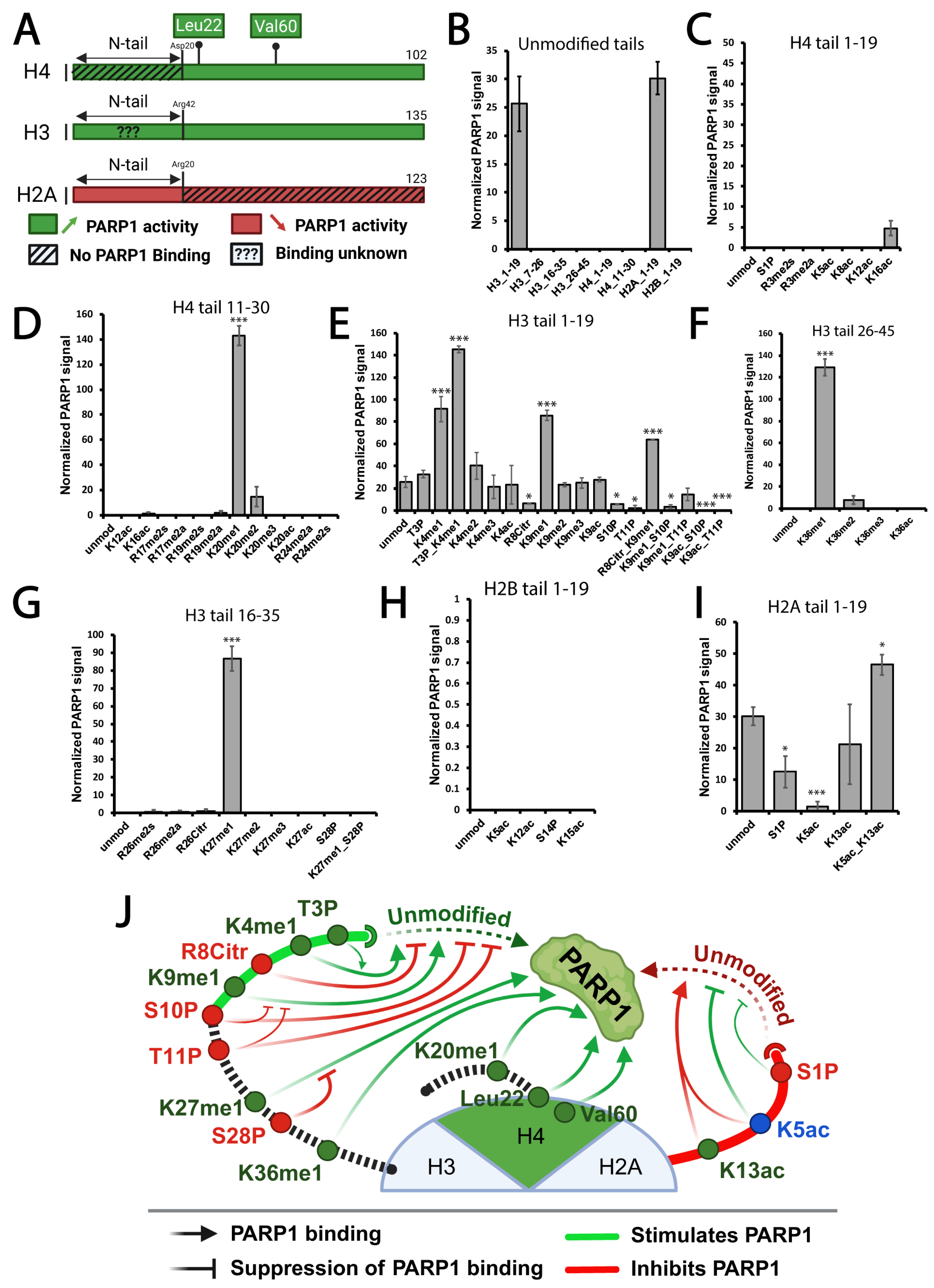
3.1. Mono-Methylated Histone Tails Bind PARP1 with the Highest Affinity In Vitro
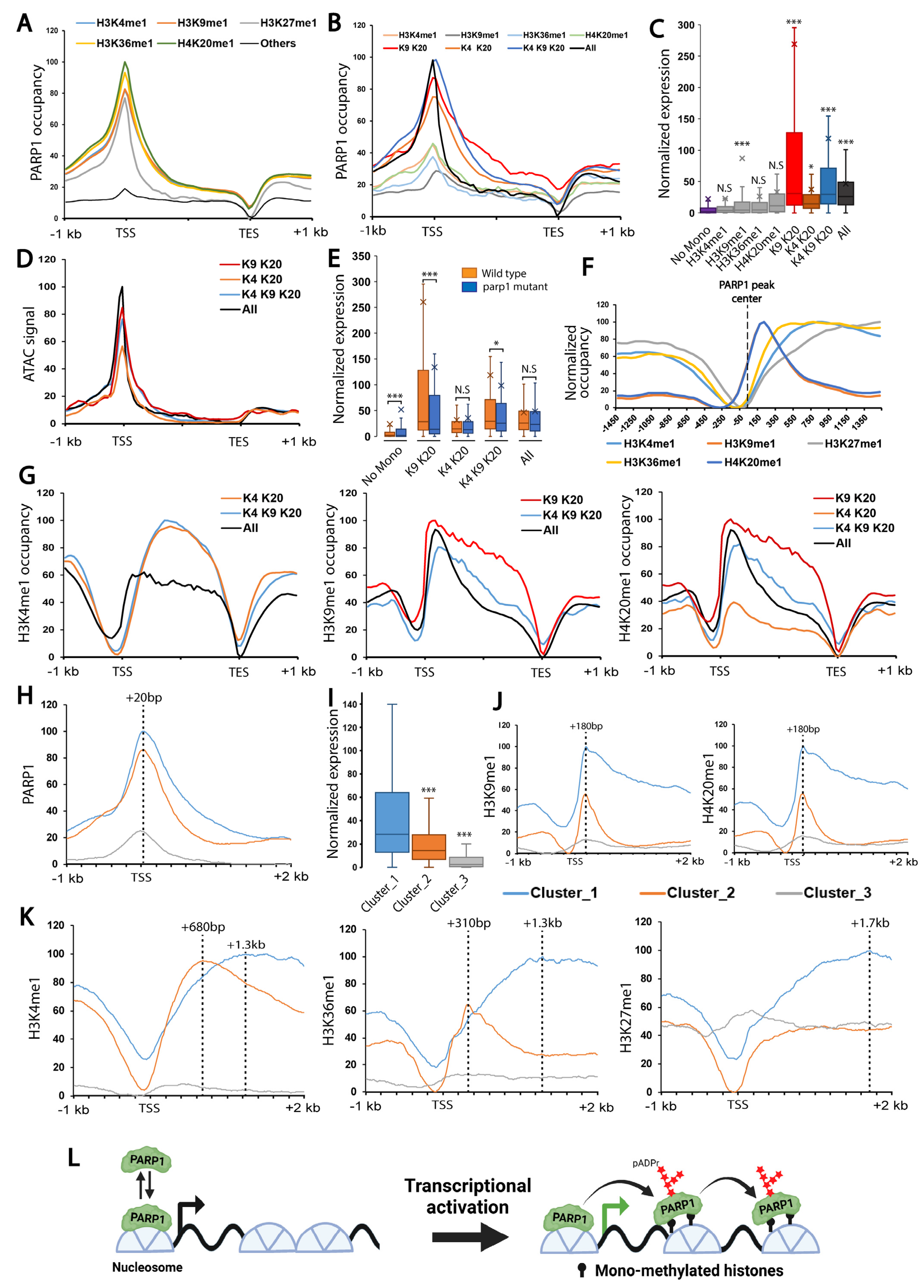
3.2. Mono-Methylated Histones Control PARP1 Binding at PARP1-Controlled Loci Genome-Wide
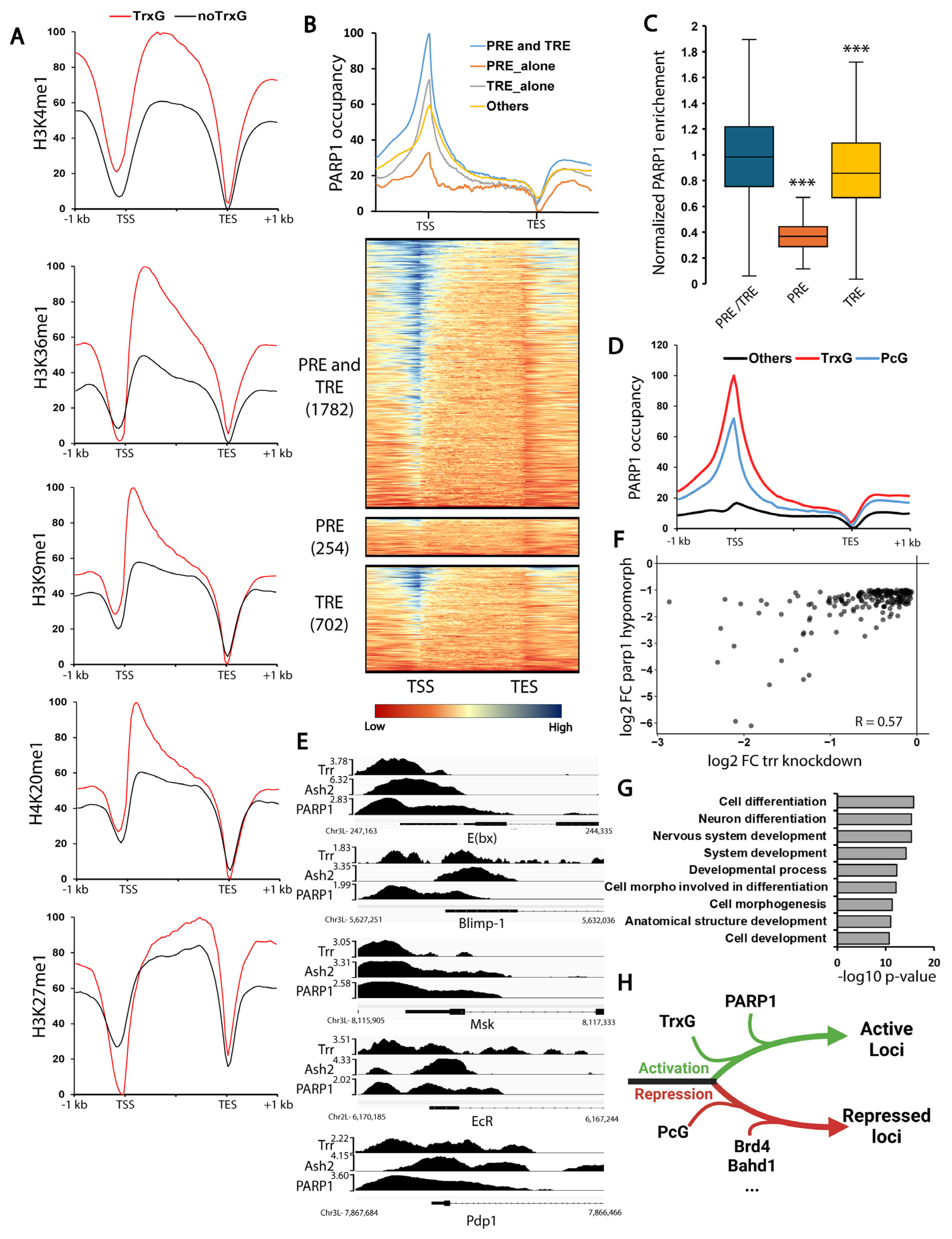
3.3. PARP1 Controls the Expression of Developmental Genes Controlled by TrxG
3.4. PARG Controls the Expression of PcG-Controlled Loci
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levine, M.; Davidson, E.H. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 2005, 102, 4936–4942. [Google Scholar] [CrossRef]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Bourbon, H.-M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Isono, K.; Kondo, K.; Endo, T.A.; Itohara, S.; Vidal, M.; Koseki, H. Polycomb Potentiates Meis2 Activation in Midbrain by Mediating Interaction of the Promoter with a Tissue-Specific Enhancer. Dev. Cell 2014, 28, 94–101. [Google Scholar] [CrossRef]
- Cavalli, G.; Paro, R. Epigenetic Inheritance of Active Chromatin After Removal of the Main Transactivator. Science 1999, 286, 955–958. [Google Scholar] [CrossRef] [PubMed]
- Ringrose, L.; Paro, R. Epigenetic Regulation of Cellular Memory by the Polycomb and Trithorax Group Proteins. Annu. Rev. Genet. 2004, 38, 413–443. [Google Scholar] [CrossRef]
- Steffen, P.A.; Ringrose, L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell. Biol. 2014, 15, 340–356. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Francis, N.J.; Kingston, R.E.; Woodcock, C.L. Chromatin Compaction by a Polycomb Group Protein Complex. Science 2004, 306, 1574–1577. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Y.; Jaganathan, A.; Sun, Y.; Ma, N.; Li, N.; Han, X.; Sun, X.; Yi, H.; Fu, S.; et al. BRD4-PRC2 represses transcription of T-helper 2-specific negative regulators during T-cell differentiation. EMBO J. 2023, 42, e111473. [Google Scholar] [CrossRef]
- Sudarsanam, P.; Winston, F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000, 16, 345–351. [Google Scholar] [CrossRef]
- Bamgbose, G.; Tulin, A. PARP-1 is a transcriptional rheostat of metabolic and bivalent genes during development. Life. Sci. Alliance 2023, 7, e202302369. [Google Scholar] [CrossRef]
- Bordet, G.; Bamgbose, G.; Tulin, A.V. Poly(ADP-ribosyl)ating enzymes coordinate changes in the expression of metabolic genes with developmental progression. Sci. Rep. 2023, 13, 20320. [Google Scholar] [CrossRef]
- Tulin, A.; Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 2003, 299, 560–562. [Google Scholar] [CrossRef] [PubMed]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Kotova, E.; Jarnik, M.; Tulin, A.V. Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc. Natl. Acad. Sci. USA 2010, 107, 6406–6411. [Google Scholar] [CrossRef]
- Petesch, S.J.; Lis, J.T. Activator-Induced Spread of Poly(ADP-Ribose) Polymerase Promotes Nucleosome Loss at Hsp70. Mol. Cell 2012, 45, 64–74. [Google Scholar] [CrossRef]
- Thomas, C.; Ji, Y.; Wu, C.; Datz, H.; Boyle, C.; MacLeod, B.; Patel, S.; Ampofo, M.; Currie, M.; Harbin, J.; et al. Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes. Proc. Natl. Acad. Sci. USA 2019, 116, 9941–9946. [Google Scholar] [CrossRef]
- Pinnola, A.; Naumova, N.; Shah, M.; Tulin, A.V. Nucleosomal Core Histones Mediate Dynamic Regulation of Poly(ADP-ribose) Polymerase 1 Protein Binding to Chromatin and Induction of Its Enzymatic Activity. J. Biol. Chem. 2007, 282, 32511–32519. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.J.; Kotova, E.; Andrake, M.; Adolf-Bryfogle, J.; Glaser, R.; Regnard, C.; Tulin, A.V. Kinase-Mediated Changes in Nucleosome Conformation Trigger Chromatin Decondensation via Poly(ADP-Ribosyl)ation. Mol. Cell 2014, 53, 831–842. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Barnett, D.W.; Garrison, E.K.; Quinlan, A.R.; Strömberg, M.P.; Marth, G.T. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 2011, 27, 1691–1692. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Liu, T.; Qin, B.; Zhang, Y.; Liu, X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012, 7, 1728–1740. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; He, Q.-Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Khan, A.; Mathelier, A. Intervene: A tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Meers, M.P.; Adelman, K.; Duronio, R.J.; Strahl, B.D.; McKay, D.J.; Matera, A.G. Transcription start site profiling uncovers divergent transcription and enhancer-associated RNAs in Drosophila melanogaster. BMC Genom. 2018, 19, 157. [Google Scholar] [CrossRef] [PubMed]
- Bamgbose, G.; Bordet, G.; Lodhi, N.; Tulin, A. Mono-methylated histones control PARP-1 in chromatin and transcription. eLife 2024, 13, RP91482. [Google Scholar] [CrossRef] [PubMed]
- Herz, H.-M.; Mohan, M.; Garrett, A.S.; Miller, C.; Casto, D.; Zhang, Y.; Seidel, C.; Haug, J.S.; Florens, L.; Washburn, M.P.; et al. Polycomb Repressive Complex 2-Dependent and -Independent Functions of Jarid2 in Transcriptional Regulation in Drosophila. Mol. Cell. Biol. 2012, 32, 1683–1693. [Google Scholar] [CrossRef]
- Erokhin, M.; Brown, J.L.; Lomaev, D.; Vorobyeva, N.E.; Zhang, L.; Fab, L.V.; Mazina, M.Y.; Kulakovskiy, I.V.; Ziganshin, R.H.; Schedl, P.; et al. Crol contributes to PRE-mediated repression and Polycomb group proteins recruitment in Drosophila. Nucleic Acids Res. 2023, 51, 6087–6100. [Google Scholar] [CrossRef] [PubMed]
- Posukh, O.V.; Maksimov, D.A.; Laktionov, P.P.; Koryakov, D.E.; Belyakin, S.N. Functional dissection of Drosophila melanogaster SUUR protein influence on H3K27me3 profile. Epigenetics Chromatin 2017, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Loubiere, V.; Papadopoulos, G.L.; Szabo, Q.; Martinez, A.-M.; Cavalli, G. Widespread activation of developmental gene expression characterized by PRC1-dependent chromatin looping. Sci. Adv. 2020, 6, eaax4001. [Google Scholar] [CrossRef]
- Koemans, T.S.; Kleefstra, T.; Chubak, M.C.; Stone, M.H.; Reijnders, M.R.F.; de Munnik, S.; Willemsen, M.H.; Fenckova, M.; Stumpel, C.T.R.M.; Bok, L.A.; et al. Functional convergence of histone methyltransferases EHMT1 and KMT2C involved in intellectual disability and autism spectrum disorder. PLoS Genet. 2017, 13, e1006864. [Google Scholar] [CrossRef]
- Pérez-Lluch, S.; Blanco, E.; Carbonell, A.; Raha, D.; Snyder, M.; Serras, F.; Corominas, M. Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res. 2011, 39, 4628–4639. [Google Scholar] [CrossRef] [PubMed]
- Vorobyeva, N.E.; Erokhin, M.; Chetverina, D.; Krasnov, A.N.; Mazina, M.Y. Su(Hw) primes 66D and 7F Drosophila chorion genes loci for amplification through chromatin decondensation. Sci. Rep. 2021, 11, 16963. [Google Scholar] [CrossRef]
- The modENCODE Consortium; Roy, S.; Ernst, J.; Kharchenko, P.V.; Kheradpour, P.; Negre, N.; Eaton, M.L.; Landolin, J.M.; Bristow, C.A.; Ma, L.; et al. Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE. Science 2010, 330, 1787–1797. [Google Scholar] [CrossRef]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2010, 471, 473–479. [Google Scholar] [CrossRef]
- Zraly, C.B.; Zakkar, A.; Perez, J.H.; Ng, J.; White, K.P.; Slattery, M.; Dingwall, A.K. The Drosophila MLR COMPASS complex is essential for programming cis-regulatory information and maintaining epigenetic memory during development. Nucleic Acids Res. 2020, 48, 3476–3495. [Google Scholar] [CrossRef]
- Blanco, E.; Pignatelli, M.; Beltran, S.; Punset, A.; Pérez-Lluch, S.; Serras, F.; Guigó, R.; Corominas, M. Conserved chromosomal clustering of genes governed by chromatin regulators in Drosophila. Genome Biol. 2008, 9, R134. [Google Scholar] [CrossRef] [PubMed]
- Bredesen, B.A.; Rehmsmeier, M. DNA sequence models of genome-wide Drosophila melanogaster Polycomb binding sites improve generalization to independent Polycomb Response Elements. Nucleic Acids Res. 2019, 47, 7781–7797. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Hallson, G.; Hollebakken, R.E.; Li, T.; Syrzycka, M.; Kim, I.; Cotsworth, S.; Fitzpatrick, K.A.; Sinclair, D.A.R.; Honda, B.M. dSet1 Is the Main H3K4 Di- and Tri-Methyltransferase Throughout Drosophila Development. Genetics 2012, 190, 91–100. [Google Scholar] [CrossRef]
- Rickels, R.; Herz, H.-M.; Sze, C.C.; Cao, K.; Morgan, M.A.; Collings, C.K.; Gause, M.; Takahashi, Y.-H.; Wang, L.; Rendleman, E.J.; et al. Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet. 2017, 49, 1647–1653. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Banerjee, R.; Saiakhova, A.R.; Howard, B.; Monteith, K.E.; Scacheri, P.C.; Cosgrove, M.S.; Harte, P.J. Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development 2014, 141, 1129–1139. [Google Scholar] [CrossRef]
- Ferrari, K.J.; Scelfo, A.; Jammula, S.; Cuomo, A.; Barozzi, I.; Stützer, A.; Fischle, W.; Bonaldi, T.; Pasini, D. Polycomb-Dependent H3K27me1 and H3K27me2 Regulate Active Transcription and Enhancer Fidelity. Mol. Cell 2014, 53, 49–62. [Google Scholar] [CrossRef]
- Lee, H.-G.; Kahn, T.G.; Simcox, A.; Schwartz, Y.B.; Pirrotta, V. Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res. 2015, 25, 1170–1181. [Google Scholar] [CrossRef]
- Simon, J.A.; Kingston, R.E. Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol. Cell 2013, 49, 808–824. [Google Scholar] [CrossRef]
- Bordet, G.; Karpova, I.; Tulin, A.V. Poly(ADP-ribosyl)ating enzymes cooperate to coordinate development. Sci. Rep. 2022, 12, 22120. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, X.; Guan, H.; Xiong, X.; Shi, X.; Deng, H.; Li, H. The BAH domain of BAHD1 is a histone H3K27me3 reader. Protein Cell 2016, 7, 222–226. [Google Scholar] [CrossRef]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.-M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef]
- Arora, M.; Packard, C.Z.; Banerjee, T.; Parvin, J.D. RING1A and BMI1 bookmark active genes via ubiquitination of chromatin-associated proteins. Nucleic Acids Res. 2015, 44, 2136–2144. [Google Scholar] [CrossRef]
- Chan, H.L.; Beckedorff, F.; Zhang, Y.; Garcia-Huidobro, J.; Jiang, H.; Colaprico, A.; Bilbao, D.; Figueroa, M.E.; LaCava, J.; Shiekhattar, R.; et al. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun. 2018, 9, 3377. [Google Scholar] [CrossRef] [PubMed]
- Pherson, M.; Misulovin, Z.; Gause, M.; Mihindukulasuriya, K.; Swain, A.; Dorsett, D. Polycomb repressive complex 1 modifies transcription of active genes. Sci. Adv. 2017, 3, e1700944. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Wang, X.; Zhao, M.; Yang, R.; Malik, R.; Qiao, Y.; Poliakov, A.; Yocum, A.K.; Li, Y.; Chen, W.; et al. The central role of EED in the orchestration of polycomb group complexes. Nat. Commun. 2014, 5, 3127. [Google Scholar] [CrossRef] [PubMed]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Saurin, A.J.; Shao, Z.; Erdjument-Bromage, H.; Tempst, P.; Kingston, R.E. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 2001, 412, 655–660. [Google Scholar] [CrossRef]
- Kahn, T.G.; Dorafshan, E.; Schultheis, D.; Zare, A.; Stenberg, P.; Reim, I.; Pirrotta, V.; Schwartz, Y.B. Interdependence of PRC1 and PRC2 for recruitment to Polycomb Response Elements. Nucleic Acids Res. 2016, 44, 10132–10149. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordet, G.; Tulin, A.V. PARP1 and PARG Are the Draft Horses for Polycomb-Trithorax Chromatin Regulator Machinery. Biomolecules 2025, 15, 1314. https://doi.org/10.3390/biom15091314
Bordet G, Tulin AV. PARP1 and PARG Are the Draft Horses for Polycomb-Trithorax Chromatin Regulator Machinery. Biomolecules. 2025; 15(9):1314. https://doi.org/10.3390/biom15091314
Chicago/Turabian StyleBordet, Guillaume, and Alexei V. Tulin. 2025. "PARP1 and PARG Are the Draft Horses for Polycomb-Trithorax Chromatin Regulator Machinery" Biomolecules 15, no. 9: 1314. https://doi.org/10.3390/biom15091314
APA StyleBordet, G., & Tulin, A. V. (2025). PARP1 and PARG Are the Draft Horses for Polycomb-Trithorax Chromatin Regulator Machinery. Biomolecules, 15(9), 1314. https://doi.org/10.3390/biom15091314





