Multi-Temperature Crystallography of S-Adenosylmethionine Decarboxylase Observes Dynamic Loop Motions
Abstract
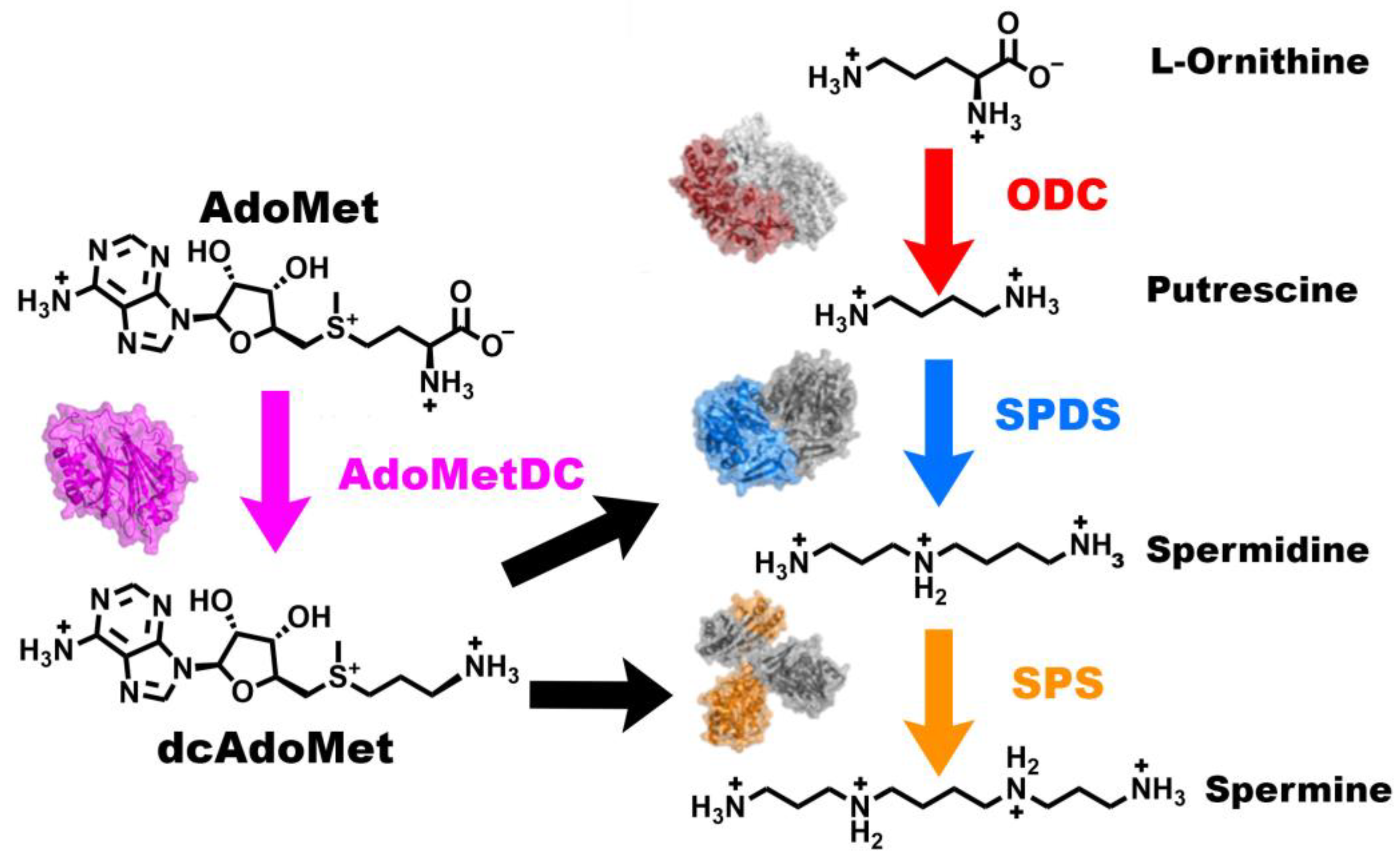
1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
2.2. Protein Crystallization
2.3. Data Collection and Processing
3. Results
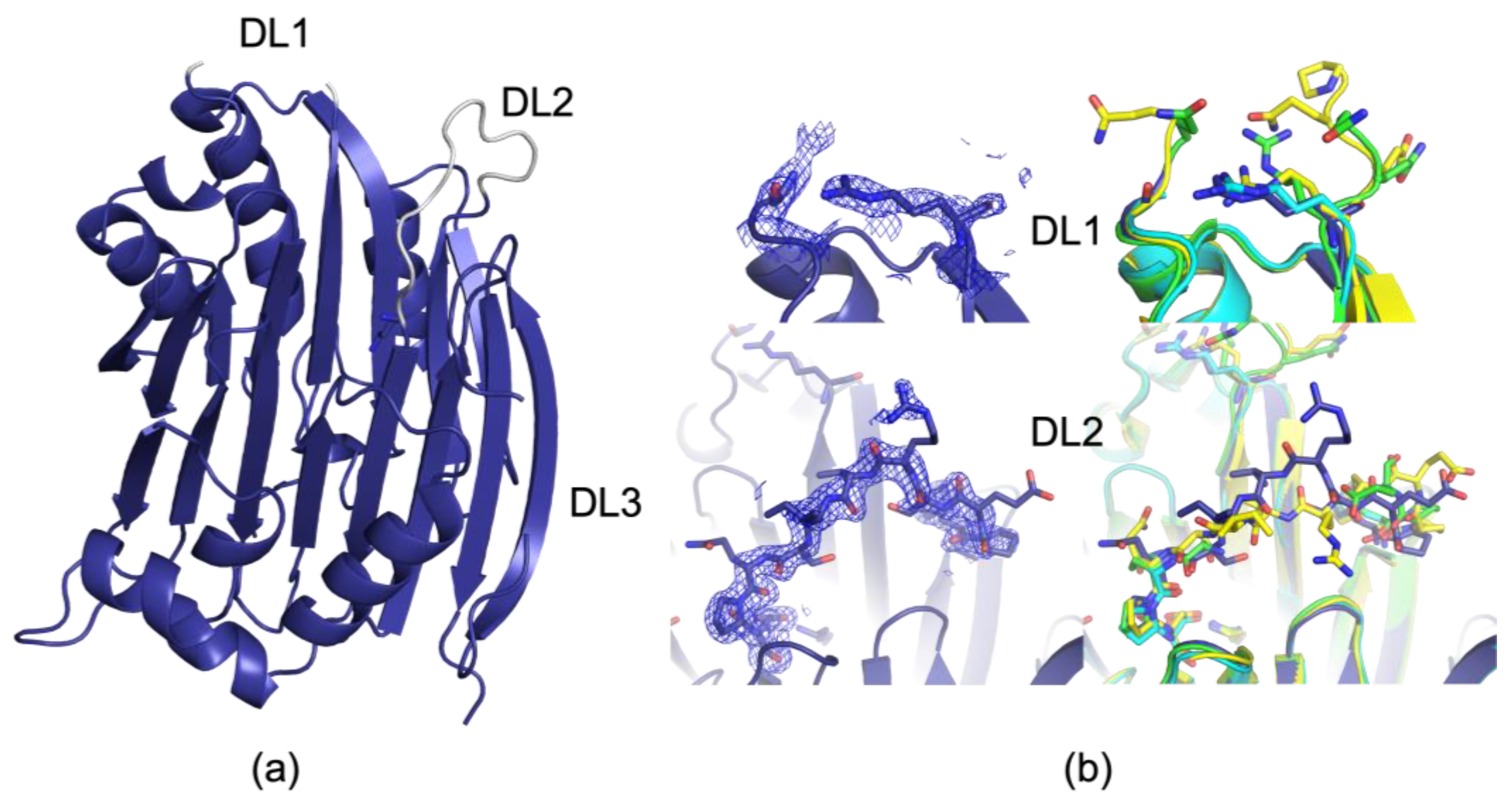
3.1. Comparison of PDB ID 9P1H, the New 100 K Apo Structure, and Previously Reported Structures
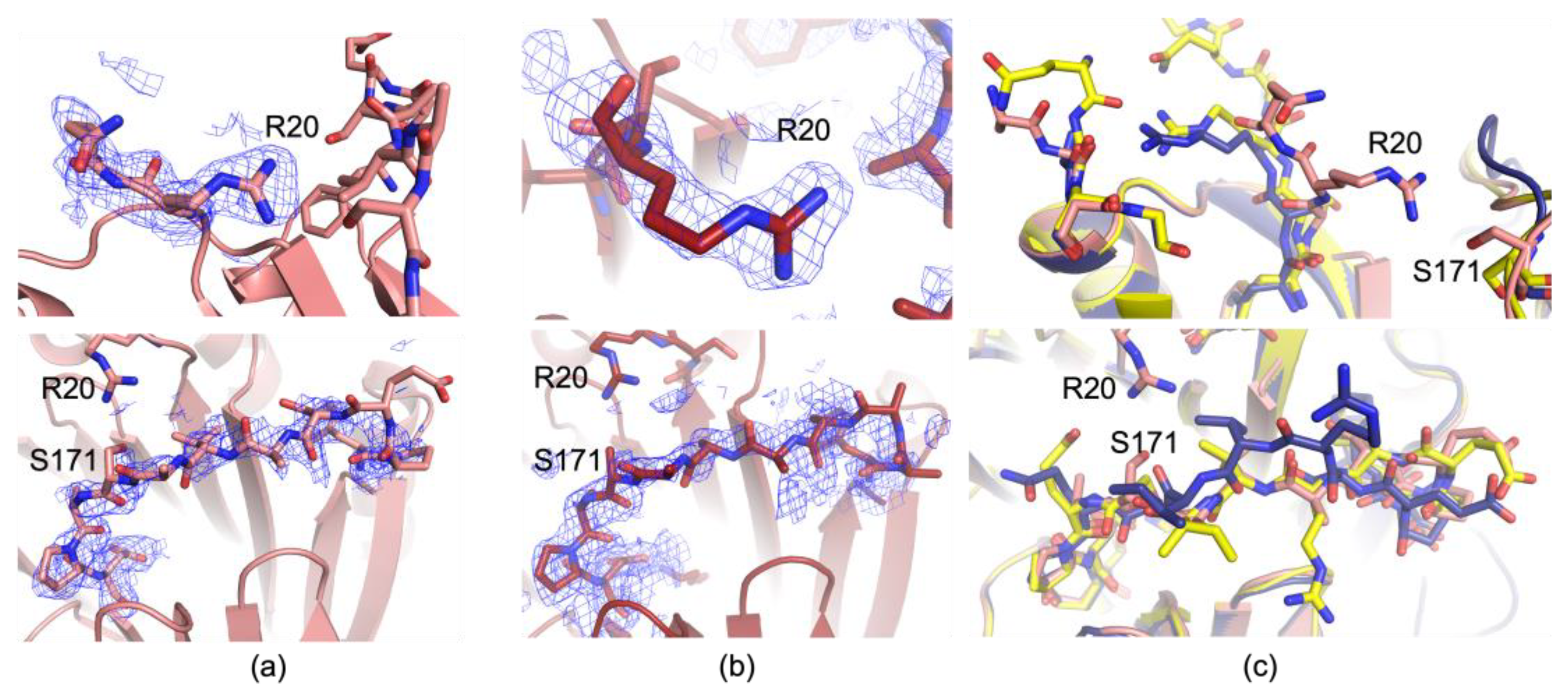
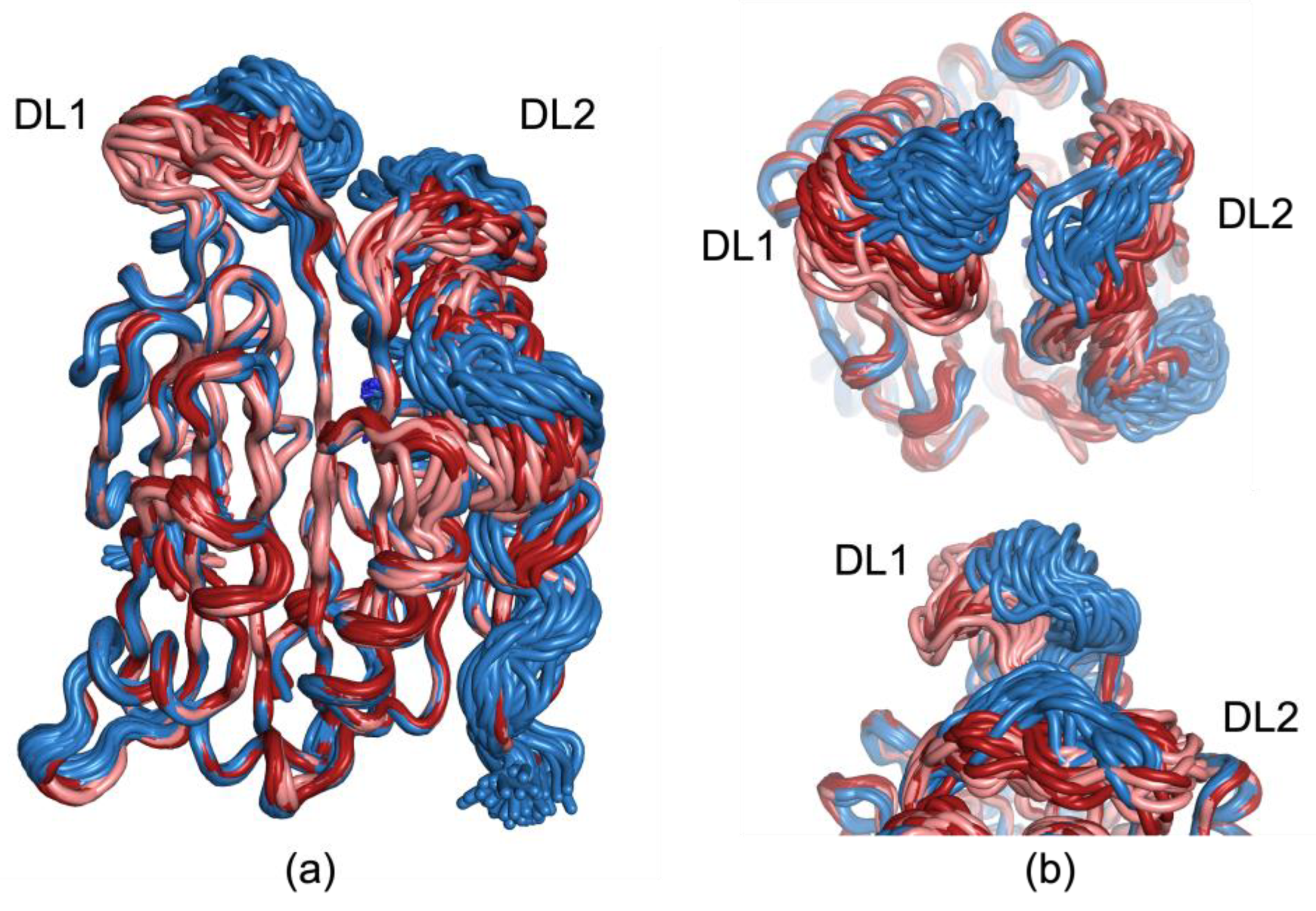
3.2. The 273 K and 293 K AdoMetDC Structures Show Loop Fluctuations
3.3. Additional Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ODC | Ornithine decarboxylase |
| AdoMetDC | S-adenosylmethionine decarboxylase |
| SPDS | Spermidine synthase |
| SPS | Spermine synthase |
| AdoMet | S-adenosylmethionine |
| dcAdoMet | Decarboxylated S-adenosylmethionine |
| hAdoMetDC | Human S-adenosylmethionine decarboxylase |
| DL1 | Disordered loop 1 |
| DL2 | Disordered loop 2 |
| DL3 | Disordered loop 3 |
| AMD1 | Human AdoMetDC gene |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| MTA | 5′-Deoxy-5′-(methylthio)adenosine |
| MGBG | Methylglyoxal bis(guanylhydrazone) |
| RoPE | Representation of Protein Entities |
| PASSer | Protein Allosteric Sites Server |
References
- Michael, A.J. Polyamines in Eukaryotes, Bacteria, and Archaea. J. Biol. Chem. 2016, 291, 14896–14903. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.J. Biosynthesis of Polyamines and Polyamine-Containing Molecules. Biochem. J. 2016, 473, 2315–2329. [Google Scholar] [CrossRef] [PubMed]
- Xuan, M.; Gu, X.; Li, J.; Huang, D.; Xue, C.; He, Y. Polyamines: Their Significance for Maintaining Health and Contributing to Diseases. Cell Commun. Signal. 2023, 21, 348. [Google Scholar] [CrossRef]
- Moinard, C.; Cynober, L.; de Bandt, J.-P. Polyamines: Metabolism and Implications in Human Diseases. Clin. Nutr. 2005, 24, 184–197. [Google Scholar] [CrossRef]
- Bae, D.-H.; Lane, D.J.R.; Jansson, P.J.; Richardson, D.R. The Old and New Biochemistry of Polyamines. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2018, 1862, 2053–2068. [Google Scholar] [CrossRef]
- Pegg, A.E. Mammalian Polyamine Metabolism and Function. IUBMB Life 2009, 61, 880–894. [Google Scholar] [CrossRef]
- Pegg, A.E.; Casero, R.A., Jr. (Eds.) Current Status of the Polyamine Research Field. In Polyamines: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2011; pp. 3–35. ISBN 978-1-61779-034-8. [Google Scholar]
- Ikeguchi, Y.; Bewley, M.C.; Pegg, A.E. Aminopropyltransferases: Function, Structure and Genetics. J. Biochem. 2006, 139, 1–9. [Google Scholar] [CrossRef]
- Wu, H.; Min, J.; Ikeguchi, Y.; Zeng, H.; Dong, A.; Loppnau, P.; Pegg, A.E.; Plotnikov, A.N. Structure and Mechanism of Spermidine Synthases. Biochemistry 2007, 46, 8331–8339. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Min, J.; Zeng, H.; McCloskey, D.E.; Ikeguchi, Y.; Loppnau, P.; Michael, A.J.; Pegg, A.E.; Plotnikov, A.N. Crystal Structure of Human Spermine Synthase: Implications of Substrate Binding and Catalytic Mechanism. J. Biol. Chem. 2008, 283, 16135–16146. [Google Scholar] [CrossRef]
- Williams-Ashman, H.G.; Schenone, A. Methyl Glyoxal Bis(Guanylhydrazone) as a Potent Inhibitor of Mammalian and Yeast S-Adenosylmethionine Decarboxylases. Biochem. Biophys. Res. Commun. 1972, 46, 288–295. [Google Scholar] [CrossRef]
- Regenass, U.; Mett, H.; Stanek, J.; Mueller, M.; Kramer, D.; Porter, C.W. CGP 48664, a New S-Adenosylmethionine Decarboxylase Inhibitor with Broad Spectrum Antiproliferative and Antitumor Activity1. Cancer Res. 1994, 54, 3210–3217. [Google Scholar] [PubMed]
- Millward, M.J.; Joshua, A.; Kefford, R.; Aamdal, S.; Thomson, D.; Hersey, P.; Toner, G.; Lynch, K. Multi-Centre Phase II Trial of the Polyamine Synthesis Inhibitor SAM486A (CGP48664) in Patients with Metastatic Melanoma. Invest. New Drugs 2005, 23, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Siu, L.L.; Rowinsky, E.K.; Hammond, L.A.; Weiss, G.R.; Hidalgo, M.; Clark, G.M.; Moczygemba, J.; Choi, L.; Linnartz, R.; Barbet, N.C.; et al. A Phase I and Pharmacokinetic Study of SAM486A, a Novel Polyamine Biosynthesis Inhibitor, Administered on a Daily-Times-Five Every-Three-Week Schedule in Patients with Advanced Solid Malignancies1. Clin. Cancer Res. 2002, 8, 2157–2166. [Google Scholar] [PubMed]
- Muthukumaran, S.; Sulochana, K.N.; Umashankar, V. Structure Based Design of Inhibitory Peptides Targeting Ornithine Decarboxylase Dimeric Interface and in Vitro Validation in Human Retinoblastoma Y79 Cells. J. Biomol. Struct. Dyn. 2021, 39, 5261–5275. [Google Scholar] [CrossRef]
- Zhou, X.E.; Suino-Powell, K.; Schultz, C.R.; Aleiwi, B.; Brunzelle, J.S.; Lamp, J.; Vega, I.E.; Ellsworth, E.; Bachmann, A.S.; Melcher, K. Structural Basis of Binding and Inhibition of Ornithine Decarboxylase by 1-Amino-Oxy-3-Aminopropane. Biochem. J. 2021, 478, 4137–4149. [Google Scholar] [CrossRef]
- Schultz, C.R.; Aleiwi, B.; Zhou, X.E.; Suino-Powell, K.; Melcher, K.; Almeida, N.M.S.; Wilson, A.K.; Ellsworth, E.L.; Bachmann, A.S. Design, Synthesis, and Biological Activity of Novel Ornithine Decarboxylase (ODC) Inhibitors. J. Med. Chem. 2025, 68, 5760–5773. [Google Scholar] [CrossRef]
- Bale, S.; Ealick, S.E. Structural Biology of S-Adenosylmethionine Decarboxylase. Amino Acids 2010, 38, 451–460. [Google Scholar] [CrossRef]
- Gallagher, T.; Rozwarski, D.A.; Ernst, S.R.; Hackert, M.L. Refined Structure of the Pyruvoyl-Dependent Histidine Decarboxylase from Lactobacillus 30a. J. Mol. Biol. 1993, 230, 516–528. [Google Scholar] [CrossRef]
- Albert, A.; Dhanaraj, V.; Genschel, U.; Khan, G.; Ramjee, M.K.; Pulido, R.; Sibanda, B.L.; von Delft, F.; Witty, M.; Blundell, T.L.; et al. Crystal Structure of Aspartate Decarboxylase at 2.2 Å Resolution Provides Evidence for an Ester in Protein Self–Processing. Nat. Struct. Mol. Biol. 1998, 5, 289–293. [Google Scholar] [CrossRef]
- Schmitzberger, F.; Kilkenny, M.L.; Lobley, C.M.C.; Webb, M.E.; Vinkovic, M.; Matak-Vinkovic, D.; Witty, M.; Chirgadze, D.Y.; Smith, A.G.; Abell, C.; et al. Structural Constraints on Protein Self-processing in L-aspartate-α-decarboxylase. EMBO J. 2003, 22, 6193–6204. [Google Scholar] [CrossRef]
- Soriano, E.V.; McCloskey, D.E.; Kinsland, C.; Pegg, A.E.; Ealick, S.E. Structures of the N47A and E109Q Mutant Proteins of Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 377–382. [Google Scholar] [CrossRef]
- Tolbert, W.D.; Graham, D.E.; White, R.H.; Ealick, S.E. Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii: Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms. Structure 2003, 11, 285–294. [Google Scholar] [CrossRef]
- Ekstrom, J.L.; Tolbert, W.D.; Xiong, H.; Pegg, A.E.; Ealick, S.E. Structure of a Human S-Adenosylmethionine Decarboxylase Self-Processing Ester Intermediate and Mechanism of Putrescine Stimulation of Processing As Revealed by the H243A Mutant. Biochemistry 2001, 40, 9495–9504. [Google Scholar] [CrossRef]
- Bale, S.; Lopez, M.M.; Makhatadze, G.I.; Fang, Q.; Pegg, A.E.; Ealick, S.E. Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase. Biochemistry 2008, 47, 13404–13417. [Google Scholar] [CrossRef]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef]
- Yordanova, M.M.; Loughran, G.; Zhdanov, A.V.; Mariotti, M.; Kiniry, S.J.; O’Connor, P.B.F.; Andreev, D.E.; Tzani, I.; Saffert, P.; Michel, A.M.; et al. AMD1 mRNA Employs Ribosome Stalling as a Mechanism for Molecular Memory Formation. Nature 2018, 553, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Ekstrom, J.L.; Mathews, I.I.; Stanley, B.A.; Pegg, A.E.; Ealick, S.E. The Crystal Structure of Human S-Adenosylmethionine Decarboxylase at 2.25 Å Resolution Reveals a Novel Fold. Structure 1999, 7, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Bale, S.; Brooks, W.; Hanes, J.W.; Mahesan, A.M.; Guida, W.C.; Ealick, S.E. Role of the Sulfonium Center in Determining the Ligand Specificity of Human S-Adenosylmethionine Decarboxylase. Biochemistry 2009, 48, 6423–6430. [Google Scholar] [CrossRef][Green Version]
- McCloskey, D.E.; Bale, S.; Secrist, J.A.I.; Tiwari, A.; Moss, T.H.I.; Valiyaveettil, J.; Brooks, W.H.; Guida, W.C.; Pegg, A.E.; Ealick, S.E. New Insights into the Design of Inhibitors of Human S-Adenosylmethionine Decarboxylase: Studies of Adenine C8 Substitution in Structural Analogues of S-Adenosylmethionine. J. Med. Chem. 2009, 52, 1388–1407. [Google Scholar] [CrossRef] [PubMed]
- Tolbert, W.D.; Ekstrom, J.L.; Mathews, I.I.; Secrist, J.A.; Kapoor, P.; Pegg, A.E.; Ealick, S.E. The Structural Basis for Substrate Specificity and Inhibition of Human S-Adenosylmethionine Decarboxylase. Biochemistry 2001, 40, 9484–9494. [Google Scholar] [CrossRef]
- Keedy, D.A.; van den Bedem, H.; Sivak, D.A.; Petsko, G.A.; Ringe, D.; Wilson, M.A.; Fraser, J.S. Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR. Structure 2014, 22, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Keedy, D.A.; Kenner, L.R.; Warkentin, M.; Woldeyes, R.A.; Hopkins, J.B.; Thompson, M.C.; Brewster, A.S.; Van Benschoten, A.H.; Baxter, E.L.; Uervirojnangkoorn, M.; et al. Mapping the Conformational Landscape of a Dynamic Enzyme by Multitemperature and XFEL Crystallography. eLife 2015, 4, 07574. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Shoichet, B.K.; Fraser, J.S. One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites. ChemBioChem 2015, 16, 1560–1564. [Google Scholar] [CrossRef]
- Keedy, D.A.; Hill, Z.B.; Biel, J.T.; Kang, E.; Rettenmaier, T.J.; Brandão-Neto, J.; Pearce, N.M.; von Delft, F.; Wells, J.A.; Fraser, J.S. An Expanded Allosteric Network in PTP1B by Multitemperature Crystallography, Fragment Screening, and Covalent Tethering. eLife 2018, 7, e36307. [Google Scholar] [CrossRef]
- Doukov, T.; Herschlag, D.; Yabukarski, F. Instrumentation and Experimental Procedures for Robust Collection of X-Ray Diffraction Data from Protein Crystals across Physiological Temperatures. J. Appl. Crystallogr. 2020, 53, 1493–1501. [Google Scholar] [CrossRef]
- Fischer, M. Macromolecular Room Temperature Crystallography. Q. Rev. Biophys. 2021, 54, e1. [Google Scholar] [CrossRef] [PubMed]
- Bradford, S.Y.C.; El Khoury, L.; Ge, Y.; Osato, M.; Mobley, D.L.; Fischer, M. Temperature Artifacts in Protein Structures Bias Ligand-Binding Predictions. Chem. Sci. 2021, 12, 11275–11293. [Google Scholar] [CrossRef]
- Ebrahim, A.; Riley, B.T.; Kumaran, D.; Andi, B.; Fuchs, M.R.; McSweeney, S.; Keedy, D.A. The Tem per ature-Dependent Conformational Ensemble of SARS-CoV-2 Main Protease (Mpro). IUCrJ 2022, 9, 682–694. [Google Scholar] [CrossRef]
- Milano, S.K.; Huang, Q.; Nguyen, T.T.T.; Ramachandran, S.; Finke, A.; Kriksunov, I.; Schuller, D.J.; Szebenyi, D.M.; Arenholz, E.; McDermott, L.A.; et al. New Insights into the Molecular Mechanisms of Glutaminase C Inhibitors in Cancer Cells Using Serial Room Temperature Crystallography. J. Biol. Chem. 2022, 298, 101535. [Google Scholar] [CrossRef] [PubMed]
- Stachowski, T.R.; Vanarotti, M.; Seetharaman, J.; Lopez, K.; Fischer, M. Water Networks Repopulate Protein–Ligand Interfaces with Temperature. Angew. Chem. 2022, 134, e202112919. [Google Scholar] [CrossRef]
- Yabukarski, F.; Doukov, T.; Mokhtari, D.A.; Du, S.; Herschlag, D. Evaluating the Impact of X-Ray Damage on Conformational Heterogeneity in Room-Temperature (277 K) and Cryo-Cooled Protein Crystals. Acta Crystallogr. Sect. D Struct. Biol. 2022, 78, 945–963. [Google Scholar] [CrossRef]
- Ayan, E.; Yuksel, B.; Destan, E.; Ertem, F.B.; Yildirim, G.; Eren, M.; Yefanov, O.M.; Barty, A.; Tolstikova, A.; Ketawala, G.K.; et al. Cooperative Allostery and Structural Dynamics of Streptavidin at Cryogenic- and Ambient-Temperature. Commun. Biol. 2022, 5, 73. [Google Scholar] [CrossRef]
- Doukov, T.; Herschlag, D.; Yabukarski, F. Obtaining Anomalous and Ensemble Information from Protein Crystals from 220 K up to Physiological Temperatures. Acta Crystallogr. Sect. D Struct. Biol. 2023, 79, 212–223. [Google Scholar] [CrossRef]
- Sharma, S.; Ebrahim, A.; Keedy, D.A. Room-Temperature Serial Synchrotron Crystallography of the Human Phosphatase PTP1B. Acta Crystallogr. Sect. F 2023, 79, 23–30. [Google Scholar] [CrossRef]
- McLeod, M.J.; Barwell, S.A.E.; Holyoak, T.; Thorne, R.E. A Structural Perspective on the Temperature Dependent Activity of Enzymes. Structure 2025, 33, 924–934.e2. [Google Scholar] [CrossRef] [PubMed]
- McPhillips, T.M.; McPhillips, S.E.; Chiu, H.-J.; Cohen, A.E.; Deacon, A.M.; Ellis, P.J.; Garman, E.; Gonzalez, A.; Sauter, N.K.; Phizackerley, R.P.; et al. Blu-Ice and the Distributed Control System: Software for Data Acquisition and Instrument Control at Macromolecular Crystallography Beamlines. J. Synchrotron Radiat. 2002, 9, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Waterman, D.G.; Parkhurst, J.M.; Brewster, A.S.; Gildea, R.J.; Gerstel, M.; Fuentes-Montero, L.; Vollmar, M.; Michels-Clark, T.; Young, I.D.; et al. DIALS: Implementation and Evaluation of a New Integration Package. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 85–97. [Google Scholar] [CrossRef]
- Evans, P.R.; Murshudov, G.N. How Good Are My Data and What Is the Resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards Automated Crystallographic Structure Refinement with Phenix.Refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Lang, P.T.; Ng, H.-L.; Fraser, J.S.; Corn, J.E.; Echols, N.; Sales, M.; Holton, J.M.; Alber, T. Automated Electron-Density Sampling Reveals Widespread Conformational Polymorphism in Proteins. Protein Sci. 2010, 19, 1420–1431. [Google Scholar] [CrossRef]
- Ginn, H.M. Torsion Angles to Map and Visualize the Conformational Space of a Protein. Protein Sci. 2023, 32, e4608. [Google Scholar] [CrossRef]
- Tian, H.; Xiao, S.; Jiang, X.; Tao, P. PASSer: Fast and Accurate Prediction of Protein Allosteric Sites. Nucleic Acids Res. 2023, 51, W427–W431. [Google Scholar] [CrossRef] [PubMed]
- Ploscariu, N.; Burnley, T.; Gros, P.; Pearce, N.M. Improving Sampling of Crystallographic Disorder in Ensemble Refinement. Acta Crystallogr. Sect. D Struct. Biol. 2021, 77, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 2.5.5; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Morin, A.; Eisenbraun, B.; Key, J.; Sanschagrin, P.C.; Timony, M.A.; Ottaviano, M.; Sliz, P. Collaboration Gets the Most out of Software. eLife 2013, 2, e01456. [Google Scholar] [CrossRef] [PubMed]
- Russi, S.; Juers, D.H.; Sanchez-Weatherby, J.; Pellegrini, E.; Mossou, E.; Forsyth, V.T.; Huet, J.; Gobbo, A.; Felisaz, F.; Moya, R.; et al. Inducing Phase Changes in Crystals of Macromolecules: Status and Perspectives for Controlled Crystal Dehydration. J. Struct. Biol. 2011, 175, 236–243. [Google Scholar] [CrossRef]
- Wheeler, M.J.; Russi, S.; Bowler, M.G.; Bowler, M.W. Measurement of the Equilibrium Relative Humidity for Common Precipitant Concentrations: Facilitating Controlled Dehydration Experiments. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 111–114. [Google Scholar] [CrossRef]
- Englich, U.; Kriksunov, I.A.; Cerione, R.A.; Cook, M.J.; Gillilan, R.; Gruner, S.M.; Huang, Q.; Kim, C.U.; Miller, W.; Nielsen, S.; et al. Microcrystallography, High-Pressure Cryocooling and BioSAXS at MacCHESS. J. Synchrotron Radiat. 2011, 18, 70–73. [Google Scholar] [CrossRef]
- Huang, Q.; Gruner, S.M.; Kim, C.U.; Mao, Y.; Wu, X.; Szebenyi, D.M.E. Reduction of Lattice Disorder in Protein Crystals by High-Pressure Cryocooling. J. Appl. Crystallogr. 2016, 49, 149–157. [Google Scholar] [CrossRef]
- Teng, T.Y.; Moffat, K. Cooling Rates During Flash Cooling. J. Appl. Crystallogr. 1998, 31, 252–257. [Google Scholar] [CrossRef]
- Kriminski, S.; Kazmierczak, M.; Thorne, R.E. Heat Transfer from Protein Crystals: Implications for Flash-Cooling and X-Ray Beam Heating. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003, 59, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Warkentin, M.; Berejnov, V.; Husseini, N.S.; Thorne, R.E. Hyperquenching for Protein Cryocrystallography. J. Appl. Crystallogr. 2006, 39, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Clinger, J.A.; Moreau, D.W.; McLeod, M.J.; Holyoak, T.; Thorne, R.E. Millisecond Mix-and-Quench Crystallography (MMQX) Enables Time-Resolved Studies of PEPCK with Remote Data Collection. IUCrJ 2021, 8, 784–792. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, J.R.; Bonzon, T.J.; Bakht, T.F.; Fagbohun, O.O.; Clinger, J.A. Multi-Temperature Crystallography of S-Adenosylmethionine Decarboxylase Observes Dynamic Loop Motions. Biomolecules 2025, 15, 1274. https://doi.org/10.3390/biom15091274
Patel JR, Bonzon TJ, Bakht TF, Fagbohun OO, Clinger JA. Multi-Temperature Crystallography of S-Adenosylmethionine Decarboxylase Observes Dynamic Loop Motions. Biomolecules. 2025; 15(9):1274. https://doi.org/10.3390/biom15091274
Chicago/Turabian StylePatel, Jenitha R., Timothy J. Bonzon, Timothy F. Bakht, Omowumi O. Fagbohun, and Jonathan A. Clinger. 2025. "Multi-Temperature Crystallography of S-Adenosylmethionine Decarboxylase Observes Dynamic Loop Motions" Biomolecules 15, no. 9: 1274. https://doi.org/10.3390/biom15091274
APA StylePatel, J. R., Bonzon, T. J., Bakht, T. F., Fagbohun, O. O., & Clinger, J. A. (2025). Multi-Temperature Crystallography of S-Adenosylmethionine Decarboxylase Observes Dynamic Loop Motions. Biomolecules, 15(9), 1274. https://doi.org/10.3390/biom15091274






